Abstract
Background
Acquisition of mesenchymal phenotype by epithelial cells by means of epithelial mesenchymal transition (EMT) is considered as an early event in the multi-step process of tumor metastasis. Therefore, inhibition of EMT might be a rational strategy to prevent metastasis.
Methods
Utilizing the global gene expression profile from a cell culture model of TGF-β-induced EMT, we identified potential EMT inhibitors. We used a publicly available database (www.broad.mit.edu/cmap) comprising gene expression profiles obtained from multiple different cell lines in response to various drugs to derive negative correlations to EMT gene expression profile using Connectivity Map (C-Map), a pattern matching tool.
Results
Experimental validation of the identified compounds showed rapamycin as a novel inhibitor of TGF-β signaling along with 17-AAG, a known modulator of TGF-β pathway. Both of these compounds completely blocked EMT and the associated migratory and invasive phenotype. The other identified compound, LY294002, demonstrated a selective inhibition of mesenchymal markers, cell migration and invasion, without affecting the loss of E-cadherin expression or Smad phosphorylation.
Conclusions
Collectively, our data reveals that rapamycin is a novel modulator of TGF-β signaling, and along with 17-AAG and LY294002, could be used as therapeutic agent for inhibiting EMT. Also, this analysis demonstrates the potential of a systems approach in identifying novel modulators of a complex biological process.
INTRODUCTION
Metastasis is the major cause of mortality in cancer-related deaths. Hence determining and targeting precise molecular mechanisms of metastasis is critical for a successful prevention strategy. During metastasis, cancer cells acquire the ability to invade surrounding tissue with subsequent dissemination to secondary organs (1). The acquisition of migratory and invasive capability by otherwise stationary epithelial cells is associated with gain of mesenchymal characteristics and concomitant loss of epithelial phenotype, a phenomenon referred to as epithelial–mesenchymal transition (EMT) (2). EMT also confers resistance to anoikis, evasion of immune surveillance, and in certain cases is associated with stem cell-like properties of the resulting mesenchymal cells, all of which may be required for a cancer cell to successfully metastasize. Therefore, inhibition of EMT might be a rational strategy to prevent metastasis.
The cytokine Transforming Growth Factor-β (TGF-β) plays a paradoxical role in cancer biology, whereby it acts as a tumor suppressor in early stages and as a tumor promoter in late stages of tumor progression. The tumor-promoting functions of TGF-β include induction of EMT in cancer cells (3-5). Depending on the cell type and context, TGF-β induces EMT via activation of multiple signaling pathways, both Smad-dependent and Smad-independent, and cross talk with developmental pathways like WNT and Notch signaling (6-9). Given the complex nature of EMT regulation, it is challenging to identify critical regulatory molecules or pathways for targeting EMT.
System-wide profiling of molecular changes offers an opportunity to understand the underlying mechanisms and design strategies to perturb the system (10). Gene expression profiling represents all the transcriptional alterations happening in a given disease state and time. Compounds that can reverse some, if not all, of these changes might serve as potential inhibitors of that particular disease state. A recently developed pattern matching tool known as Connectivity Map (C-Map) has demonstrated its utility in identifying potential inhibitors using gene expression profiles of a given biological state. The C-Map tool is built on a database comprised of 564 gene expression profiles derived from multiple cell lines after treatment with 164 different compounds at different doses (453 profiles, or “instances”), along with 111 corresponding controls (11). Using C-Map, one can derive negative correlations between the gene expression perturbations of the biological state of interest and the perturbations of each drug instance in the database. The drugs whose instances are most significantly correlated are ones that may serve as potential inhibitors of that particular state; in this case it is EMT. Utilizing C-Map we analyzed the global gene expression profile obtained from TGF-β-induced EMT in the A549 lung adenocarcinoma cell line to identify potential inhibitors of EMT. We identified known as well as new potential EMT inhibitors. Validation of these compounds for EMT inhibition revealed their novel mechanism of action and the potential of targeting mTOR, HSP90 and PI3K pathways for inhibiting EMT, tumor cell migration and invasion.
EXPERIMENTAL PROCEDURES
EMT experiment with test compounds
A549 (human lung adenocarcinoma) and H358 (human bronchioalveolar carcinoma) cell lines were obtained from the American Type Culture Collection (Manassas, VA) and maintained in RPMI-1640 medium with supplemented with 10% FBS, glutamine, penicillin and streptomycin at 37 ° in 5% CO2. The authentication of cell lines was not performed by authors. In all experiments cells at 40-50% confluency in complete medium were serum starved for 24 h and treated with TGF-β (5 ng/ml) for 72 h in the presence and absence of compounds at indicated concentrations. Test compounds were added to the cultures 30 min prior to TGF-β stimulation. After 72 h cells were either lysed for assessing protein expression or trypsinized for re-plating in the transwell chambers for assessing migration and invasion. The conditioned media was collected for estimation of MMPs. All the test compounds used in this study were purchased from Tocris Biosciences, USA.
