Abstract
Acrolein (2,3-propenal) is a major indoor and outdoor air pollutant originating largely from tobacco smoke or organic combustion. Given its high reactivity, the adverse effects of inhaled acrolein are likely due to direct interactions with the airway epithelium, resulting in altered epithelial function, but only limited information exists to date regarding the primary direct cellular targets for acrolein. Here, we describe a global proteomics approach to characterize the spectrum of airway epithelial protein targets for Michael adduction in acrolein-exposed bronchial epithelial (HBE1) cells, based on biotin hydrazide labeling and avidin purification of biotinylated proteins or peptides for analysis by LC-MS/MS. Identified protein targets included a number of stress proteins, cytoskeletal proteins, and several key proteins involved in redox signaling, including thioredoxin reductase, thioredoxin, peroxiredoxins, and glutathione S-transferase π. Because of the central role of thioredoxin reductase in cellular redox regulation, additional LC-MS/MS characterization was performed on purified mitochondrial thioredoxin reductase to identify the specific site of acrolein adduction, revealing the catalytic selenocysteine residue as the target responsible for enzyme inactivation. Our findings indicate that these approaches are useful in characterizing major protein targets for acrolein, and will enhance mechanistic understanding of the impact of acrolein on cell biology.
1. Introduction
Acrolein (2,3-propenal) is a common indoor and outdoor pollutant, produced by combustion of fossil fuels, wood burning, heating cooking oils, and cigarette smoking, and is considered one of the greatest non-cancer health risks of all organic air pollutants [1–3]. Cigarette smoking represents the primary source of acrolein exposure to humans, and mainstream cigarette smoke contains up to 90 ppm acrolein [4]. Acrolein can also be produced endogenously during several biological processes, such as lipid or amino acid oxidation during conditions of inflammation [5, 6], oxidative deamination of spermine [7, 8], or drug metabolism [9]. Measurements of acrolein in airway secretions from smoking subjects indicate its presence at concentrations of 1–10 μM [10, 11], concentrations that exert major effects on cellular function [12, 13]. Because of its considerable chemical reactivity, acrolein is thought to play an important role in many of the adverse health effects associated with smoking, such as chronic bronchitis or lung cancer [13], [14].
The biological effects of acrolein are related to the presence of two reactive groups: a carbonyl group which can form Schiff bases with primary amines, and an adjacent α,β-unsaturated bond with strong electrophilic character, which is highly reactive with nucleophilic cellular targets by Michael addition at the β-carbon. Primary reactions of acrolein with biological molecules are predominated by reactions with nucleophilic amino acid side chains in proteins, primarily cysteine thiol residues or the ε-amino group lysine, and the imidazole nitrogen of histidine [9, 12], as well as DNA bases [14], and small nucleophiles such as GSH [15]. Many of the reported cellular effects of acrolein, such as the activation of stress responses, (in)activation of transcription factors, and effects of cell proliferation and cell death pathways, are closely associated with changes in cellular GSH [16–18] and are often attributed to increased oxidative stress as a result of GSH depletion [19–22]. Because the acrolein reaction with GSH is an important detoxification mechanism, catalyzed by glutathione S-transferases (GSTs) [23, 24], and serves to minimize interactions of acrolein with other critical cell constituents, the biological effects of acrolein are most likely mediated by direct interactions with critical protein targets or DNA bases. Examples of these include proteins involved in activation of transcription factors such as NF-κB or Nrf2 [25-28], caspases that regulate apoptotic cell death [16, 18, 19], protein tyrosine phosphatases [29], or proteins involved in regulating redox signaling [30, 31]. Such chemical interactions of acrolein are analogous to those of other biologically relevant α,β-unsaturated carbonyls, such as the lipid oxidation product 4-hydroxynonenal (HNE) [32, 33] or the cyclopentenone prostaglandins [34], [35, 36], which have been extensively studied [37, 38]. However, differences in reactivity and cellular distribution may account for some observed differences in cell signaling effects of acrolein compared to more lipid-soluble electrophiles such as HNE [39].
In an attempt to understand the general mechanisms by which acrolein affects cell function, various global genomic and proteomic approaches have been employed to delineate alterations in gene or protein expression levels in response to acrolein [40, 41]. However, such approaches have as yet not been applied to identify direct protein targets of acrolein, information that would be critical to better understand the impact of acrolein on specific biological processes or signaling pathways. Several previous reports have described global strategies to identify protein targets of other electrophiles such as HNE, using immunopurification of HNE-protein adducts with specific antibodies [42]. Such approaches have the limitation that they would only recognize specific stable adducts that are selectively recognized by these antibodies [5, 42–46], and ignore other potentially important adducts. More recent strategies to identify protein targets for electrophiles have employed the use of e.g. biotin-tagged electrophiles [47–50], or the use of click chemistry to label proteins for their purification and analysis by MS [51]. Such approaches are, however, not applicable to acrolein because of its small molecular size and the absence of suitable sites for labeling without significant impact on its chemical properties or cellular distribution.
Alternative strategies based on derivatization of protein-bound carbonyls with hydrazides, such as dinitrophenylhydrazide (DNPH) [52, 53] or biotin hydrazide [19, 54], have been described. In the present study, we describe the use of biotin hydrazide labeling as a means to characterize protein targets for acrolein in intact bronchial epithelial cells, based on avidin purification of labeled proteins and proteomic analysis using LC-MS/MS. Secondly, we demonstrate more detailed proteomic analysis of one prominent target, thioredoxin reductase (TrxR), in order to identify the consequence of specific modifications within this selenoprotein for enzymatic activity. Overall, these combined approaches are useful to demonstrate direct targeting by acrolein of specific proteins in complex biological samples, and will guide more directed approaches to link specific protein modifications to the biological effects associated with acrolein exposure.
2. Materials and methods
2.1. Reagents
Cell culture materials and media were purchased from Invitrogen. Biotin hydrazide and monomeric avidin were obtained from Pierce. Sequencing grade trypsin was purchased from Promega. Streptavidin-HRP, TrxR1 primary antibody and all secondary antibodies were obtained from Sigma. Primary antibodies against Trx1 and Prx1 were purchased from Abcam and GSTπ from Abnova. Full length murine mitochondrial TrxR (mTrxR-GCUG) and a truncated form (mTrxRΔ3, lacking the terminal three amino acids: cysteine (C), selenocysteine (U) and glycine (G)) were generated and purified as previously described [55]. All other chemicals were purchased from either Sigma or Fisher.
2.2. Cell Culture and Acrolein Treatment
Experiments were performed with human bronchial epithelial HBE1 cells, which were cultured at 37°C under 5% CO2 in DMEM/F12 medium supplemented with 50 units/mL penicillin, 50 μg/mL streptomycin, 10 ng/mL cholera toxin, 10 ng/mL epidermal growth factor, 5 μg/mL insulin, 5 μg/mL transferrin, 0.1 μM dexamethasone, 15 μg/mL bovine pituitary extract and 0.5 mg/mL BSA, as described previously [56]. Cells were plated at a density of 500,000 cells per well in a 6 well plate and grown to 80–90% confluence prior to acrolein treatment. At least 30 min prior to acrolein treatment, media was replaced by Hank’s balanced salt solution (HBSS) containing 1.3 mM CaCl2 and 0.5 mM MgCl. Acrolein was prepared as a stock solution in water (immediately prior to use) and added to the HBSS to give a final concentration up to 30 μM (equivalent to 60 nmoles/106 cells). Following incubation with acrolein, HBSS was removed and cells were washed on ice with 5% dextrose and subsequently lysed in 500 mL lysis buffer containing 1% Triton X-100, 50 mM HEPES, 250 mM NaCl, 10% glycerol, 1.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 1 mM ethylene glycol-bis[β-aminoethyl ether] N,N,N’N’-tetraacetic acid (EDTA), 2 mM Na3VO4 and 10 μg/mL aprotinin and leupeptin (pH 7.4). Following 10 min on ice, cells were sonicated for 30 sec and centrifuged at 14,000 rpm for 5 min to remove cell debris. Protein concentrations were determined using the BCA reagent kit (Pierce).
2.3. Biotin Labeling of Acrolein Adducts
Cell lysates from untreated or acrolein-treated HBE1 cells were subjected to biotin hydrazide derivatization to label acrolein-protein adducts (Figure 1). Aliquots of cell lysate containing 550 μg protein were diluted in water (to a final protein concentration of 1.1 μg/μL), and reacted with 5 mM biotin hydrazide (at pH 5.0), added from a stock solution of 50 mM biotin hydrazide. Reaction mixtures were incubated at room temperature for 2 hrs with constant rotation. The samples were then cooled on ice, and an equal volume of 30 mM NaCNBH4 in 1X PBS was added to each mixture to stop the reaction. The samples were then incubated on ice for 1 hr with occasional vortex mixing prior to further processing.
