Abstract
Most T cell-based HIV-1 vaccine candidates induce responses of limited breadth for reasons that are unclear. We evaluated vaccine-induced T-cell responses in individuals receiving an HIV-1 recombinant adenoviral vaccine. Certain HLA alleles (B27, B57, B35 and B14) are preferentially utilized to mount HIV-specific responses, whereas other alleles (A02 and B07) are rarely utilized (p<0.001). This preference appears due to four factors individually or in combination: higher affinity of specific peptides to specific HLA alleles; higher avidity of TCR; HLA and peptide interaction; and/or higher surface expression of certain HLA. Thus, HLA immunodominance plays a substantial role in vaccine-induced T-cell responses.
Keywords: HIV vaccine, immunodominance, preferential HLA usage
The global HIV pandemic continues to take a staggering toll, with an estimated 33.4 million individuals currently infected (UNAIDS, 2008 Epidemic Update), prompting incredible efforts to develop an efficacious vaccine. Difficulty producing immunogens capable of eliciting broadly neutralizing antibodies has resulted in emphasis on T cell-based immunogen approaches. However, the failure of a recombinant Ad5-vectored HIV vaccine to either prevent transmission or reduce viral load following infection in the Phase IIb proof-of-concept Step study has dampened enthusiasm for T cell-based approaches [1]. Despite this disappointing result, recent studies in rhesus macaques suggest that appropriately designed T cell-based vaccines may have efficacy in reducing viral load or causing abortive infection [2-3]. These studies suggest that the breadth, specificity and phenotype of T-cell effectors induced may determine efficacy.
In humans it is difficult to predict which T-cell responses will be induced after vaccination because they are governed by the phenomenon of immunodominance [4]: most pathogen-specific CD8+ T cells are directed towards a few epitopes, despite the presence of thousands of potential epitopes. Factors governing immunodominance are complex and include 1) positive and negative selection of the T-cell receptor (TCR) repertoire in the thymus, 2) abundance of intracellular antigens, 3) efficiency of protease generation of class I epitopes [5], 4) efficiency of transporter associated with antigen processing (TAP)-dependent or -independent transport of peptides into the endoplasmic reticulum, 5) affinity of peptide binding to class I molecules, and 6) naïve T-cell precursor frequency [6]. It is important to determine which factors select the specificities and restrictions of vaccine-induced T-cell responses since some T-cell specificities seem more effective at controlling viral replication [7-8]. Here we investigated the epitopes and HLA molecules utilized to mount HIV-specific CD8+ T-cell responses in healthy HIV-seronegative individuals who participated in clinical trials of candidate recombinant adenoviral vaccines. These findings have profound implications for vaccine design as they show that immunodominance plays a substantial role in vaccine-induced responses.
Methods
All experiments utilized peripheral blood mononuclear cells (PBMC) from 40 Ad5-naïve participants who received a tetravalent recombinant adenovirus vaccine (VRC-HIVADV014-00-VP) previously described in [9], which encoded a clade B HIV-1 Gag-Pol polyprotein (HXB2/NL4-3) and HIV-1 Env glycoproteins from clades A (92RW020), B (HXB2/BaL) and C (97ZA012), in a 3:1:1:1 ratio. By ELISpot, 85% had a detectable vaccine response. Avidity and HLA-B35-restricted CD8+ T-cell response experiments also utilized PBMC from 74 participants who received a recombinant adenovirus vaccine (MRK-Gag) previously described in [10], which encoded Gag from HIV-1CAM-1. By ELISpot, 65% had a detectable vaccine response. All PBMC samples were from visits within 6 months of final vaccination. Subjects were recruited and enrolled at HIV Vaccine Trials Units and Merck sites; appropriate Institutional Review Boards approved the studies and volunteers provided written consent. The HLA class I alleles of all subjects are listed in Supplemental Table 1.
Two peptide sets, potential T-cell epitopes (PTE) and Consensus B (ConB), were employed to detect HIV-specific T-cell responses. PTE peptides were 15-mers designed according to published methods [11] (synthesized by BioSyn, Lewisville, TX). ConB peptides were 15-mers overlapping by 11 a.a. based on HIV-1 clade B consensus sequence (provided by NIH AIDS Research and Reference Reagent Program, Bethesda, MD). Additionally, 8-11-mers corresponding to described class I HLA-restricted Cytotoxic T Lymphocyte (CTL) epitopes were tested according to an individual's HLA type (synthesized by BioSyn, New England Peptide (Gardner, MA), and Mimitopes (Minneapolis, MN)). When PBMC were limiting, only known CTL epitopes that corresponded to the individual's HLA type were tested.