Gene expression and C-Map analysis
A549 lung-cancer cells were treated with 5 ng/mL of TGF-β and harvested at various time points in 3 separate experiments, and the resulting RNA collected, assayed using Affymetrix HG-U133_plus_2 arrays, and analyzed as previously described (12). We used probe set annotation from Affymetrix web sites (version na30, dated Nov 13, 2009). Using two-way ANOVA models with terms for the 3 experiments and 9 time points, we selected probe sets that gave p<0.001 for each time point compared to the 0 h control samples and also gave average fold-differences of at least 1.5-fold. This data set, and the p-values and fold-differences obtained are publicly available as GEO series GSE17708 (12). We formed the union of the selected probe sets for the 0.5, 1, and 2 h time points as a representative list of early-responding genes, and the union of 4 and 8 h as representative list of intermediate-responding genes. For the 3 early time points this yielded 478 probe sets increased with TGF-β and 244 decreased, of which 237 and 113, respectively, were also on the smaller Affymetrix U133A (or similar HT_HG-U133A) arrays, which are the arrays used to generate data on the effects of various compounds by Lamb and coworkers in their work on “the connectivity map” (13). For the union of 4 and 8h time points we obtained 1884 increased and 1254 decreased probe sets, of which 1006 and 703 were on U133A arrays. The connectivity map data consist of 164 compounds tested on several cell lines (MCF7, PC-3, HL-60 & SKMEL5), with a total of 453 treatments, called instances, as well as 111 arrays of appropriate control treatments (564 total arrays). We input our list of up and down probe sets, given values of 1 and −1, into the software of Zhang and Gant (14) which computes Cscores that are similar to correlation coefficients between our values and the ranks of the ratios of treatment to the average of controls for the instances of Lamb et al. Negative Cscores indicate the compound altered probe sets in an opposite-correlated way compared to the differences we observed with TGF-β treatment. The software computes similar scores in 10000 additional runs in which the probe set labels are randomly permuted, and computes two-sided permutation test p-values as the fraction of scores from permuted data sets with larger absolute value than the one actually obtained for the instance. To judge the significance of compounds, Cscores for the instances of the compound are averaged to compute the SetCscore for each compound, and this averaging is also performed on the permuted data sets, and the software again computes permutation test p-values. With only 10000 permuted data sets, these p-values can be no smaller than 0.0001 however, the means and standard deviations of the SetCscores from permutations are also reported, enabling us to obtain a finer-grained test of significance by dividing the SetCscore by this standard deviation and computing two-sided tests by referring this standardized SetCscore to standard Normal (Gaussian) distributions. For p-values of approximately 0.0001 and larger the two methods agreed fairly well, but for the largest SetCscores the p-values from standardized SetCscores were much smaller, as expected, and enabled us to better judge the relative evidence in favor of the top-scoring compounds.
Fluorescence microscopy
Cells treated in 48 well tissue culture plates were fixed in 4% formalin, blocked with 5% horse serum and 0.3% Triton-X 100 and stained with FITC conjugated E-cadherin antibody overnight at 4°C. Cells were washed with PBS and stained sequentially for F-actin with Rhodamine Phalloidin and for nuclei with DAPI. Images were captured using a fluorescent microscope (EVOSfl, Bothell, WA 98021, U.S.A) at 20x magnification. Images were processed by Adobe Photoshop (Adobe Systems Inc., San Jose, CA, U.S.A.).
Cell migration and invasion assays
In vitro migration assays were performed as previously described (15). Briefly, cells were seeded in the top chamber of the 8.0μ pore size cell culture inserts that were either coated or uncoated with matrigel for migration and invasion assays respectively. Then the inserts were placed in a 24 well plate filled with RPMI 1640 medium with 5% FBS. Cells that penetrated to the underside surfaces of the inserts were fixed and stained with the Diff-Quick (Fisher Scientific, Pittsburgh, PA) method, and counted under the microscope. The mean of three high power fields for each condition run in triplicates was calculated.
Western blot
Samples containing 20 μg of total protein were electrophoresed on SDS–polyacrylamide gels and transferred onto a polyvinyldifluoride membrane by electroblotting. Membranes were probed with primary antibodies with overnight incubation at 4 °, followed by horseradish peroxidase–conjugated secondary antibodies. Finally the immunoblots were visualized by using ECL reagents.
Smad Transcriptional Activity
Effect of test compounds on Smad transcriptional activity was determined in A549-SBE-Luc cells as previously described (15). Briefly, cells were serum starved overnight and treated with TGF-β (5 ng/ml) in presence and absence of compounds pretreatment. After 4 hours luciferase activity was measured using the steady-glo luciferase kit (from Promega, USA) as per the manufacturer’s instructions. Luciferse counts were normalized to the total protein concentrations in the respective samples.
Statistical analysis
Data are represented as mean ± standard deviations and were analysed with the Prism 4.0 statistical program (GraphPad Software, San Diego, CA). Groups were compared using one-way ANOVA or student t-test. Differences were considered significant if P < 0.05
RESULTS
C-Map analysis using early gene expression changes during EMT identified potential inhibitors of EMT
Stimulation of cells with TGF-β induces activation and nuclear translocation of transcription factors Smad2 and Smad3 (16). This results in the subsequent robust transcriptional regulation of the target genes. These transcriptional changes are critical for the regulation of TGF-β-induced complex biological responses including EMT (16). Reversal of these transcriptional changes that occur in the context of a biological process may be critical for inhibiting that particular process. Therefore, to identify inhibitors of EMT, we derived a list of TGF-β-responding (increased and decrease) probe sets in EMT, from the union of 3 time points (0.5, 1 h and 2 h) from a time course gene expression analysis of TGF-β-induced EMT in the A549 lung adenocarcinoma cell line (12). Using the C-Map tool, we computed connectivity scores (Cscores) between this EMT profile and the 453 instances in Lamb et al. data base from 164 compounds (13). Cscores are similar to correlation coefficients, and a negative Cscore indicates that the compound from which that instance is derived potentially reverses the gene expression changes in the input profile, which in this case was EMT. The Cscores for the instances were averaged to obtain SetCscores for each compound, and we standardized these by dividing the standard deviation of the SetCscores for the same compound, obtained from 10000 data sets in which the probe set labels were randomly permuted. We identified 49 negatively correlated compounds with p<0.01, of which 30 gave p<0.0001. In order to focus on the most reliable findings we reduced these 30 candidates to 21 compounds that had at least 2 instances, which are shown in Table 1. Since a total of 95 compounds had at least two instances, we expect only about 0.01 (=0.0001*95) false positive compounds using this selection criterion. Compounds identified include inhibitors of HSP90, PI3K, mTOR, cycloxygenase, prostaglandin synthetase, DNA gyrase, Rho-Kinase, Calcineurin, purine synthesis, α-estradiol and aromatase. Interestingly, for all 21 compounds, either the compounds themselves or the primary pathways that the compounds are known to inhibit were implicated in cancer (Table 1). This includes the unanticipated, antipsychotic compounds Chlorpromazine and Clozapine, which have also shown to inhibit cancer cell growth (17, 18). Complete analysis and the Cscores derived for all the instances are presented in supplementary table-1. Similar analysis with the gene profile derived from the union of 4 h and 8h time points also largely identified the same compounds (supplementary table-1) with compound scores for two temporal profiles being highly correlated (r=0.90 for 95 compounds, p=1×10−34)
Table 1. Top scoring compounds in C-Map analysis.