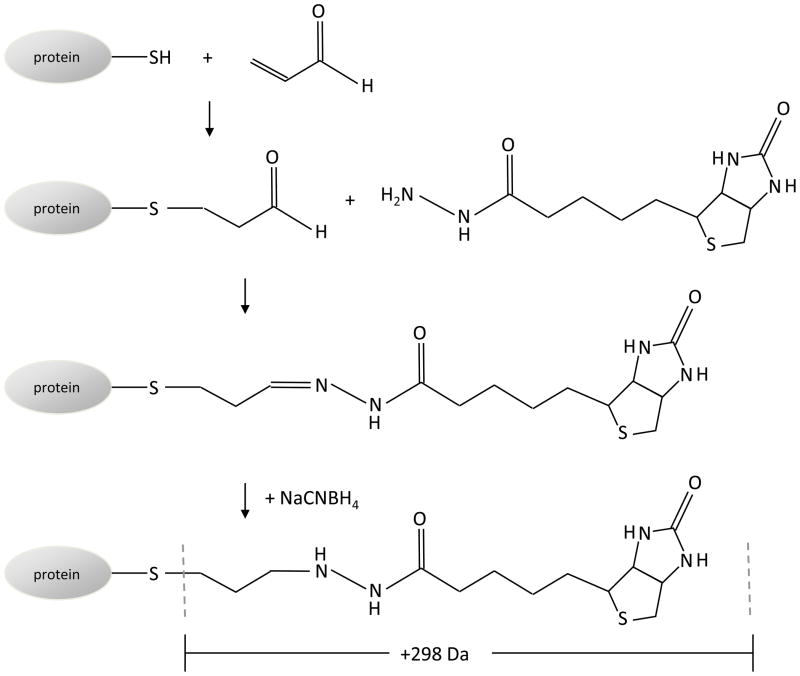
Figure 1. Biotin hydrazide labeling of protein Michael adducts of acrolein.
Acrolein reacts with cysteine, histidine and lysine amino acids via Michael addition to form a protein-bound aldehyde adduct. The aldehyde is labeled with biotin hydrazide, and the hydrazone bond is stabilized by reduction with NaCNBH4. Adduction of acrolein and derivatization with biotin hydrazide modification results in a mass increase of 298 Da.
2.4. Acrolein Adduct Confirmation
One microgram of total protein from each sample was boiled for 5 min with Laemmli sample buffer and was separated on a 10% Tris-Glycine reducing gel (Invitrogen), transferred to nitrocellulose (BioRad) in a wet transfer unit (BioRad) and blocked for 1 hr with 3% bovine serum albumin in Tris-buffered saline containing 0.05% Tween-20. The membrane was probed for biotin-labeled proteins with streptavidin-peroxidase (1/5000 dilution) and visualized using Pico Chemiluminescent Substrate (Pierce).
2.5. Purification of Biotin-Tagged Proteins and/or Peptides
For analysis of acrolein-modified proteins by bottom-up proteomics approaches, sample complexity was reduced in two ways. First, intact biotin-labeled proteins were isolated using avidin chromatography (see below), separated by 1-D gel electrophoresis for fractionation, and subjected to in-gel digestion of gel fragments and analysis by LC-MS/MS. In a second “peptide” approach, biotin-labeled cell lysates were first subjected to trypsinolysis, and biotin-tagged tryptic peptides were then purified by avidin column isolation for analysis by LC-MS/MS (Figure 2). Ideally, these two approaches provide complementary information, as the first method may include false-positive identification of proteins that were unlabeled but co-purified by avidin chromatography, whereas the second method may be more selective but may limit detection of certain acrolein-modified proteins.
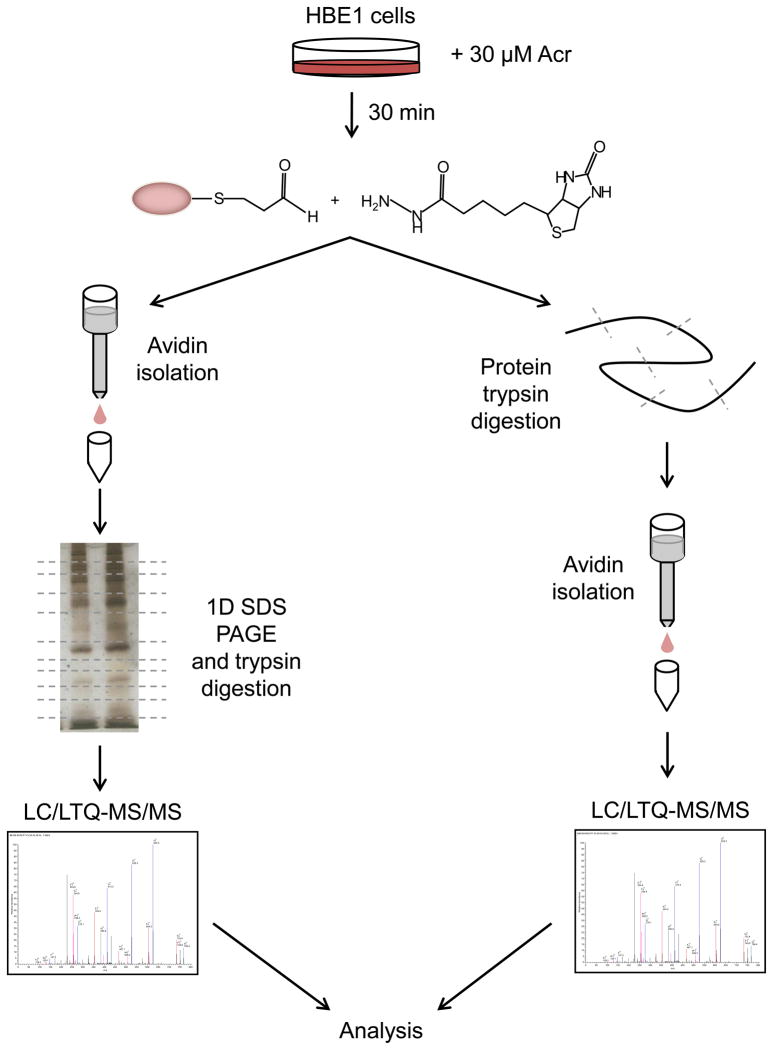
Figure 2. General strategy for proteomic analysis of acrolein-protein adducts.
HBE1 cells are treated with acrolein and lysed for labeling of protein-bound aldehydes with biotin hydrazide. Resulting biotinylated proteins are then isolated as intact proteins using avidin column chromatography, followed by trypsin digestion and LC-MS/MS analysis, or by initial trypsin digestion of the labeled proteins, followed by avidin column isolation of biotin-labeled peptides and detection by LC-MS/MS. Detected peptides are subsequently analyzed by database searching.
2.5.1. Avidin Chromatography
In the whole protein approach, cell lysate proteins were precipitated using 10% TCA for 20 min, and precipitated proteins were collected by centrifugation (14,000 rpm; 10 min; 4 °C) and washed three times by resuspension in 1 mL of 50% ethanol/50% ethylacetate and centrifugation. Following the last wash, protein pellets were allowed to dry and resuspended in 1 mL of lysis buffer. Biotin-labeled proteins were isolated as described previously [19]. Briefly, each sample was applied to a column packed with 2 mL Ultralink Immobilized Monomeric Avidin (Pierce) equilibrated with Buffer A (0.1 M Na2HPO4, 0.15 M NaCl; pH 7.2). After sample application, columns were washed with Buffer A until the absorbance at 280 nm reached baseline, and biotinylated proteins were eluted with 4 mM D-biotin in Buffer A. Collected protein fractions were concentrated to about 100 μL using Amicon Ultra 3 kDa molecular weight cutoff centrifugal units (Millipore).
For the “peptide” approach, 1.0 mg of the biotin hydrazide labeled samples were dried by SpeedVac and redissolved in 20 μL of 100 mM ammonium bicarbonate (NH4HCO3) and 10 mM dithiothreitol for 1 h at 56 °C. Then 5 μL of 55 mM iodoacetamide in 100 mM NH4HCO3 was added for 30 min in the dark, followed by SpeedVac drying again. The dried protein was dissolved in 30 μL of trypsin solution (10 ng/μL) in 50 mM NH4HCO3 and 4% acetonitrile, and incubated overnight at 37 °C. The resulting peptide solution was separated by avidin column isolation, as described above. Eluted peptides were concentrated and cleaned using PepClean C-18 Spin Columns according to manufactures specifications (Pierce). Peptides were eluted from the columns in 70% acetonitrile for analysis by ESI LC-MS/MS.
2.5.2. Separation and Digestion of Adducted Proteins
Purified biotin-tagged proteins from control samples and acrolein-treated samples were separated by 1-D SDS-PAGE, and stained with Silver Stain Plus (Bio-Rad). Each lane of the gel was cut into 28 individual slices, and each band was then cut into 1 mm2 cubes and further destained with three washes of 25 mM NH4HCO3 in 50% CH3CN with 10 min incubations followed by a 30 min incubation in water. Each group of gel cubes was then dehydrated in CH3CN for 10 min and dried in a Speed Vac. Protein samples were reduced by incubation of the gel pieces for 1 hr at 56 °C in 10 mM dithiothreitol (DTT) in 100 mM NH4HCO3. After removal of DTT, newly exposed cysteines were alkylated by the addition of 55 mM iodoacetamide in CH3CN to a solution of 100 mM NH4HCO3. The total volume of solution was large enough to fully submerge the gel pieces, and samples were incubated for 45 min in the dark. After removal of supernatant, gel pieces were washed with 100 mM NH4HCO3 for 10 min, and dehydrated in acetonitrile for 10 min. These re-swelling/dehydration steps were repeated once more followed by removal of the remaining solvents in a Speed Vac. A solution of 10 ng/μL trypsin in 10% acetonitrile/25 mM NH4HCO3 was used to re-swell the gel pieces completely at 4 °C for 30 min, followed by a 37 °C digestion overnight. A small amount of 10% formic acid was then added to stop the digestion, and samples were centrifuged at 2,800 x g and supernatants were collected for LC-MS/MS.