IFN-γ ELISpot was performed as previously described [8]. Results were considered positive if the number of spot forming cells (SFC) was at least twice background and ≥50 SFC/106 PBMC. Positive ELISpot results to a 15-mer from either peptide set were confirmed by testing positive for one of the following: IFN-γ ELISpot to a corresponding 15-mer from the other peptide set, IFN-γ ELISpot to an embedded optimal epitope (8-11-mers), or an Intracellular Cytokine Staining (ICS) assay detecting IFN-γ, TNF-α, IL-2, and/or CD107a. Additionally this ICS assay was used to discriminate T-cell responses as either CD4+ or CD8+; CD4+ T-cell responses were not included in this analysis. HLA restrictions for defined optimal epitopes were predicted based on published class I HLA-restricted CTL epitopes.
To measure T-cell avidity, IFN-γ ELISpot was performed using peptide concentrations titrated from 20 μg/ml to 0.0002 μg/ml. Functional avidity values (EC50) were calculated based on regression curves drawn with the Sigmoidal Fit tool in Origin 6.0 software.
To measure HLA surface expression, CD4+ T-cells were enriched by negative selection (StemCell Technologies, Vancouver, BC) and incubated with mouse anti-human HLA monoclonal antibodies (One Lambda, Canoga Park, CA) for 30 minutes before washing and staining with goat anti-mouse IgG Fluorescein-conjugated antibody (Beckman Coulter, Fullerton, CA). Data acquisition was performed on an LSRII flow cytometer (BD) and analyzed with FlowJo (TreeStar, Ashland, OR). The mean fluorescent intensity (MFI) of samples was compared to QIFI beads (DakoCytomation, Glostrup, Denmark) coated with well-defined quantities of mouse monoclonal antibody stained with the same secondary antibody. Antibody-binding capacity (ABC) was calculated based on the QIFI bead MFI curve.
Quantitative assays to measure the binding of peptides to HLA class I molecules were based on the inhibition of binding of a radiolabeled standard peptide, as described previously [12].
To compare the HLA restriction of T-cell responses among HLA types, Fisher's exact test was used with Bonferroni correction for multiple comparisons. For avidity analysis, functional avidity values were tested between groups with the Wilcoxon-Mann-Whitney test. For HLA-expression analysis, ABC ratios of HLA-B27 expression relative to A11 or A24 expression (for one individual, mean of A11 and A24 expression) were compared by a Wilcoxon signed-rank test versus the hypothetical value of 1.0 (equal expression). Tests were two-sided with a threshold of p=0.05 unless stated otherwise, and were calculated using Prism version 4.02 (GraphPad Software, San Diego, CA). Affinity analysis was limited to a set of 69 A-list defined optimal epitopes with available affinity data (N. Frahm, personal communication; see Supplemental Table 2) that where conserved within the vaccine insert. A Mann-Whitney test was used to compare affinity values from epitopes recognized or not recognized by vaccination.
Results and Discussion
Certain HLA alleles appear to be preferentially utilized to mount HIV-1-specific optimal epitope responses during vaccination (Table 1). Vaccine-induced responses restricted by A03, A03/11, B14, B27, B35, B44/49, and B57/58 were all significantly overrepresented (p<0.0016). Similar findings were previously reported for an ALVAC-HIV vaccine within individuals possessing a B27 or B57 HLA allele [13]. Notably, all individuals with HLA-B27 (present in 10% of our population), B14 (present in 13%), B57/58 (present in 10%), and 7/9 (78%) with B35 (present in 23%) possessed vaccine-induced T-cell responses restricted by these alleles, while other HLA alleles were infrequently utilized to restrict vaccine-induced responses. In particular, only 4/18 (22%) of HLA-A02 (present in 45%) vaccinees mounted an A02-restricted response, in agreement with other studies showing little contribution of HLA-A02 during primary infection or vaccination [14]. B07 (1/9; 11%) and B08 (0/8; 0%) were also infrequent or non-contributors to vaccine-induced T-cell responses. The over representation of responses restricted by B27, B57/B58, and B35 are of particular interest due to previously published data which correlated these alleles with differences in rate of progression to AIDS [15]. B27 and B57/58 HLA types have been shown to be protective alleles, while a subset of B35 HLA types (Px), in contrast, have been shown as risk alleles.