| Compound | Set Size |
Standardized SetCscore (0.5, 1 &2 h) |
p-value from standardized SetCscore (Z-score) |
Mode of action | Relevance in cancer (Reference) |
|---|---|---|---|---|---|
| 17-allylamino-geldanamycin | 18 | −7.73 | 1.07E-14 | HSP90 inhibitor | (34, 35) |
| LY294002 | 17 | −5.91 | 3.41E-09 | PI3K inhibitor | (36, 37) |
| Sirolimus (Rapamycin) | 10 | −4.72 | 2.36E-06 | mTOR inhibitor | (36, 38) |
| Geldanamycin | 6 | −5.33 | 1.01E-07 | HSP90 inhibitor | (34, 35) |
| Novobiocin | 6 | −4.17 | 3.10E-05 | DNA Gyrase inhibitor | (39, 40) |
| Nordihydroguaiaretic acid | 5 | −5.20 | 1.96E-07 | Cyclooxygenase inhibitor | (41, 42) |
| Staurosporine | 4 | −5.42 | 5.87E-08 | Protein Kinase C inhibitor | (43) |
| Chlorpromazine | 4 | −4.84 | 1.31E-06 | Dopamine & seretonin receptor antagonist | (17) |
| SC-58125 | 4 | −4.01 | 5.96E-05 | Cyclooxygenase inhibitor | (44) |
| LM-1685 | 3 | −5.50 | 3.81E-08 | Cyclooxygenease inhibitor | (42, 45) |
| Acetylsalicylic acid | 3 | −5.36 | 8.46E-08 | Cyclooxygenase inhibitor | (42, 46) |
| 17-dimethylamino-geldanamycin | 2 | −6.39 | 1.67E-10 | HSP90 inhibitor | (34, 35) |
| Cyclosporin | 2 | −5.41 | 6.40E-08 | Calcineurin inhibitor | (47, 48) |
| Fasudil | 2 | −5.23 | 1.71E-07 | Rho- Kinase inhibitor | (49) |
| Mercaptopurine | 2 | −5.17 | 2.40E-07 | Purine synthesis inhibitor | (50) |
| Oxaprozin | 2 | −5.16 | 2.52E-07 | Prostaglandin synthetase inhibitor | (51) |
| Diclofenac | 2 | −4.85 | 1.26E-06 | Cyclooxygenase inhibitor | (42) |
| 4,5-dianilinophthalimide | 2 | −4.68 | 2.88E-06 | Multikinase inhibitor | (52) |
| NU-1025 | 2 | −4.62 | 3.77E-06 | PARP inhibitor | (53) |
| Butein | 2 | −4.41 | 1.03E-05 | Aromatase inhibitor | (54) |
| Clozapine | 2 | −3.91 | 9.05E-05 | Dopamine & seretonin receptor antagonist | (18) |
The 21 compounds with probe set alterations negatively correlated to early (0.5, 1 h and 2 h) alterations observed with TGF-β treatment are given. Each was represented by at least two instances (SetSize) and gave p<0.0001 for a Z-score test of the average Cscores for all the instances of the compound (SetCscores). The p-values are two-sided tests based on the standardized SetCscore, which were the SetCscores divided by the standard deviation of this score computed from 10000 random permutations of the probe set labels. Results from testing alterations from 4 and 8h TGF-β are also given. Detailed results for all 164 compounds are given in Supplementary Table.
Experimental validation of compounds identified by the C-Map analysis
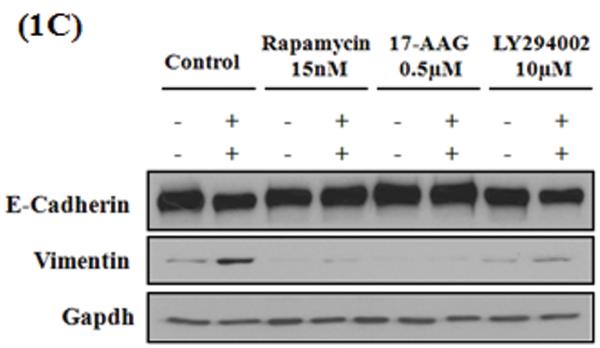
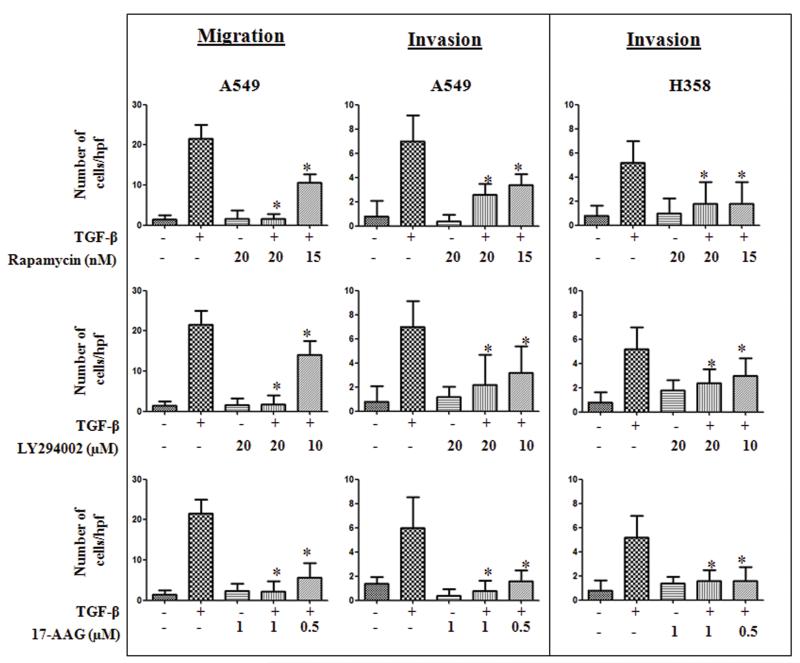
EMT is characterized by loss of epithelial markers (E-cadherin) and gain of mesenchymal markers (N-cadherin, Vimentin) resulting in the acquisition of migratory and invasive phenotype. Hence, to test the ability of the compounds identified by C-Map analysis, to inhibit EMT, we assessed their effects on biochemical markers as well as functional attributes of EMT in two different cell culture models, A549 and H358. A549 Cells were stimulated with TGF-β (5 ng/ml) in the presence and absence of test compounds at indicated concentrations and assessed stress-fiber formation, expression of epithelial and mesenchymal markers by immunofluorescence microscopy and western immunoblotting (Figure 1A&B). Consistent with EMT, 72 h TGF-β treatment significantly suppressed the E-cadherin expression compared to the untreated controls (Figure 1A & B). However, the presence of rapamycin (Sirolimus, mTOR inhibitor) or 17-AAG (HSP90 inhibitor) completely reversed TGF-β-induced suppression of