2.6. LC-MS/MS Analysis
A fused silica microcapillary LC column (15 cm long x 75 μm id) packed with C18 reversed-phase resin (5 μm particle size; 20 nm pore size; Magic C18AQ, Michrom Bioresources Inc.) was used with nanospray ESI. The nanospray ESI was fitted onto a linear quadrupole ion trap mass spectrometer (Thermo Electron) that was operated in CID mode to obtain both MS and tandem MS (MS/MS) spectra. Four μL of tryptic peptide samples were loaded onto the microcapillary column and separated by applying a gradient of 3–60% acetonitrile in 0.1% formic acid at a flow rate of 250 nL/min for 45 min. Mass spectrometry data were acquired in a data-dependent acquisition mode, in which a full MS scan was followed by 10 MS/MS scans of the most abundant ions.
2.6.1 Protein Identification
Obtained MS spectra were searched against the IPI Human protein sequence database (v3.75) using SEQUEST (Bioworks software, v3.3.1; Thermo Electron). The search parameters permitted a 2.0 Da peptide MS tolerance and a 1.4 Da MS/MS tolerance. Oxidation of methionine (M) and carboxymethylation of cysteines (C) were allowed as variable modifications. The other variable modifications of acrolein plus biotin hydrazide were set up as +298.0 on cysteines (C), histidines (H), and lysines (K) to find possible acrolein-reacted sites that had been subsequently labeled with biotin hydrazide. Up to two missed tryptic peptide cleavages were considered. The cutoffs for SEQUEST assignment were cross-correlation (Xcorr) scores greater than 1.9, 2.5, and 3.0 for peptide charge states of 1, 2, and 3, respectively, and a delta-correlation ( Cn) score >0.1. All proteins scores ≥20.0 (at least two unique peptides matched) were used in the following analysis.
2.7. Validation of Identified Proteins
Identification of proteins as targets for acrolein by LC-MS/MS was validated by analysis of purified biotin-hydrazide derivatized proteins by SDS-PAGE and Western blotting for proteins of interest, including TrxR1 (polyclonal rabbit primary 1/1000 in 5% milk, HRP-linked anti-rabbit secondary 1/8000 in 5% milk), Trx1 (polyclonal rabbit primary 1/5000 in 5% milk, HRP-linked anti-rabbit secondary 1/2000 in 5% milk), Prx1 (polyclonal rabbit primary 1/5000 in 5% milk, HRP-linked anti-rabbit secondary 1/2000 in 5% milk), and GSTπ (polyclonal rabbit primary 1/1000 in 5% milk, HRP-linked anti-rabbit secondary 1/1000 in 5% milk).
2.8. Functional and Network Analysis
In order to focus only on newly adducted proteins in response to acrolein exposure, identified biotin-labeled proteins from each of the gel slices obtained from acrolein-treated cell lysates were not included if they were also identified in corresponding gel slices from control samples. A list of acrolein adducted proteins was compiled and their corresponding gene ontogeny (GO) terms were determined using the Rosetta ID Converter (BABELOMICS, v3.20) [57]. A functional enrichment analysis (FatiGO) was performed and reported as the molecular function at level 3 (BABELOMICS).
2.9. Acrolein Inhibition of TrxR
Because the selenoprotein TrxR is highly susceptible to inactivation by electrophiles such as acrolein [31, 58], we performed additional biochemical and proteomic analyses of murine mTrxR to determine the specific modification sites that are associated with its inactivation. First, in an attempt to determine the importance of the C-terminal selenocysteine (U) as the primary target for acrolein, full-length mTrxR-GCUG or a truncated form lacking the 3 C-terminal CUG residues (mTrxRΔ3) was reacted with acrolein, and mTrxR activity was subsequently determined as described [59]. Reaction mixtures containing either mTrxR-GCUG or mTrxRΔ3 (200 nM) in reaction buffer containing 100 mM each of sodium acetate, MES, Tris, and dibasic potassium phosphate was first incubated with 100 μM NADPH for 5 min to reduce the mTrxR enzyme, and then treated with acrolein (0.1–30 μM). In attempt to distinguish between the C-terminal C or U residues, these reactions were carried out at varying pH (ranging from 5.5–8.5). Aliquots (20 μL) of this reaction mixture were then mixed with 1 mL reaction buffer (pH 7.0), containing 200 μM NADPH, 5 mM EDTA, and 3 mM DTNB, in a cuvette and absorbance increase at 412 nm was monitored for 1 min using a BioMate 5 spectrophotometer (Thermo Fisher). In addition, aliquots of these reaction mixtures were labeled with 50 μM biotin hydrazide (pH 5) and stopped with the addition of NaCNBH4 in 1X PBS, as described above. Protein samples were run on SDS-PAGE, transferred to nitrocellulose and probed with streptavidin-HRP (1/5000).
2.9.1 LC-MS/MS analysis of acrolein-mTrxR adducts
Full-length mTrxR was allowed to react with acrolein in a 1:50 ratio for 30 min at room temp. The reaction was terminated by addition of 5 mM NaBH4 and placing the reaction on ice for 60 min. The resultant solution was dialyzed against water to remove excess reagents. Tryptic peptides were generated as described above followed by LC separation and generation of tandem mass spectra, also as described previously. A single mammalian mitochondrial TrxR protein database was used to search the resulting spectra using the parameters described above, and the variable modification of +58 (acrolein) on C/H/K. All matched peptides were confirmed by manual inspection of their corresponding MS/MS spectra.
3. Results
3.1. Validation of Biotin Hydrazide Labeling of Acrolein-Modified Proteins
Derivatization of protein-acrolein adducts with biotin hydrazide allows for convenient purification of biotinylated proteins using avidin chromatography for analysis of labeled proteins by bottom-up proteomic approaches. To verify that this labeling procedure allows for specific dose-dependent labeling of acrolein adducts, HBE1 cells were exposed to increasing doses of acrolein, biotin hydrazide-labeled lysates were analyzed by SDS-PAGE and probed with streptavidin-peroxidase. As shown in Figure 3A, acrolein exposure resulted in increased formation of a broad range of biotin labeled proteins adducts, in a dose-dependent manner. Biotin hydrazide labeled proteins were detected immediately after acrolein exposure (< 30 min after acrolein exposure) and persisted for several hours after which they gradually disappeared due to reversal of acrolein adduction or proteolytic degradation of modified proteins (Fig. 3B). Although biotin hydrazide derivatization is clearly not specific for protein Michael adducts of acrolein, as it also derivatizes other protein carbonyl moieties, these findings suggest that this biotin labeling procedure is suitable for detecting acrolein adducts in response to acute exposure to acrolein.
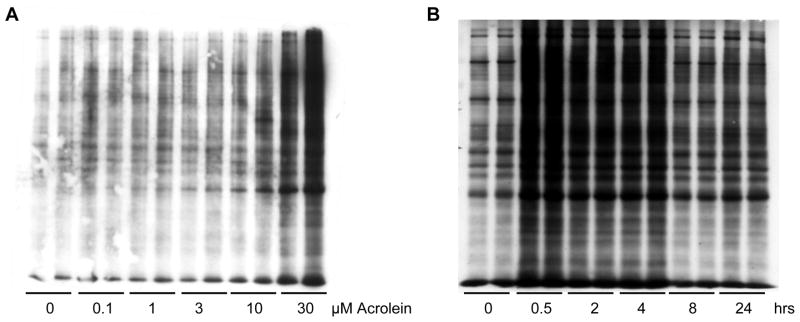
Figure 3. Visualization of biotin hydrazide labeled proteins in acrolein-exposed HBE1 cells.
(A) HBE1 cells were treated with 0-30 μM acrolein for 30 min and cell lysates were labeled with biotin hydrazide. (B) HBE1 cells were treated with 30 μM acrolein for 0-24 hrs and lysates were labeled with biotin hydrazide. One μg of total protein was separated by SDS-PAGE and proteins were transferred to a nitrocellulose membrane. Biotin bound proteins were detected by probing the membrane with streptavidin-HRP.