Table 1. ELISpot response frequency and epitopes restricted by HLA alleles in vaccinated individuals.
| MHC alleles | Epitopes | Conservation (%)a | ||||
|---|---|---|---|---|---|---|
| Vaccinees (%) | Locationb | Sequence | Abbrev. | M Clade | B Clade | |
| A03 | 5/11 (45) | Gag p1720-28 | RLRPGGKKK | RK9 | 45 | 65 |
| A03/11c | 9/16 (56) | Env gp16037-46 | TVYYGVPVWK | TK10 | 68 | 92 |
| B14 | 5/5 (100) | Env gp4173-81 | ERYLKDQQL | EL9 | 51 | 51 |
| B2705d | 4/4 (100) | Pol Int185-194 | FKRKGGIGGY | FY10 | 86 | 80 |
| 4/4 (100) | Gag p24131-140 | KRWIILGLNK | KK10 | 87 | 82 | |
| B35Pxe | 4/4 (100) | Env gp16078-86 | DPNPQEVVL | DL9 | 12 | 38 |
| 4/4 (100) | Pol RT293-301 | IPLTEEAEL | IL9 | 9 | 34 | |
| B35PYe | 5/7 (71) | Pol RT175-183 | NPDIVIYQY | NY9 | 24 | 66 |
| 3/7 (43) | Pol GagPolTF8-16 | FPQGKAREF | FF9 | 9 | 30 | |
| B44/49f | 6/7 (86) | Pol RT397-406 | TWEAWWTEYW | TWW10 | 6 | 25 |
| B57/58g | 2/4 (50) | Pol RT244-252 | IVLPEKDSW | IVW9 | 14 | 62 |
| 4/4 (100) | Gag p24108-117 | TSTLQEQIGW | TW10 | 40 | 73 | |
Calculated as the frequency of a complete epitope match within all aligned M or B clade sequences from the LANL database.
Location of epitope within specified gene and protein, based on HXB2 sequence.
TK10 can be restricted by HLA A03 or A11 this group Includes two subjects with both A03 and A11 alleles and one subject homozygous for A11.
One additional subject with a B15-FY10 response not shown.
Includes two subjects with combined B35Px/PY alleles with responses in both alleles, one subject with combined B*2705/35Px alleles with responses in both alleles, and one subject with combined B57/35PY alleles with only B57-restricted responses.
TWW10 can be restricted by HLA-B44 or -B49.
IVW9 and TW10 can be restricted by HLA-B57 or -B58; this group includes only one subject with a B58 alelle.
Our data and others have shown in addition to specific HLA alleles preferentially utilized to mount vaccine-induced responses, specific HIV-1 epitopes were targeted (Table 1) [16]. All HLA-B27+ vaccinees mounted a B27-restricted Gag p24 response against KRWIILGLNK (KK10) in conjunction with a Pol response against FKRKGGIGGY (FY10). The latter epitope is likely KRKGGIGGY (KY9), which was a recently reported B27-restricted epitope found in chronic infection [17]. Similarly, all HLA-B57+ vaccinees mounted a B57-restricted Gag p24 response against TSTLQEQIGW (TW10) [12]. The preferential selection of certain epitopes may be because they have significantly higher affinity for their HLA alleles, as assessed by HLA binding assays (Figure 1A). The HLA-B35-restricted responses were split between the Px and PY alleles, which differ in their binding motif anchor residues. Both motifs bind peptides with a proline at position two, but peptides bound by PY alleles have a tyrosine at the C-terminus position, whereas peptides bound by Px alleles have other C-terminal residues. All B35Px vaccinees recognized the B35-restricted Env DPNPQEVVL (DL9) epitope in conjunction with the B35-restricted Pol IPLTEEAEL (IL9) epitope. Conversely, 71% of B35PY vaccinees recognized the B35-restricted Pol NPDIVIYQY (NY9) epitope (Table 1). Interestingly, B35+ participants receiving the VRC vaccine only mounted four responses to Gag and 28 responses to other HIV proteins regardless of HLA restriction. In addition, among 21 B35+ MRK-Gag vaccinees, only one mounted a B35-restricted response. These data suggest that in the absence of highly mutable Env and Pol epitopes (present within the VRC vaccine) HLA class I alleles other than B35 are utilized to mount a response.
Figure 1. Epitope affinity, TCR avidity, and HLA surface expression play a role in determining CD8+ T-cell immunodominance.