E-cadherin expression, at all concentrations tested (Figure 1A & B). Further, both the compounds also blocked basal and TGF-β-induced up-regulation of mesenchymal marker N-cadherin. Treatment of Rapamycin and 17-AAG alone induced a slight increase in the basal vimentin levels in the control cells but it was not statistically significant (Figure S1). While rapamycin had no effect, 17-AAG completely abrogated the TGF-β-induced vimentin expression (Figure 1B). Interestingly, LY294002 had no effect on TGF-β-induced E-cadherin suppression (Figure 1A & B), but attenuated both the basal and TGF-β-induced up-regulation of N-cadherin and vimentin, suggesting a selective effect on mesenchymal phenotype (Figure 1B). Consistent with their effect on mesenchymal phenotype, all the three compounds inhibited TGF-β-induced change in morphology as well as stress fiber formation in A549 cells (Figure 1A). Reflecting their effect on epithelial and mesenchymal markers, rapamycin and 17-AAG inhibited EMT-induced cellular migration and invasion in A549 cells (Figure 2). These two compounds also blocked concomitant secretion of MMP2 and MMP9 during EMT (Data not shown). Interestingly, LY294002, which only inhibited mesenchymal markers, also inhibited EMT-induced cellular migration, invasion (Figure 2A) as well as MMP secretion (Data not shown). All the above three compounds, demonstrated similar effects on expression of E-cadherin and vimentin (Figure 1C), and cellular invasion (Figure 2) during TGF-β-induced EMT in H358 cells, another non-small cell lung cancer cell line. This demonstrates that the observed effects of these compounds are not specific to a single cell line (Figure S1 and Table S2).
Figure 1. Effect of rapamycin, 17-AAG and LY 294002 on Epithelial and mesenchymal markers during TGF-β induced EMT.
A549 or H358 cells were serum starved for 24 h, pretreated with inhibitors and stimulated with or without TGF-β (5ng/ml). After 72 h (A) E-cadherin and stress fibers (F-actin by Phalloidin staining) were assessed in A549 cells by immunofluorescence (scale bar 100μ), (B) epithelial (E-cadherin) and mesenchymal (N-cadherin, vimentin) markers were assessed in A549 cells (C) epithelial (E-cadherin) and mesenchymal (vimentin) markers in H358 cells were assessed by western immunoblotting. Quantitation of western immunoblotting data from three independent experiments was done by ImageJ software and presented in supplementary figure S1.
Figure 2. Effect of rapamycin, 17-AAG and LY 294002 on migration and invasion during TGF-β induced EMT.
A549 or H358 cells were serum starved for 24 h, stimulated with TGF-ß (5 ng/ml) in the presence or absence of inhibitors at indicated concentrations. After 72 h cells were trypsinized and plated in uncoated or matrigel coated transwell chambers to assess cellular migration and invasion respectively. Error bars represent the standard deviation (SD) from three independent replicates. (*) denotes statistically significant (P<0.05) difference when compared to TGF-β treated cells.
From the list of compounds identified, we also assessed the effect of acetylsalicyclic acid and novobiocin on TGF-β-induced EMT. At the concentrations tested, both these compounds showed no significant effects on either biochemical or functional markers (migration and invasion) of EMT (data not shown). However, we have not ruled out the effect of these two compounds on the other functional phenotypes conferred by EMT, including growth inhibition, resistance to apoptosis, evasion of immune surveillance and, in certain cases, stem cell-like properties (2, 19).