3.2. Global Analysis of Acrolein-Protein Adducts
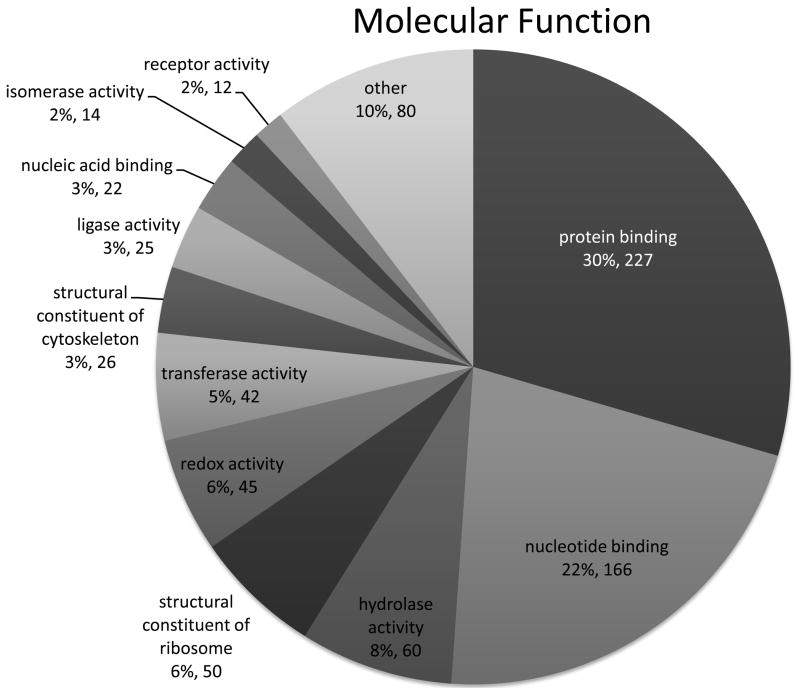
Intact purified biotinylated proteins were digested by trypsin and analysed by LC-MS/MS, which led to identification of 3468 proteins that differed between the control and 30 μM acrolein treated samples. Of these, 2178 proteins had corresponding Entrez gene identification numbers that were used for molecular function and network analysis. Because many proteins were identified in multiple gel bands, ultimately 769 unique proteins were determined to be adducted by acrolein (supplemental Table S1). For a fraction of these proteins (161), we were able to identify peptides that contained an acrolein-biotin hydrazide tag (characterized by mass increase of +298; Supplemental Table S2). Of the total number of 769 identified proteins, 629 had documented molecular functions. Examination of the molecular function of the 769 proteins demonstrated that 227 (30%) are involved in protein binding, 166 (22%) nucleotide binding, 60 (8%) contained hydrolase activity, 50 (6%) are a structural constituent of the ribosome, and 46 (6%) contain oxidoreductase activity (Fig. 4). According to the gene ontogeny curators, the definition of “protein binding” as a functional term is “Interacting selectively and non-covalently with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules)" (GO database release 2010-11-20). This indicates that many of the proteins identified as acrolein adducts are involved in protein-protein interactions, lending a layer of complexity when determining if the acrolein adduct disrupts not only individual protein activity/function but also that of complexes and signaling pathways.
Figure 4. Functional categorization of acrolein-adducted proteins.
A total of 769 proteins were identified as targets for adduction by acrolein, and 90% of these proteins fall within 11 major functional categories. 227 (30%) are involved in protein binding, 166 (22%) nucleotide binding, 60 (8%) contained hydrolase activity, 50 (6%) are a structural constituent of the ribosome, and 46 (6%) contain oxidoreductase activity.
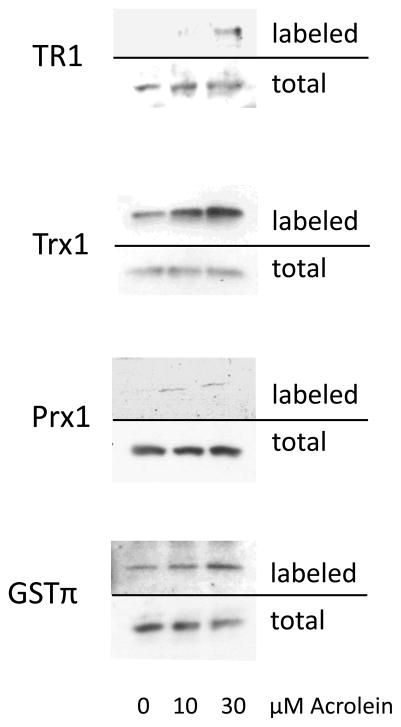
Another prominent group of proteins containing acrolein adducts comprise those with redox activity. Many of the 46 proteins identified in this group contain highly reactive cysteines within their active sites. Table 1 lists these proteins, the gel band(s) they were found in, along with the protein score and the number of unique peptides used for identification. Western blots analysis of purified biotin hydrazide-derivatized proteins for the presence of selected redox enzymes identified in the protein screen confirms increased biotin labeling of these proteins in response to acrolein, in a dose-dependent manner, consistent with direct adduction of these proteins by Michael addition (Fig. 5).
Table 1.
Redox proteins identified in global analysis. Forty six proteins were identified in the global analysis as having redox activity. These proteins were identified over a wide molecular weight range and many were found in multiple gel slices.
| Protein | gel sliceA | Protein Score | peptidesB |
|---|---|---|---|
| Glycerol-3-phosphate dehydrogenase, mitochondrial | 14 | 20.14 | 2 |
| Glutamate dehydrogenase 1&2, mitochondrial | 9 | 20.12 | 2 |
| Squalene synthetase | 11 | 30.18 | 3 |
| 13 | 28.18 | 3 | |
| 18 | 20.17 | 2 | |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial | 23 | 20.14 | 2 |
| MDH1 Malate dehydrogenase, cytoplasmic | 14 | 50.20 | 5 |
| 20 | 20.16 | 2 | |
| Quinone oxidoreductase | 14 | 20.19 | 2 |
| prostaglandin-endoperoxide synthase 2 | 27 | 20.16 | 2 |
| 28 | 20.18 | 2 | |
| Inosine-5′-monophosphate dehydrogenase 2 | 5 | 20.13 | 2 |
| 17 | 20.13 | 2 | |
| 26 | 20.15 | 2 | |
| 28 | 30.15 | 3 | |
| Aldo-keto reductase family 1, member C1 | 23 | 20.13 | 2 |
| 25 | 20.13 | 2 | |
| Amine oxidase A | 3 | 20.12 | 2 |
| 15 | 20.11 | 2 | |
| 16 | 28.13 | 3 | |
| Sorbitol dehydrogenase | 3 | 28.15 | 3 |
| Vinculin | 22 | 30.23 | 3 |
| 28 | 20.13 | 2 | |
| Aldehyde dehydrogenase X, mitochondrial | 1 | 20.16 | 2 |
| 3 | 30.17 | 3 | |
| 4 | 20.15 | 2 | |
| Aldehyde dehydrogenase, dimeric NADP-preferring | 14 | 30.15 | 3 |
| 17 | 20.19 | 2 | |
| 22 | 20.17 | 2 | |
| Glucose-6-phosphate dehydrogenase | 16 | 30.16 | 3 |
| 25 | 20.13 | 2 | |
| Cytochrome b-c1 complex subunit 1, mitochondrial | 21 | 28.12 | 3 |
| Superoxide dismutase [Mn], mitochondrial | 26 | 20.13 | 2 |
| Aldo-keto reductase family 1, member C2 | 23 | 20.13 | 2 |
| 25 | 20.13 | 2 | |
| Amine oxidase B | 11 | 20.12 | 2 |
| 16 | 20.13 | 2 | |
| ERO1-like protein alpha | 11 | 30.18 | 3 |
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | 14 | 20.19 | 2 |
| 17 | 20.17 | 2 | |
| 6-phosphogluconate dehydrogenase, decarboxylating | 15 | 20.18 | 2 |
| 23 | 20.20 | 2 | |
| Isocitrate dehydrogenase [NADP], cytoplasmic | 19 | 20.20 | 2 |
| 24-dehydrocholesterol reductase | 1 | 30.11 | 3 |
| Peroxiredoxin-1 | 19 | 20.12 | 2 |
| 26 | 30.12 | 3 | |
| 27 | 20.12 | 2 | |
| Peroxiredoxin-2 | 27 | 20.18 | 2 |
| Peroxiredoxin-6 | 28 | 30.16 | 3 |
| Aldehyde dehydrogenase, mitochondrial | 14 | 30.15 | 3 |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | 8 | 20.14 | 2 |
| Estradiol 17-b-dehydrogenase 12 | 19 | 20.15 | 2 |
| Prostaglandin Reductase 1 | 20 | 20.19 | 2 |
| Epidermal retinal dehydrogenase 2 | 16 | 40.20 | 4 |
| 21 | 30.19 | 3 | |
| 23 | 30.23 | 3 | |
| Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | 25 | 20.18 | 2 |
| Sulfide:quinone oxidoreductase, mitochondrial | 2 | 20.11 | 2 |
| NADH-cytochrome b5 reductase 3 | 15 | 20.15 | 2 |
| Isoform 1 of Fatty acid desaturase 2 | 19 | 20.16 | 2 |
| Isoform 1 of Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | 15 | 22.12 | 2 |
| DNA-(apurinic or apyrimidinic site) lyase | 16 | 20.12 | 2 |
| aldehyde dehydrogenase 7 family, member A1 | 13 | 20.15 | 2 |
| 21 | 20.11 | 2 | |
| Isoform 5 of Thioredoxin reductase 1, cytoplasmic | 6 | 22.11 | 2 |
| 13 | 20.13 | 2 | |
| Trifunctional enzyme subunit alpha, mitochondrial | 1 | 20.10 | 2 |
| 19 | 20.20 | 2 | |
| 1 2,4-dienoyl-CoA reductase, mitochondrial Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 14 | 20.18 | 2 |
| 10 | 20.14 | 2 | |
| 17 | 20.17 | 2 | |
| 20 | 30.18 | 3 | |
| Carbonyl reductase [NADPH] 1 | 28 | 20.21 | 2 |
| Thioredoxin | 2 | 20.11 | 2 |
| 10 | 20.14 | 2 | |
| 11 | 20.23 | 2 | |
| 22 | 20.15 | 2 | |
| 23 | 20.15 | 2 | |
| 27 | 30.17 | 3 |
The gel slice that the protein was detected in;
The number of peptides used for identification.
Figure 5. Validation of acrolein adducted proteins from HBE1 cells by Western blot.