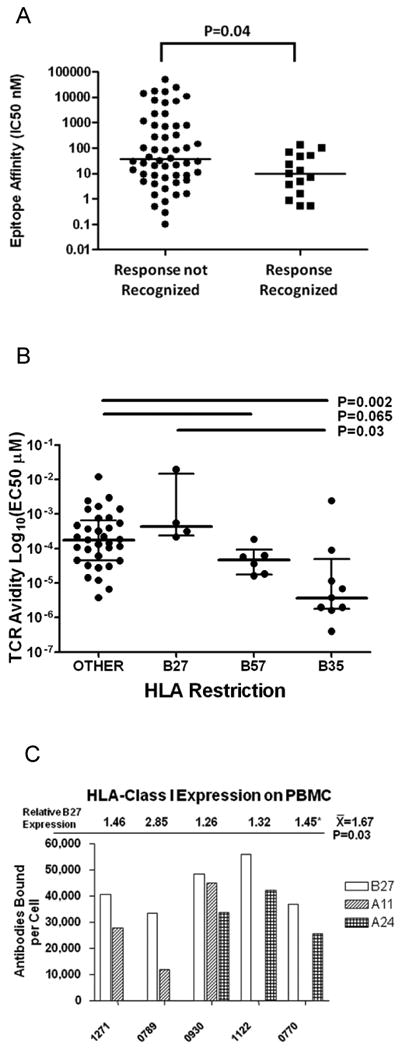
(A) Epitope affinity IC50 nM values were measured for peptides to HLA class I molecules for 69 A-list defined optimal CD8+ T-cell epitopes conserved within the vaccine insert. TCR affinity values were separated by whether or not the responses were recognized by participants who received VRC-HIVADV014-00-VP. (B) TCR avidity log10 EC50 μM values were measured for 52 optimal CD8+ T-cell epitopes recognized by participants who received either VRC-HIVADV014-00-VP or MRK-Gag. Responses were divided by HLA restriction: B27, B57, B35, or other. (C) HLA surface expression as measured by number of antibodies bound per cell for five participants with a B27 HLA type and A24 and/or an A11 HLA type who received VRC-HIVADV014-00-VP.
Other data from our group suggest that epitopes targeted early in infection determine disease progression [8]. However, that study did not address why specific epitopes/HLA were immunodominant in both early infection and vaccination. It has been postulated that high-avidity T-cell responses dominate in early infection [18]. In this study we found vaccine-induced B27 KK10-specific T cells to be of low avidity. Of note, vaccine-induced B27 KK10-specific T cells were of lower avidity than KK10-specific T cells from HIV-1 infected individuals. In contrast B35-specific T cells were of higher avidity than those restricted by other alleles (p=0.0017; Figure 1B; see Supplemental Table 3 for complete list). This suggests that B35 may be preferentially utilized after vaccination, because T cells restricted by this allele are of higher avidity than T cells restricted by other alleles.
However, it remained unclear why T cells restricted by B27 dominate in early infection and after vaccination despite their low avidity. We hypothesized that this may be due to higher cell surface density of B27 molecules. To address this we quantitatively measured cell surface expression of HLA-B27 in relation to HLA-A24 and HLA-A11 alleles (Figure 1C) and found that HLA-B27 is expressed at a 45% higher level on the cell surface than other HLA alleles tested. Thus, T cells restricted by HLA alleles which are expressed at high levels at the cell surface can dominate even if they have low avidity TCRs. Unfortunately it was not possible to assess the surface expression of a larger set of HLA alleles including HLA-B57 due to the unavailability of specific IgG antibodies.
In addition to TCR avidity, epitope affinity, and HLA surface expression, factors such as intracellular abundance and stability of epitopes and proteosome/tapasin-dependence may also play a role in determining immunodominance [19-20], although these were outside the scope of this study.
In conclusion, our data suggest that upon vaccination of healthy seronegative individuals certain HLA alleles are preferentially utilized to mount HIV-1-specific T cell responses (B27, B57, B35, B14), whereas other alleles are rarely utilized (A02, B07). Furthermore, the epitopes targeted by these alleles are highly predictable and appear to be selected based on affinity for their cognate HLA. Finally we show data suggesting that B35Px DL9/IL9-specific T cells likely dominate after vaccination because they possess high avidity TCRs, whereas B27-restricted T cells likely dominate because of the higher cell surface expression of HLA-B27. Our data suggest that immunodominance and immune focusing play a substantial role in vaccine-induced responses. This has obvious consequences for vaccine design since it appears that responses can be predicted based on HLA genotype and immunogen sequence.