Effect of rapamycin, 17-AAG and LY294002 on Smad phosphorylation and transcriptional activation
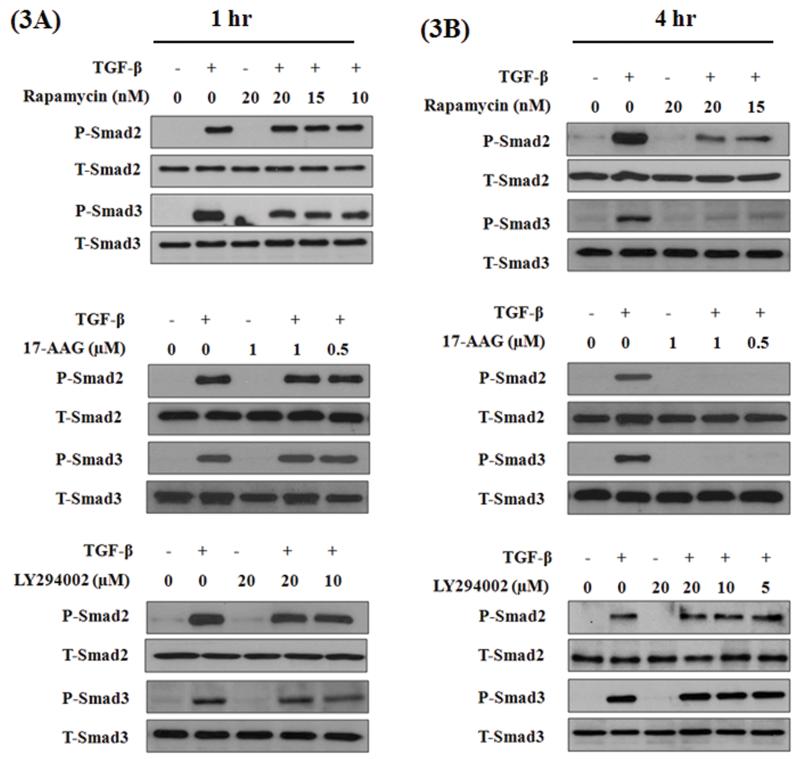
TGF-β induces robust phosphorylation of Smad 2 and 3, by TGF-β-receptor-I kinase, within one hour and persists beyond 4 hours. Both Smad-dependent and independent signaling pathways were implicated in TGF-β-induced EMT (8). However, in different cells we and others (20) have shown that activation of Smad3 is indispensible for TGF-β-induced EMT, including in A549 cells (21). We tested the above three compounds for their potential effects on TGF-β-induced Smad phosphorylation. A549 cells were stimulated with TGF-β for 1 h in the presence and absence of LY-294002 or rapamycin or 17-AAG at indicated concentrations and assessed for Smad2 and Smad3 phosphorylation by western immuno-blotting. All three compounds had no effect on Smad2 or Smad3 phosphorylation after 1 h of TGF-β stimulation (Figure 3A). This demonstrates that none of these three compounds have any non-specific effect on the TGF-β-receptor-I kinase.
Figure 3. Effect of rapamycin, 17-AAG and LY 294002 on TGF-β induced Smad phosphorylation.
A549 cells were serum starved for 24 h, stimulated with TGF-β (5 ng/ml) in the presence or absence of inhibitors for 1h or 4 h. Cell lysates were assessed for phospho-Smad 2 and 3 and total Smad 2 and 3 protein levels by western immunoblotting.
In a recent study, HSP90 was shown to be critical for the stability of TGF-β receptors, after stimulation with TGF-β, for a sustained Smad phosphorylation. As a result, inhibitors of HSP90 had no effect on immediate Smad phosphorylation within an hour, but blocked sustained Smad phosphorylation as they triggered slow degradation of TGF-β-receptors (22). Consistent with these findings we observed a total inhibition of Smad phosphorylation after 4 h of TGF-β stimulation (Figure 3B). Interestingly, in contrast to its effect at 1 h time point, rapamycin also blocked Smad phosphorylation at 4 h after TGF-β stimulation (Figure 3B). Whereas, LY294002 had no effect on Smad phosphorylation at either time points (Figure 3A & B).
Effect of rapamycin, 17-AAG and LY294002 on Smad transcriptional activity
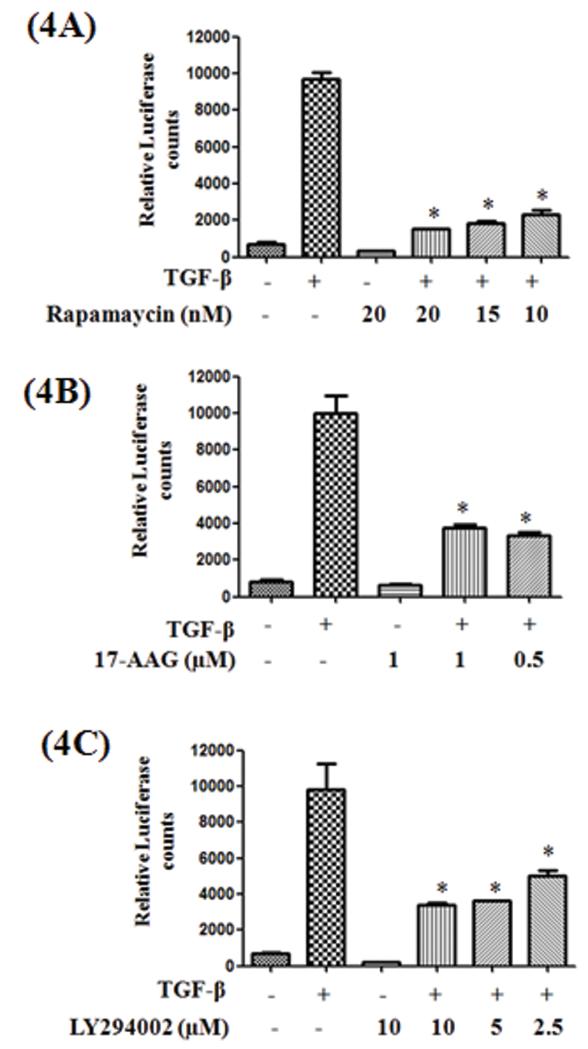
Following TGF-β stimulation, phosphorylated Smad 2 or 3 translocate into the nucleus as Smad 2/4 or Smad 3/4 heterodimers, bind to the Smad Binding Elements (SBE) in the promoters of their target genes and trigger gene transcription. To determine whether these compounds had any effect on TGF-β-induced Smad transcriptional activity, we tested the effect of these compounds in the presence and absence of TGF-β in A549 cells stably transfected with a Lentiviral based SBE-Luciferase reporter plasmid. Consistent with the inhibition of Smad phosphorylation, both 17-AAG and rapamycin significantly inhibited the TGF-β induced Smad transcriptional activity (Figure 4A & B). Surprisingly, although LY294002 had no effect on smad phosphorylation, it inhibited the TGF-β-induced transcriptional activation (Figure 4C).
Figure 4. Effect of rapamycin, 17-AAG and LY 294002 on TGF-β-induced Smad functional activity.
A549-SBE-Luc cells were serum starved for 24 h, stimulated with TGF-β (5ng/ml) in the presence or absence of inhibitors at indicated concentrations. After 4 h luciferase expression was measured and normalized to the protein concentrations. Error bars represent the standard deviation (SD) from three independent replicates. (*) denotes statistically significant (P<0.05) difference when compared to TGF-β treated cells.