Avidin-purified biotinylated proteins from untreated or acrolein-treated HBE1 cells were separated by SDS-PAGE and analyzed by Western blots using antibodies against TrxR, Trx, Prx or GSTπ. Whole cell lysates were analyzed similarly for comparison.
Our more restrictive “peptide” approach, in which biotin-labeled peptides were purified and analyzed after protein digestion, led to identification of 142 proteins, each containing at least one acrolein/biotin hydrazide modification (Supplemental Table S3). Of these 142 proteins, 33 were also identified in our analysis of intact biotinylated proteins. Of all biotin hydrazide labels detected in isolated labeled peptides, 48% were cysteine adducts, 36% were lysine adducts and 16% were histidine adducts. Biotin-labeled peptides were also detected in control samples, although none of them were found to contain a detectable acrolein/biotin hydrazide modification (+298). Biotin hydrazide labeling of control samples would reflect endogenous protein carbonylation resulting from normal cell metabolism. Indeed a number of diverse mechanisms can contribute to endogenous formation of protein carbonyls, which may include Michael addition by endogenously generated α,β-unsaturated aldehydes (e.g. HNE) but also metal-catalyzed oxidation or glycation/glycoxidation of lysine residues [60]. These adducts would not be identified using our restricted search of variable modification of +298, specific for acrolein adducts. Because of the highly variable nature of potential endogenous protein carbonylation mechanisms, we did not attempt to expand our search to identify other modifications. The main point of the present study was to evaluate the use of biotin hydrazide labeling to detect protein modifications by exogenous acrolein exposure.
3.3. Acrolein adduction of mTrxR Sec
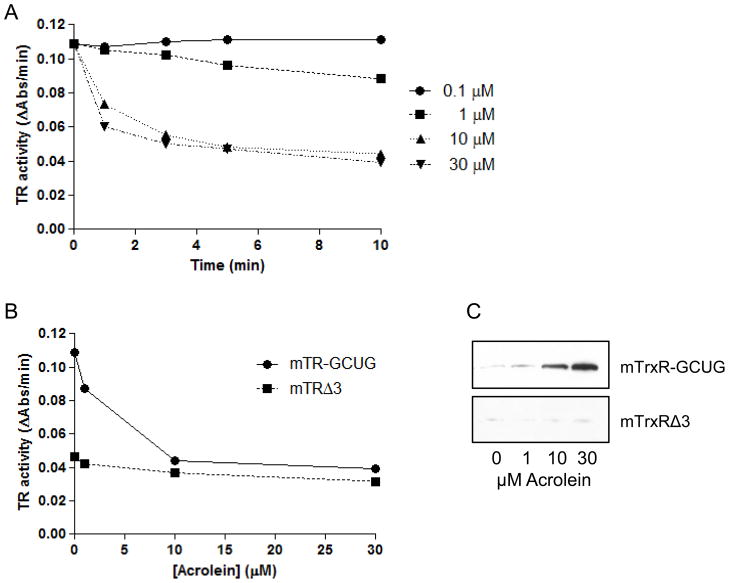
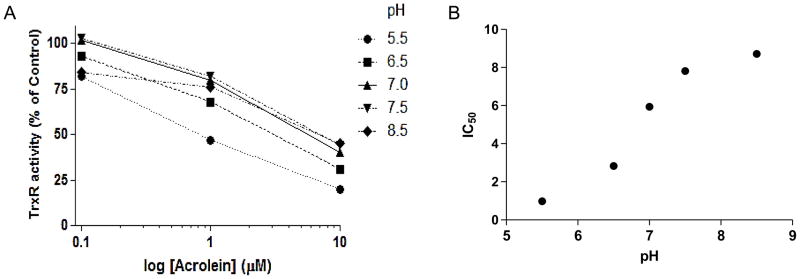
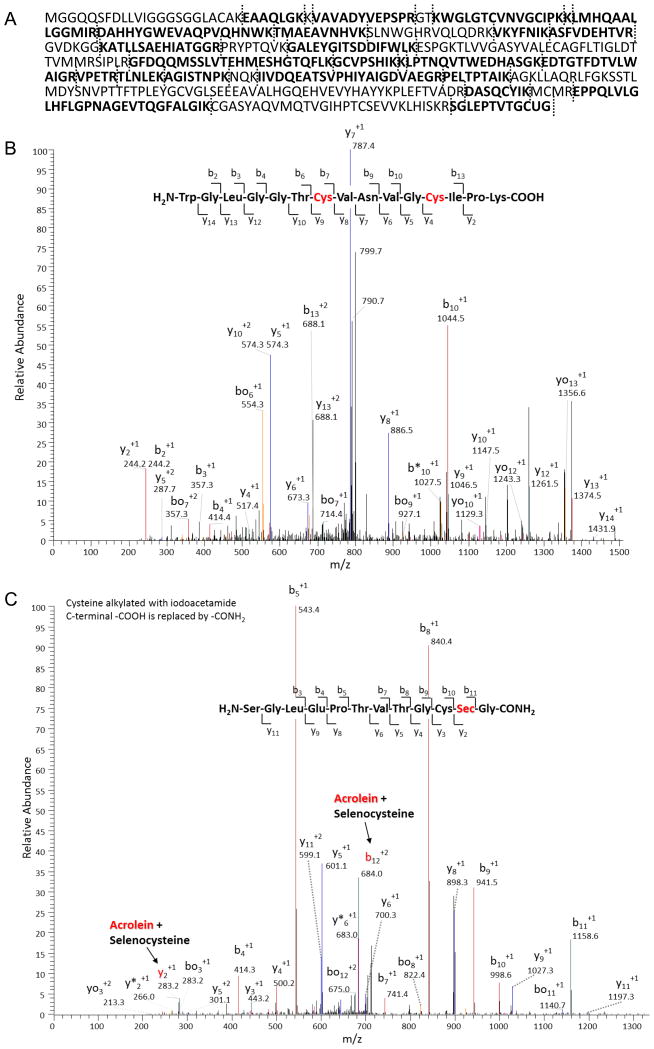
To investigate the impact of acrolein adduction on TrxR activity, follow-up studies were performed using purified mTrxR. As expected, exposure of mTrxR-GCUG to increasing concentrations of acrolein resulted in a dose- and time-dependent decrease in enzymatic activity (Fig. 6AB). A truncated form of mTrxR, lacking the C-terminal penultimate selenocysteine residue (mTrxRΔ3), had reduced specific activity compared to the full-length form, but this residual activity was relatively resistant to further inactivation by similar concentrations of acrolein (Fig. 6B). This suggested that inactivation of mTrxR by acrolein was due to binding to either the cysteine or selenocysteine dyad residues at the C-terminus. Accordingly, streptavidin blotting of biotin hydrazide labeled reaction mixtures showed alkylation of mTrxR-GCUG by acrolein (Fig. 6C), and the extent of biotin hydrazide binding corresponds with loss of enzyme activity. Furthermore, markedly less biotin labeling was observed in the truncated mTrxRΔ3 under similar conditions (Fig. 6C), suggesting that alkylation of full-length mTrxR occurred primarily at the C-terminal CUG sequence. In a further attempt to distinguish between the C-terminal C or U residue as targets for acrolein, we performed a reaction of acrolein with mTrxR-GCUG at pH 5.5-8.5, based on difference in pKa values of C (usually about 8.5) and U (typically around 5.2) [61]. Our results demonstrate that mTrxR-GCUG was more sensitive to inactivation at lower pH (Fig. 7), even though at this pH most C residues are likely to be protonated and significantly less nucleophilic. These findings point to U as the primary target for acrolein, consistent with its stronger nucleophilic character [61]. To directly determine the location of acrolein adduction in the full length mTrxR, the enzyme was incubated with a 50x molar excess of acrolein followed by trypsinization and analysis by LC-MS/MS. Matching of identified peptides resulted in 60% sequence coverage (Table 2 and Figure 8A). All C residues within identified peptides were found to be alkylated, that is, they did not react with acrolein. This can be seen in the N-terminal peptide (fig 8B). The only amino acid found to be adducted by acrolein was the C-terminal Sec (+58) (Fig. 8C). This identification is confirmed by the presence of both the b12+2 and the y2+1 ions in the spectrum of the C-terminal peptide. Interestingly, Table S2 indicates detection of an acrolein adduct on a cysteine in peptide KCYAKIICNTKDNER, representing aa 570–584 of TrxR1, but we did not detect a similar modification within a corresponding peptide from mTrxR-GCUG, QCUIKMVCMREPPQL (representing aa 445-459). This is likely due to marked sequence differences between human TrxR1 (cytosolic) and murine mitochondrial TrxR. Also, the importance of C570 of TrxR1 for enzymatic activity is unknown.
Figure 6. Inactivation of TrxR by acrolein.
(A) Time-dependent inhibition of enzyme activity of 200 nM mTrxR-GCUG during incubation with acrolein (0.1–30 μM) at pH 7.0. (B) Comparison of enzyme dose-dependent inactivation by acrolein (10 min, pH 7.0) of full length mTrxR-GCUG and truncated mTrxRΔ3 (200 nM each). (C) Reaction mixtures from panel B were derivatized with biotin hydrazide, for detection of biotin incorporation in mTrxR-GCUG and mTrxRΔ3 using streptavidin blotting.
Figure 7. Acrolein-induced inactivation of mTrxR as a function of pH.