Supplementary Material
Acknowledgments
We thank Nicole Frahm for providing epitope affinity data.
We thank the James B. Pendleton Charitable Trust for their generous equipment donation.
Funding: This work was supported by the National Institutes of Health, Public Health Service grants R01 AI065328, U01 AI046747, and U01 AI046725.
Footnotes
Conflicts of interest: None declared
References
- 1.Buchbinder SP, Hehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blinded, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372(9653):1881–93. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson NA, Keele BF, Reed JS, et al. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol. 2009;83:6508–21. doi: 10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen SG, Vieville C, Whizin N, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293–9. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 5.Tenzer S, Wee E, Burgevin A, et al. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat Immunol. 2009;10:636–46. doi: 10.1038/ni.1728. [DOI] [PubMed] [Google Scholar]
- 6.Kotturi MF, Scott I, Wolfe T, et al. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008;181:2124–33. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton H, Frank I, Baydo R, et al. Preservation of T cell proliferation restricted by protective HLA alleles is critical for immune control of HIV-1 infection. J Immunol. 2006;177:7406–15. doi: 10.4049/jimmunol.177.10.7406. [DOI] [PubMed] [Google Scholar]
- 8.Dinges WL, Richardt J, Friedrich D, et al. Virus specific CD8+ T cell responses better define HIV disease progression than HLA genotype. J Virol. 2010;84:4461–4468. doi: 10.1128/JVI.02438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peiperl L, Morgan C, Moodie Z, et al. Safety and Immunogenicity of a Replication-Defective Adenovirus Type 5 HIV Vaccine in Ad5-Seronegative Persons: A Randomized Clinical Trial (HVTN 054) PLoS ONE. 2010;5(10):e13579. doi: 10.1371/journal.pone.0013579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholson O, Dicandilo F, Kublin J, et al. Safety and Immunogenicity of the MRKAd5 gag HIV-1 Vaccine in a Worldwide Phase 1 Study of healthy Adults. AIDS Res Hum Retroviruses. 2010 Nov 23; doi: 10.1089/aid.2010.0151. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F, Malhotra U, Gilbert PB, et al. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24:6893–6904. doi: 10.1016/j.vaccine.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Sidney J, Southwood S, Oseroff C, et al. Current protocols in immunology. Wiley; New York: 1998. Measurements of MHC/Peptide interactions by gel filtration; pp. 18.13.11–18.13.19. [Google Scholar]
- 13.Kaslow RA, Rivers C, Tang J, et al. Polymorphisms in HLA Class I Genes Associated with both Favorable Prognosis of Human Immunodeficiency Virus (HIV) Type 1 Infection and Positive Cytotoxic T-Lymphocyte Responses to ALVAC-HIV Recombinant Canarypox Vaccines. J Virology. 2001;75(18):8681–9. doi: 10.1128/JVI.75.18.8681-8689.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goulder PJ, Altfeld MA, Rosenberg ES, et al. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J Exp Med. 2001;193:181–94. doi: 10.1084/jem.193.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao X, Nelson GW, Karacki P, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med. 2001;344:1668–75. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 16.Koup RA, Roederer M, Lamoreaux L, et al. Priming immunization with DNA Augments Immunogenicity of Recombinant Adenoviral Vectors for Both HIV-1 Specific Antibody and T-Cell Responses. PLoS ONE. 2010;5(2):e9015. doi: 10.1371/journal.pone.0009015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne R, Kloverpris H, Sacha J, et al. Efficacious early antiviral activity of HIV Gag- and Pol-specific HLA-B*2705-restricted CD8+ T-cells. J Virology. 2010;84(20):10543–57. doi: 10.1128/JVI.00793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lichterfeld M, Yu XG, Mui SK, et al. Selective depletion of high-avidity human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T cells after early HIV-1 infection. J Virol. 2007;81:4199–214. doi: 10.1128/JVI.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, Norbury CC, Cho Y, et al. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J Exp Med. 2001;193:1319–26. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thirdborough SM, Roddick JS, Radcliffe JN, et al. Tapasin shapes immunodominance hierarchies according to the kinetic stability of peptide - MHC class I complexes. Eur J Immunol. 2008;38(2):364–9. doi: 10.1002/eji.200737832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