DISCUSSION
Recently several groups successfully identified and validated potential modulators of different biological processes by analyzing the gene expression profiles using C-Map approach (11, 23-25). C-Map analysis does not require prior knowledge of the molecules or pathways involved in a biological process. Instead, by simply utilizing the pattern of gene expression alterations under study, compounds that can potentially reverse those alterations and therefore can serve as potential inhibitors of the process can be identified. Utilizing this approach we identified 21 compounds with various mechanisms of action as potential inhibitors of EMT and validated their affects in two independent TGF-β induced EMT models.
Experimental validation of hits from C-Map analysis identified rapamycin as a novel inhibitor of TGF-β signaling and a potent inhibitor of EMT. Rapamycin in complex with FKBP12 interacts with mTOR and inhibits its activity in the mTORC1 complex (26). mTOR activity is increased in many tumors, including lung cancer (27); inhibition of mTOR function through rapamycin analogues is considered as promising therapeutic strategy. Earlier reports have suggested that activation of mTOR is a Smad-independent TGF-β pathway that regulates protein synthesis, complementing the Smad-mediated transcriptional regulation (28). Studies with NMuMG mouse mammary epithelial cells and HaCat human keratinocytes showed no effect of rapamycin on TGF-β-induced EMT; however, rapamycin blocked EMT-associated increase in cell size and invasion in these cells (28). In contrast, we observed a potent inhibition of TGF-β-induced EMT by rapamycin in both A549 and H358 models of EMT. The effect of rapamycin on EMT was evident at the level of both biochemical markers (preventing the loss of E-cadherin and gain of N-cadherin proteins) as well as at the resulting functional phenotype (inhibition of migration and invasion). This discrepancy might be indicative of a potential difference in TGF-β signaling between malignant (A549) and non-malignant (NMuMG and HaCat) cells.
The most surprising observation was the effect of rapamycin on TGF-β-induced Smad phosphorylation. Rapamycin significantly inhibited phosphorylation of Smad2 and Smad3 at 4 h, but not at 1h, after TGF-β stimulation. This clearly indicates that the effect of rapamycin on Smad phosphorylation is not due to a non-specific or off-target effect on TGF-β receptor-I kinase. The HSP90 inhibitor 17-AAG demonstrated similar kinetics in inhibiting Smad phosphorylation (Figure 3A and Figure 3B). This is consistent with the recent finding that HSP90 is critical for the stability of TGF-β receptors and required longer duration of drug treatment to observe significant degradation of TGF-β receptors (22). Accordingly, 17-AAG was also a potent inhibitor of EMT in this study in both cell types tested. Given the similarity between the effects of rapamycin and 17-AAG, it may be important to investigate the role of rapamycin and potentially mTOR in regulating the stability of TGF-β receptors, particularly in cancer cells. As opposed to our observations, earlier studies have reported potentiation of TGF-β signaling with rapamycin (29). FKBP12, the protein to which rapamycin binds, interacts with TGFβRI to inhibit activation of Smads (29, 30). It was suggested that presence of rapamycin sequesters FKBP12 from TGFβRI to potentiate TGF-β signaling (31). These observations were primarily made in non-malignant epithelial cells and predominantly from the NMuMG mouse mammary epithelial cell line. It would be interesting to investigate whether the FKBP12 pathway is still functional in cancer cells and, if it is, then how rapamycin is modulating TGF-β signaling. In contrast to rapamycin and 17-AAG, LY294002 had no effect on Smad phosphorylation. Interestingly, LY294002 did significantly inhibit TGF-β-induced Smad transcriptional activity, suggesting a role for the PI3K pathway in the transcriptional regulation of TGF-β signaling.
Earlier reports showed cross-talk between PI3K and mTOR pathways where inhibition of one pathway modulates the other, depending on the cell type and the context. Hence, it was expected that inhibition of PI3K or mTOR may result in similar effects. On the contrary, we observed that rapamycin attenuated both E-cadherin loss and N-cadherin gain, whereas LY294002 selectively inhibited EMT-induced N-cadherin and vimentin expression without affecting the loss of E-cadherin. This suggests that both these compounds have effects that are independent of the cross-talk between them, such as modulation of TGF-β signaling by rapamycin. However, both compounds equally blocked EMT-induced migration, invasion and MMP secretion which strongly suggests a role for both cross-talk dependent and independent pathways.
In addition to these three compounds, we also assessed the effect of acetylsalicyclic acid and novobiocin on TGF-β-induced EMT. At the concentrations tested, both these compounds showed no significant effects on either biochemical or functional markers (migration and invasion) of EMT (data not shown). Apart from migratory and invasive phenotype, EMT is known to confer other functional phenotypes to cancer cells, including growth inhibition, resistance to apoptosis, evasion of immune surveillance and, in certain cases, stem cell-like properties (2, 19). Therefore, it is possible that the compounds that showed no effect on the markers we tested may still affect the other functional phenotypes described above to justify their identification as potential EMT inhibitors.
In summary, despite the prevalent notion that rapamycin either potentiates TGF-β signaling (29) or has no effect on EMT (28), we identified rapamycin as a candidate inhibitor of TGF-β signaling and EMT. Also, in contrast to previous reports (32), we identified LY294002 as a selective inhibitor of mesenchymal phenotype during EMT. In addition, 17-AAG was identified as a potent EMT inhibitor which was consistent with the role of HSP90 in the stability of TGF-β receptors (22). Collectively, these results demonstrate the need for such system-wide approaches (33) to look beyond the bias of prior information for gaining new insights.
Supplementary Material
Acknowledgments
This research is funded by NIH/NCI (CA132571-01), and American Cancer Society (RSG - CSM-116801) grants to V.G.K. Proteomics Alliance for Cancer Research grants from the Michigan Technology Tri-Corridor (MEDC-GR238, MTTC-GR687) to G.S.O.