(A) mTrxR- GCUG was incubated with acrolein (0.1–10 μM) at various pH (ranging from 5.5–8.5) for 10 min, and TrxR activity was determined using the DTNB assay. (B) Plot of IC50 values for acrolein as a function of buffer pH.
Table 2.
Matched peptides of mTrxR-GCUC reacted with acrolein. This peptide list was generated following LC-MS/MS analysis of acrolein adducted mTrxR-GCUG and resulted in a 60% sequence coverage.
| Peptide | MH+ | z |
|---|---|---|
| R.VPETR.T | 601.33040 | 1 |
| K.EAAQLGK.K | 716.39373 | 1 |
| K.AGISTNPK.N | 787.43084 | 1 |
| K.GC# VPSHIK.K | 897.44463 | 2 |
| K.TM* AEAVQNHVK.S | 1243.60903 | 2 |
| R.DASQC# YIK.M | 984.42904 | 1 |
| K.ASFVDEHTVR.G | 1160.56946 | 2 |
| K.RSGLEPTVTGC# U@ G.- | 1441.58193 | 2 |
| R.VPETRTLNLEK.A | 1299.72669 | 2 |
| K.VKYFNIK.A | 911.53491 | 2 |
| K.VAVADYVEPSPR.G | 1302.66884 | 2 |
| R.DAHHYGWEVAQPVQHNWK.T | 2202.02640 | 3 |
| K.KLM* HQAALLGGMIR.D | 1554.85978 | 3 |
| K.KLPTNQLQVTWEDHASGK.E | 2052.05088 | 3 |
| K.KLMHQAALLGGMIR.D | 1538.86578 | 2 |
| K.ATLLSAEHIVIATGGR.P | 1608.90678 | 2 |
| K.WGLGGTC# VNVGC# IPK.K | 1617.75467 | 2 |
| R.GFDQQMSSLVTEHMESHGTQFLK.G | 2637.20720 | 3 |
| K.IIVDAQEATSVPHIYAIGDVAEGRPELTPTAIK.A | 3474.84788 | 3 |
| K.EDTGTFDTVLWAIGR.V | 1680.82278 | 3 |
| K.GALEYGITSDDIFWLK.E | 1827.91634 | 2 |
| R.EPPQLVLGLHFLGPNAGEVTQGFALGIK.C | 2902.58254 | 3 |
+57 carboxymethylation on cysteine
+16 oxidation of methionine
+58 acrolein adduction to Selenocysteine
Figure 8. Identification of acrolein adducts in mTrxR by LC-MS/MS.
(A) Matched peptides of mTrxR detected by LC-MS/MS are indicated in bold, and represent a sequence coverage of 60%. Vertical dashed lines indicate trypsin cuts. (B) MS/MS spectrum for the N-terminal peptide containing two catalytic cysteine residues (indicated in red), indicating that both cysteines were only detected as alkylated adduct with iodoacetamide, and no corresponding acrolein-adducts were found. Similarly, no acrolein-adducts were detected in any of the other Cys-containing peptides. (C) MS/MS spectrum of C-terminal peptide containing the GCUG catalytic site, indicating acrolein adduction of the SeCys. Analysis of b12 and y2 ions indicate acrolein adduction of SeCys but not Cys.
4. Discussion
The objective of the present study was to characterize the target proteome for the common electrophile, acrolein, a major component of cigarette smoke and other indoor pollutants. In spite of many studies towards its biological and molecular effects [12, 13] and previous global analyses of transcriptome and proteome changes in response to acrolein, such a global survey of direct protein targets for acrolein has to date not been performed. Our strategy was to reduce sample complexity by employing affinity capture of acrolein-modified proteins using biotin hydrazide labeling and avidin chromatography, and bottom-up shotgun proteomic analysis of captured proteins or peptides, purified either before or after trypsinolysis. Results from acrolein-treated HBE1 cells were corrected for those of untreated cells to ensure that only newly modified proteins in response to acrolein were analyzed. Our results indicate that acrolein can directly alkylate a number of stress proteins, cytoskeletal proteins, and proteins that are critical in redox signaling. Among the latter are several peroxiredoxins (Prxs), thioredoxin (Trx) and thioredoxin reductase (TrxR), important antioxidant enzymes that regulate redox signaling, and GSTπ, a critical enzyme involved in acrolein detoxification [62]. A similar biotin hydrazide labeling strategy was recently used to determine protein adduction by a similar α,β-unsaturated aldehyde, HNE [47]. Perhaps not surprisingly, many of the proteins that were identified as targets for acrolein in the present study, were also identified as targets for HNE and were largely C-containing proteins, consistent with reactive C residues as the primary target for Michael addition by these aldehydes. Forty eight percent of the isolated biotin hydrazide labeled peptides contained a +298 modification on C, indicating acrolein adduction followed by biotin hydrazide labeling. Such preferential C targeting would be in accordance with quantum mechanical and kinetic data indicating that C is the primary target for acrolein [15, 63]. Many of these C-containing target proteins have also been identified in proteomic analysis of targets for other C-modifications such as S-nitrosylation [64] or S-glutathionylation [65, 66]. Because alkylations of these proteins are generally considered less reversible compared to these oxidative modifications, it follows that such alkylation may impair protein functions and disrupt dynamic redox control of these proteins by biologically relevant oxidants.
In our initial screen of avidin-captured biotinylated proteins, we identified the presence of acrolein-biotin hydrazide labeled peptides in only a small percentage (less than 10%) of the 769 identified proteins. One potential reason for this is the fact that fragmentation of the biotin label produces spectral noise which subsequently increases the expectation value [67]. Another potential reason is the fact that non-specific binding of proteins to the NeutrAvidin column during purification may have resulted in false positive protein identification. Because NeutrAvidin resin has the lowest nonspecific binding of the known biotin-binding proteins (Dr. F. Suleman, Thermo Fisher Scientific, personal communication), this problem should be minimal. Also, the fact that identified proteins were corrected for those found in control untreated samples suggests that the non-specific binding did not contribute significantly to our positive protein identification in acrolein-exposed cells, as such proteins would also be identified in control samples. For many proteins listed in Table 1 and Table S1, we only detected 2 unique peptides, which might raise some concern of false-positive protein identification. However, a reverse human protein database search of selected data indicated that the extent of such false-positive identification was less than 3%.
Another potential source of false-positive identification may originate from co-purification of non-modified proteins with biotinylated proteins due to formation of protein complexes. Indeed, the fact that a large proportion of identified proteins were in the category of “protein binding” strongly argues in favor of this possibility. More stringent washing produces could be used during avidin purification to minimize such non-specific binding. To avoid this problem, we used a secondary approach to identify acrolein-modified proteins, by first trypsinising biotin hydrazide labeled proteins prior to neutravidin column isolation and LC-MS/MS analysis. The advantage of this approach is that non-specific protein-protein interactions are minimized during avidin purification, reducing the incidence of detecting false positives. As expected, this led to identification of a smaller number of adducted proteins, although this may also be due in part to limited positive identification of biotin-tagged peptides due to spectral noise produced by fragmentation of the biotin label. Interestingly, all positively identified peptides contained at least one biotin-acrolein modification (+298), whereas none of the positively identified proteins in control samples did. Biotin hydrazide labeled proteins in control samples most likely reflect the presence of proteins carbonyls formed by alternative mechanisms such as oxidation [68] or Michael adduction by endogenously generated α,β-unsaturated aldehydes such as HNE.
While a significant portion of identified proteins (33 out of 142) in our second screen of biotinylated peptides overlapped with identified acrolein targets in our initial screen of intact biotinylated proteins, the spectrum of identified proteins also differs significantly between both approaches. The ion signals of acrolein-biotin modified peptides are significantly lower in the mass spectrum compared with their controls, which results in decreased ability to detect acrolein modified proteins, a common challenge in global analysis of post-translational modification of proteins. Our MS was set up to automatically pick up the 10 most intense ions for MS2 (Top 10 mode), which may have contributed to lower rate of detected acrolein-adducted peptides in our global analysis. Conversely, it is possible that avidin purification of digested proteins may have resulted in capture of biotin-tagged peptides that may not have been detected in during analysis of avidi-purified intact proteins, because structural or steric factors may have prevented successful collection of certain proteins. Because either approach has its advantages as well as limitations, we feel that a combination of both approaches is preferable for a more definitive proteome coverage affected by post-translational modifications, such as Michael adduction by acrolein. In either case, it is prudent to perform a secondary approach such as Western blot analysis to validate positively identified proteins of interest, as was done in this study for certain proteins involved in redox signaling.
Another important shortcoming in analysis of protein-acrolein adducts using such biotin hydrazide labeling approaches stems from the fact that Michael adducts of acrolein (and other α,β-unsaturated aldehydes) contain a carbonyl group capable of undergoing secondary reactions to form Schiff bases with amino groups, thereby preventing its recognition by hydrazide labeling. In fact, reactivity of the carbonyl moiety of acrolein towards nearby amino groups is enhanced after Michael addition due to loss of the electron donating adjacent double bond [15]. Such secondary reactions may lead to inter- and intra-protein cross-linking [63], and could contribute to misidentification of certain peptides during proteomic analysis.
Using our proteomic screening for acrolein-modified proteins, we observed that the main direct targets for acrolein include various heat shock proteins, cytoskeletal proteins, as well as proteins involved in redox regulation. Thus, in addition to readily depleting cellular GSH [18, 19, 23], and including indirect oxidative stress by reducing GSH status, acrolein could also enhance oxidative stress by directly interfering with redox-regulating proteins. One protein of critical importance in regulating cellular redox signaling is thioredoxin reductase (TrxR), which is uniquely sensitive to inactivation by various electrophiles due to its highly nucleophilic selenocysteine [61, 69]. Alkylation of TrxR by several electrophiles was found to not only inhibit its Trx reductase activity, which may affect cell signaling pathways that are controlled by thioredoxin (Trx), but in some cases also induces a gain-of-function due to increased NADPH oxidase and pro-oxidant activity, which may contribute to induction of apoptotic cell death [61, 70–72]. Since TrxR is typically overexpressed in various cancers, alkylation of TrxR has been proposed as a potential therapeutic anti-cancer strategy.
Our proteomic screen revealed TrxR as a target for alkylation by acrolein in intact HBE1 cells (Table S1 and S2), which was confirmed by Western blotting (Fig. 5). We performed follow-up studies using semi-synthetic purified mTrxR to identify the sites of modification and its relation to enzyme activity. As expected, exposure of mTrxR to acrolein resulted in a dose-dependent decrease in reductase activity (Fig. 6). Reductase activity of TrxR is largely dependent on the presence of the C-terminal GCUG sequence, containing a C-U dyad which is critical for Trx reduction, as illustrated by reduced overall reductase activity of a truncated form of mTrxR lacking this GCUG sequence. Because C or U typically have different pKa values (~8.3 and 5.2 respectively) [73, 74] and these amino acids have strongest nucleophilic character in their deprotonated forms, we explored the pH-dependent inactivation of TrxR, which indicated that acrolein is readily capable of inactivating TrxR at pH as low as 5 (which would likely protonate all reduced C residues), implicating the more nucleophilic U as the target for acrolein [63, 75, 76]. Direct proof of acrolein adduction to this U in mTrxR-GCUG was obtained by LC-MS/MS analysis of acrolein-modified TrxR, demonstrating acrolein adduction only at the U residue (Fig. 8). Interestingly, no acrolein adduct was detected at the N-terminal dithiol redox center, which is responsible for maintaining the reduced status of the C-U dyad at the C-terminal end. This is in agreement with the relative lack of biotin hydrazide labeling of acrolein-treated truncated TrxR detected by streptavidin blotting, and the relative inability of acrolein inhibits its remaining reductase activity (resulting from the N-terminal dithiol center) (Fig. 6). Since we did not search for other potential modifications of e.g. lysine or histidine residues, we cannot completely rule out the potential contribution of such modifications in altered TrxR activity, but our findings indicate that loss of TrxR activity by acrolein is primarily due to alkylation of the C-terminal selenocysteine residue.
In summary, the present study shows that biotin hydrazide labeling followed by avidin isolation strategies are a useful approach in characterizing protein targets for acrolein using bottom-up proteomics approaches, in spite of some important limitations. Because of its small chemical size, alternative strategies using tagged electrophiles or click chemistry approaches may not be readily applicable to acrolein. Our studies confirm that acrolein has the potential to modify a wide variety of cysteine-containing proteins within airway epithelial cells, which would not be detectable using available antibodies against protein-acrolein adducts [45]. Because of the importance of cysteine within various redox-dependent signaling networks, direct alkylation of these cysteines by acrolein may have important consequences for these signaling networks and be largely responsible for its ability to induce oxidative stress, cell structure dysregulation, alterations in cellular signaling, and toxicity.
Supplementary Material
Acknowledgments
This work has been supported by NIH research grants (R01-HL068865 and P20- RR16462) and a grant from the Flight Attendant Medical Research Institute (FAMRI) to AvdV. PCS was supported by a NIEHS postdoctoral training fellowship (T-28ES007122) and by a Young Clinical Scientist Award from FAMRI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leikauf GD. Hazardous air pollutants and asthma. Environ Health Perspect. 2002;110(Suppl 4):505–26. doi: 10.1289/ehp.02110s4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seaman VY, Charles MJ, Cahill TM. A sensitive method for the quantification of acrolein and other volatile carbonyls in ambient air. Anal Chem. 2006;78(7):2405–12. doi: 10.1021/ac051947s. [DOI] [PubMed] [Google Scholar]
- 3.Woodruff TJ, et al. Estimating risk from ambient concentrations of acrolein across the United States. Environ Health Perspect. 2007;115(3):410–5. doi: 10.1289/ehp.9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 5.Uchida K. Current staus of acrolein as a lipid peroxidation product. Trends Cardiovasc Med. 1999;9:109–113. doi: 10.1016/s1050-1738(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 6.Anderson MM, et al. Human neutrophils employ the myeloperoxidase-hydrogen peroxide-chloride system to convert hydroxy-amino acids into glycolaldehyde, 2-hydroxypropanal, and acrolein. A mechanism for the generation of highly reactive alpha-hydroxy and alpha,beta-unsaturated aldehydes by phagocytes at sites of inflammation. J Clin Invest. 1997;99(3):424–32. doi: 10.1172/JCI119176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwak MK, Kensler TW, Casero RA., Jr Induction of phase 2 enzymes by serum oxidized polyamines through activation of Nrf2: effect of the polyamine metabolite acrolein. Biochem Biophys Res Commun. 2003;305(3):662–70. doi: 10.1016/s0006-291x(03)00834-9. [DOI] [PubMed] [Google Scholar]
- 8.Tanel A, Averill-Bates DA. The aldehyde acrolein induces apoptosis via activation of the mitochondrial pathway. Biochim Biophys Acta. 2005;1743(3):255–67. doi: 10.1016/j.bbamcr.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Holian A. Acrolein: a respiratory toxin that suppresses pulmonary host defense. Rev Environ Health. 1998;13(1–2):99–108. [PubMed] [Google Scholar]
- 10.Andreoli R, et al. Determination of patterns of biologically relevant aldehydes in exhaled breath condensate of healthy subjects by liquid chromatography/atmospheric chemical ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17(7):637–45. doi: 10.1002/rcm.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Annovazzi L, et al. High-performance liquid chromatography and capillary electrophoresis: methodological challenges for the determination of biologically relevant low-aliphatic aldehydes in human saliva. Electrophoresis. 2004;25(9):1255–63. doi: 10.1002/elps.200305843. [DOI] [PubMed] [Google Scholar]
- 12.Kehrer JP, Biswal SS. The molecular effects of acrolein. Toxicological Sciences. 2000;57:6–15. doi: 10.1093/toxsci/57.1.6. [DOI] [PubMed] [Google Scholar]
- 13.Stevens JF, Maier CS. Acrolein: sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol Nutr Food Res. 2008;52:7–25. doi: 10.1002/mnfr.200700412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Z, et al. Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc Natl Acad Sci U S A. 2006;103(42):15404–9. doi: 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai J, Bhatnagar A, Pierce WM., Jr Protein Modification by acrolein: formation and stability of cysteine adducts. Chem Res Toxicol. 2009;22:708–16. doi: 10.1021/tx800465m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kern JC, Kehrer JP. Acrolein-induced cell death: a caspase-influenced decision between apoptosis and oncosis/necrosis. Chem Biol Interact. 2002;139(1):79–95. doi: 10.1016/s0009-2797(01)00295-2. [DOI] [PubMed] [Google Scholar]
- 17.Biswal S, et al. Inhibition of cell proliferation and AP-1 activity by acrolein in human A549 lung adenocarcinoma cells due to thiol imbalance and covalent modifications. Chem Res Toxicol. 2002;15(2):180–6. doi: 10.1021/tx015552p. [DOI] [PubMed] [Google Scholar]
- 18.Finkelstein EI, et al. Regulation of constitutive neutrophil apoptosis by the α,Bunsaturated aldehydes acrolein and 4-hydroxynonenal. Am J Physiol Lung Cell Mol Physiol. 2005;289:L1019–L1028. doi: 10.1152/ajplung.00227.2005. [DOI] [PubMed] [Google Scholar]
- 19.Hristova M, Heuvelmans S, van der Vliet A. GSH-dependent regulation of Fas-mediated caspase-8 activation by acrolein. FEBS Lett. 2007;581(3):361–7. doi: 10.1016/j.febslet.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaimes EA, et al. Stable compounds of cigarette smoke induce endothelial superoxide anion production via NADPH oxidase activation. Arterioscler Thromb Vasc Biol. 2004;24(6):1031–6. doi: 10.1161/01.ATV.0000127083.88549.58. [DOI] [PubMed] [Google Scholar]
- 21.Nardini M, et al. Acrolein-induced cytotoxicity in cultured human bronchial epithelial cells. Modulation by alpha-tocopherol and ascorbic acid. Toxicology. 2002;170(3):173–85. doi: 10.1016/s0300-483x(01)00540-6. [DOI] [PubMed] [Google Scholar]
- 22.Luo J, Robinson JP, Shi R. Acrolein-induced cell death in PC12 cells: role of mitochondria-mediated oxidative stress. Neurochem Int. 2005;47(7):449–57. doi: 10.1016/j.neuint.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Berhane K, et al. Detoxication of base propenals and other alpha, beta-unsaturated aldehyde products of radical reactions and lipid peroxidation by human glutathione transferases. Proc Natl Acad Sci U S A. 1994;91(4):1480–4. doi: 10.1073/pnas.91.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tjalkens RB, et al. Alpha,beta-unsaturated aldehydes increase glutathione S-transferase mRNA and protein: correlation with activation of the antioxidant response element. Arch Biochem Biophys. 1998;359(1):42–50. doi: 10.1006/abbi.1998.0895. [DOI] [PubMed] [Google Scholar]