Abbreviations
- EMT
epithelial-mesenchymal transition
- TGF-β
transforming growth factor-β
- MMPs
matrix metallo proteases
- SBE
Smad binding Element
- C-Map
Connectivity Map
Footnotes
The authors declare no conflict of interest.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003 May;3(5):362–74. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009 Jun;119(6):1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006 Jul;6(7):506–20. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 4.Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT) Respir Res. 2005;6:56. doi: 10.1186/1465-9921-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muraoka RS, Dumont N, Ritter CA, et al. Blockade of TGF-beta inhibits mammary tumor cell viability, migration, and metastases. J Clin Invest. 2002 Jun;109(12):1551–9. doi: 10.1172/JCI15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies M, Robinson M, Smith E, Huntley S, Prime S, Paterson I. Induction of an epithelial to mesenchymal transition in human immortal and malignant keratinocytes by TGF-beta1 involves MAPK, Smad and AP-1 signalling pathways. J Cell Biochem. 2005 Aug 1;95(5):918–31. doi: 10.1002/jcb.20458. [DOI] [PubMed] [Google Scholar]
- 7.Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003 Aug;4(8):657–65. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 8.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006 Feb;7(2):131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 9.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006 Mar 27;172(7):973–81. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerhold DL, Jensen RV, Gullans SR. Better therapeutics through microarrays. Nat Genet. 2002 Dec;32(Suppl):547–51. doi: 10.1038/ng1042. [DOI] [PubMed] [Google Scholar]
- 11.Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007 Jan;7(1):54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 12.Sartor MA, Mahavisno V, Keshamouni VG, et al. ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics. 2010 February 15;26(4):456–63. doi: 10.1093/bioinformatics/btp683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006 Sep 29;313(5795):1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 14.Zhang SD, Gant TW. A simple and robust method for connecting small-molecule drugs using gene-expression signatures. BMC Bioinformatics. 2008;9:258. doi: 10.1186/1471-2105-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reka Ajaya Kumar, Kurapati Himabindu, Narala Venkata R, et al. Peroxisome Proliferator Activated Receptor-γ Activation Inhibits Tumor Metastasis by Antagonizing Smad3 Mediated Epithelial Mesenchymal Transition. 2010. Revision Submitted to Molecular Cancer Therapeutics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000 Dec;1(3):169–78. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 17.Lee MS, Johansen L, Zhang Y, et al. The novel combination of chlorpromazine and pentamidine exerts synergistic antiproliferative effects through dual mitotic action. Cancer Res. 2007 Dec 1;67(23):11359–67. doi: 10.1158/0008-5472.CAN-07-2235. [DOI] [PubMed] [Google Scholar]
- 18.Shin SY, Choi BH, Ko J, Kim SH, Kim YS, Lee YH. Clozapine, a neuroleptic agent, inhibits Akt by counteracting Ca2+/calmodulin in PTEN-negative U-87MG human glioblastoma cells. Cell Signal. 2006 Nov;18(11):1876–86. doi: 10.1016/j.cellsig.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002 Jun;2(6):442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 20.Roberts AB, Tian F, Byfield SD, et al. Smad3 is key to TGF-beta-mediated epithelial-to mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev. 2006 Feb-Apr;17(1-2):19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Reka AK, Kurapati H, Narala VR, et al. Peroxisome Proliferator-Activated Receptor-{gamma} Activation Inhibits Tumor Metastasis by Antagonizing Smad3-Mediated Epithelial- Mesenchymal Transition. Mol Cancer Ther. Dec;9(12):3221–32. doi: 10.1158/1535-7163.MCT-10-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrighton KH, Lin X, Feng XH. Critical regulation of TGFbeta signaling by Hsp90. Proc Natl Acad Sci U S A. 2008 Jul 8;105(27):9244–9. doi: 10.1073/pnas.0800163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vilar E, Mukherjee B, Kuick R, et al. Gene expression patterns in mismatch repair- deficient colorectal cancers highlight the potential therapeutic role of inhibitors of the phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin pathway. Clin Cancer Res. 2009 Apr 15;15(8):2829–39. doi: 10.1158/1078-0432.CCR-08-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Preter K, De Brouwer S, Van Maerken T, et al. Meta-mining of neuroblastoma and neuroblast gene expression profiles reveals candidate therapeutic compounds. Clin Cancer Res. 2009 Jun 1;15(11):3690–6. doi: 10.1158/1078-0432.CCR-08-2699. [DOI] [PubMed] [Google Scholar]
- 25.Hassan SB, Gali-Muhtasib H, Goransson H, Larsson R. Alpha terpineol: a potential anticancer agent which acts through suppressing NF-kappaB signalling. Anticancer Res. Jun;30(6):1911–9. [PubMed] [Google Scholar]
- 26.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006 Feb 10;124(3):471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Ebi H, Tomida S, Takeuchi T, et al. Relationship of deregulated signaling converging onto mTOR with prognosis and classification of lung adenocarcinoma shown by two independent in silico analyses. Cancer Res. 2009 May 1;69(9):4027–35. doi: 10.1158/0008-5472.CAN-08-3403. [DOI] [PubMed] [Google Scholar]
- 28.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007 Jul 30;178(3):437–51. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y-G, Liu F, Massague J. Mechanism of TGF[beta] receptor inhibition by FKBP12. EMBO J. 1997;16(13):3866–76. doi: 10.1093/emboj/16.13.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massague J. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell. 2001 Sep;8(3):671–82. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 31.Wang T, Donahoe PK, Zervos AS. Specific interaction of type I receptors of the TGF- beta family with the immunophilin FKBP-12. Science. 1994 Jul 29;265(5172):674–6. doi: 10.1126/science.7518616. [DOI] [PubMed] [Google Scholar]
- 32.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000 Nov 24;275(47):36803–10. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 33.Peter J. Woolf, Angel Alvarez, Keshamouni Venkateshwar G. Systems Approach for Understanding Metastasis. In: Keshamouni Venkateshwar G., Arenberg Douglas A., Kalemkerian GP., editors. Lung Cancer Metastasis: Novel Biological Mechanisms and Impact on clinical Practice. Springer; NewYork: 2009. pp. 383–94. [Google Scholar]
- 34.Rodina A, Vilenchik M, Moulick K, et al. Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat Chem Biol. 2007;3(8):498–507. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- 35.Maloney A, Workman P. HSP90 as a new therapeutic target for cancer therapy: the story unfolds. Expert Opin Biol Ther. 2002 Jan;2(1):3–24. doi: 10.1517/14712598.2.1.3. [DOI] [PubMed] [Google Scholar]
- 36.Papadimitrakopoulou V, Adjei AA. The Akt/mTOR and mitogen-activated protein kinase pathways in lung cancer therapy. J Thorac Oncol. 2006 Sep;1(7):749–51. [PubMed] [Google Scholar]
- 37.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Cancer Res. Mar 15;70(6):2146–57. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gridelli C, Maione P, Rossi A. The potential role of mTOR inhibitors in non-small cell lung cancer. Oncologist. 2008 Feb;13(2):139–47. doi: 10.1634/theoncologist.2007-0171. [DOI] [PubMed] [Google Scholar]
- 39.Donnelly A, Blagg BS. Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket. Curr Med Chem. 2008;15(26):2702–17. doi: 10.2174/092986708786242895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burlison JA, Neckers L, Smith AB, Maxwell A, Blagg BS. Novobiocin: redesigning a DNA gyrase inhibitor for selective inhibition of hsp90. J Am Chem Soc. 2006 Dec 6;128(48):15529–36. doi: 10.1021/ja065793p. [DOI] [PubMed] [Google Scholar]
- 41.Chen Q. Nordihydroguaiaretic acid analogues: their chemical synthesis and biological activities. Curr Top Med Chem. 2009;9(17):1636–59. doi: 10.2174/156802609789941915. [DOI] [PubMed] [Google Scholar]
- 42.Krysan K, Reckamp KL, Sharma S, Dubinett SM. The potential and rationale for COX-2 inhibitors in lung cancer. Anticancer Agents Med Chem. 2006 May;6(3):209–20. doi: 10.2174/187152006776930882. [DOI] [PubMed] [Google Scholar]
- 43.Swannie HC, Kaye SB. Protein kinase C inhibitors. Curr Oncol Rep. 2002 Jan;4(1):37–46. doi: 10.1007/s11912-002-0046-7. [DOI] [PubMed] [Google Scholar]
- 44.Williams CS, Sheng H, Brockman JA, et al. A cyclooxygenase-2 inhibitor (SC-58125) blocks growth of established human colon cancer xenografts. Neoplasia. 2001 Sep-Oct;3(5):428–36. doi: 10.1038/sj.neo.7900177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winters ME, Mehta AI, Petricoin EF, III, Kohn EC, Liotta LA. Supra-additive Growth Inhibition by a Celecoxib Analogue and Carboxyamido-triazole Is Primarily Mediated through Apoptosis. Cancer Res. 2005 May 1;65(9):3853–60. doi: 10.1158/0008-5472.CAN-04-1989. 2005. [DOI] [PubMed] [Google Scholar]
- 46.Fontaine E, McShane J, Page R, et al. Aspirin and non-small cell lung cancer resections: effect on long-term survival. Eur J Cardiothorac Surg. Jul;38(1):21–6. doi: 10.1016/j.ejcts.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Zhang Y, Min J, et al. Calcineurin promotes proliferation, migration, and invasion of small cell lung cancer. Tumour Biol. Jun;31(3):199–207. doi: 10.1007/s13277-010-0031-y. [DOI] [PubMed] [Google Scholar]
- 48.Eckstein LA, Van Quill KR, Bui SK, Uusitalo MS, O’Brien JM. Cyclosporin A Inhibits Calcineurin/Nuclear Factor of Activated T-Cells Signaling and Induces Apoptosis in Retinoblastoma Cells. Invest Ophthalmol Vis Sci. 2005 March 1;46(3):782–90. doi: 10.1167/iovs.04-1022. 2005. [DOI] [PubMed] [Google Scholar]
- 49.Zhu F, Zhang Z, Wu G, et al. Rho kinase inhibitor fasudil suppresses migration and invasion though down-regulating the expression of VEGF in lung cancer cell line A549. Med Oncol. Mar 19; doi: 10.1007/s12032-010-9468-5. [DOI] [PubMed] [Google Scholar]
- 50.Stork LC, Matloub Y, Broxson E, et al. Oral 6-mercaptopurine versus oral 6-thioguanine and veno-occlusive disease in children with standard-risk acute lymphoblastic leukemia: report of the Children’s Oncology Group CCG-1952 clinical trial. Blood. Apr 8;115(14):2740–8. doi: 10.1182/blood-2009-07-230656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quann EJ, Khwaja F, Zavitz KH, Djakiew D. The aryl propionic acid R-flurbiprofen selectively induces p75NTR-dependent decreased survival of prostate tumor cells. Cancer Res. 2007 Apr 1;67(7):3254–62. doi: 10.1158/0008-5472.CAN-06-3657. [DOI] [PubMed] [Google Scholar]
- 52.Dinney CP, Parker C, Dong Z, et al. Therapy of human transitional cell carcinoma of the bladder by oral administration of the epidermal growth factor receptor protein tyrosine kinase inhibitor 4,5-dianilinophthalimide. Clinical Cancer Research. 1997 February;3(2):161–8. 1997. [PubMed] [Google Scholar]
- 53.Haince JF, Rouleau M, Hendzel MJ, Masson JY, Poirier GG. Targeting poly(ADP-ribosyl)ation: a promising approach in cancer therapy. Trends Mol Med. 2005 Oct;11(10):456–63. doi: 10.1016/j.molmed.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Moon D-O, Kim M-O, Choi YH, Kim G-Y. Butein Sensitizes Human Hepatoma Cells to TRAIL-Induced Apoptosis via Extracellular Signal-Regulated Kinase/Sp1–Dependent DR5 Upregulation and NF-ΰB Inactivation. Molecular Cancer Therapeutics. 2010 June 1;9(6):1583–95. doi: 10.1158/1535-7163.MCT-09-0942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.