- 25.Valacchi G, et al. Inhibition of NFkappaB activation and IL-8 expression in human bronchial epithelial cells by acrolein. Antioxid Redox Signal. 2005;7(1–2):25–31. doi: 10.1089/ars.2005.7.25. [DOI] [PubMed] [Google Scholar]
- 26.Horton ND, et al. Acrolein causes inhibitor kappaB-independent decreases in nuclear factor kappaB activation in human lung adenocarcinoma (A549) cells. J Biol Chem. 1999;274(14):9200–6. doi: 10.1074/jbc.274.14.9200. [DOI] [PubMed] [Google Scholar]
- 27.Lambert C, et al. Acrolein inhibits cytokine gene expression by alkylating cysteine and arginine residues in the NF-κB1 DNA binding domain. The Journal of Biological Chemistry. 2007;282(27):19666–19675. doi: 10.1074/jbc.M611527200. [DOI] [PubMed] [Google Scholar]
- 28.Wu RP, et al. Nrf2 responses and the therapeutic selectivity of electrophilic compounds in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2010;107(16):7479–84. doi: 10.1073/pnas.1002890107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seiner DR, LaButti JN, Gates KS. Kinetics and mechanism of protein tyrosine phosphatase 1B inactivation by acrolein. Chem Res Toxicol. 2007;20:1315–1320. doi: 10.1021/tx700213s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Go YM, et al. Reactive aldehyde modification of thioredoxin-1 activates early steps of inflammation and cell adhesion. The American Journal of Pathology. 2007;171(5):1670–1681. doi: 10.2353/ajpath.2007.070218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers CR, Myers JM. The effects of acrolein on peroxiredoxins, thioredoxins, and thioredoxin reductase in human bronchial epithelial cells. Toxicology. 2009;257:95–104. doi: 10.1016/j.tox.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen DR, Doorn JA. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free Radic Biol Med. 2004;37(7):937–45. doi: 10.1016/j.freeradbiomed.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Leonarduzzi G, Robbesyn F, Poli G. Signaling kinases modulated by 4-hydroxynonenal. Free Radic Biol Med. 2004;37(11):1694–702. doi: 10.1016/j.freeradbiomed.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, et al. Modification of ubiquitin-C-terminal hydrolase-L1 by cyclopentenone prostaglandins exacerbates hypoxic injury. Neurobiol Dis. 2011;41(2):318–28. doi: 10.1016/j.nbd.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle K, Fitzpatrick FA. Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J Biol Chem. 2010;285(23):17417–24. doi: 10.1074/jbc.M109.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim DH, et al. 15-Deoxy-Delta(12,14)-prostaglandin J(2) stabilizes, but functionally inactivates p53 by binding to the cysteine 277 residue. Oncogene. 2010;29(17):2560–76. doi: 10.1038/onc.2010.8. [DOI] [PubMed] [Google Scholar]
- 37.Uchida K, Shibata T. 15-Deoxy-Delta(12,14)-prostaglandin J2: an electrophilic trigger of cellular responses. Chem Res Toxicol. 2008;21(1):138–44. doi: 10.1021/tx700177j. [DOI] [PubMed] [Google Scholar]
- 38.Kim EH, Surh YJ. 15-deoxy-Delta12,14-prostaglandin J2 as a potential endogenous regulator of redox-sensitive transcription factors. Biochem Pharmacol. 2006;72(11):1516–28. doi: 10.1016/j.bcp.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Forman HJ. Signaling pathways involved in phase II gene induction by alpha, beta-unsaturated aldehydes. Toxicol Ind Health. 2009;25(4–5):269–78. doi: 10.1177/0748233709102209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar P, Hayes BE. Proteomic profiling of rat lung epithelial cells induced by acrolein. Life Sciences. 2009;85:188–95. doi: 10.1016/j.lfs.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson CA, Burcham PC. Genome-wide transcriptional responses to acrolein. Chemical Research in Toxicology. 2008;21:2245–56. doi: 10.1021/tx8001934. [DOI] [PubMed] [Google Scholar]
- 42.Uchida K, et al. Characterization of epitopes recognized by 4-hydroxy-2-nonenal specific antibodies. Arch Biochem Biophys. 1995;324:241–248. doi: 10.1006/abbi.1995.0036. [DOI] [PubMed] [Google Scholar]
- 43.Levonen AL, et al. Cellular mechanisms of redox cell signaling: role of cysteine modification controlling antioxidant defenses in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parola M, et al. HNE interacts directly with JNK isoforms in human hepatic stellate cells. J Clin Invest. 1998;102:1942–1950. doi: 10.1172/JCI1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furuhata A, et al. N(epsilon)-(3-methylpyridinium) lysine, a major antigenic adduct generated in acrolein-modified protein. J Biol Chem. 2003;278:48658–48665. doi: 10.1074/jbc.M309401200. [DOI] [PubMed] [Google Scholar]
- 46.Shao B, et al. Acrolein impairs ATP binding cassette transporter A1-dependent cholesterol export from cells through site-specific modification of apolipoprotein A-I. J Biol Chem. 2005;280(43):36386–96. doi: 10.1074/jbc.M508169200. [DOI] [PubMed] [Google Scholar]
- 47.Codreanu SG, et al. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Molecular and Cellular Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirzaei M, Regnier FE. Affinity chromatographic selection of carbonylated proteins followed by identification of oxidation sites using tandem mass spectrometry. Anal Chem. 2005;77(8):2386–2392. doi: 10.1021/ac0484373. [DOI] [PubMed] [Google Scholar]
- 49.Landar A, et al. A sensitive method for the quantitative measurement of protein thiol modification in response to oxidative stress. Free Radical Biology & Medicine. 2006;40:459–468. doi: 10.1016/j.freeradbiomed.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 50.Gayarre J, et al. Differential selectivity of protein modification by the cyclopentenone prostaglandins PGA1 and 15-deoxy-Delta12,14-PGJ2: role of glutathione. FEBS Lett. 2005;579(25):5803–8. doi: 10.1016/j.febslet.2005.09.069. [DOI] [PubMed] [Google Scholar]
- 51.Jacobs AT, Marnett LJ. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res. 2010;43(5):673–83. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conrad CC, et al. Identification of protein carbonyls after two-dimensional electrophoresis. Proteomics. 2001;1(7):829–34. doi: 10.1002/1615-9861(200107)1:7<829::AID-PROT829>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 53.Mello CF, et al. Acrolein induces selective protein carbonylation in synaptosomes. Neuroscience. 2007;147:674–679. doi: 10.1016/j.neuroscience.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roe MR, et al. Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Anal Biochem. 2007;79:3747–3756. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 55.Eckenroth B, et al. Semisynthesis and characterization of mammalian thioredoxin reductase. Biochemistry. 2006;45:5158–5170. doi: 10.1021/bi0517887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu R, Zhao YH, Chang MM. Growth and differentiation of conducting airway epithelial cells in culture. Eur Respir J. 1997;10(10):2398–403. doi: 10.1183/09031936.97.10102398. [DOI] [PubMed] [Google Scholar]
- 57.Medina I, et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010;38:210–3. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szadkowski A, Myers CR. Acrolein oxidizes the cytosolic and mitochondiral thioredoxins in human endothelial cells. Toxicology. 2008;243:164–176. doi: 10.1016/j.tox.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holmgren A, Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods in Enzymology. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 60.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10(5):485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 61.Arner ES. Selenoproteins-What unique properties can arise with selenocysteine in place of cysteine? Exp Cell Res. 2010;316(8):1296–303. doi: 10.1016/j.yexcr.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 62.Conklin DJ, et al. Glutathione-S-transferase P protects against endothelial dysfunction induced by exposure to tobacco smoke. Am J Physiol Heart Circ Physiol. 2009;296:H1586–H1597. doi: 10.1152/ajpheart.00867.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LoPachin RM, et al. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: nucleophilic targets and adduct formation. Chem Res Toxicol. 2009;22(9):1499–508. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao G, et al. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci. 2006;103(4):1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dalle-Donne I, et al. Actin Cys374 as a nucleophilic target of α,β-unsaturated aldehydes. Free Radic Biol Med. 2007;42:583–598. doi: 10.1016/j.freeradbiomed.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 66.Dalle-Donne I, et al. Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass Spectrometry Reviews. 2005;24:55–99. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- 67.Borisov OV, et al. Low-energy collision-induced dissociation fragmentation analysis of cysteinyl-modified peptides. Anal Chem. 2002;74(10):2284–92. doi: 10.1021/ac010974p. [DOI] [PubMed] [Google Scholar]
- 68.Yoo BS, Regnier FE. Proteomic analysis of carbonylated proteins in two-dimensional electrophoresis using avidin-fluorescein afiinity staining. Electrophoresis. 2004;25:1334–1341. doi: 10.1002/elps.200405890. [DOI] [PubMed] [Google Scholar]
- 69.Nordberg J, et al. Mammalian thioredoxin reductase is irreversibly inhibited by dinitrohalobenzenes by alkylation of both the redox active selenocysteine and its neighboring cysteine residue. J Biol Chem. 1998;273:10835–10842. doi: 10.1074/jbc.273.18.10835. [DOI] [PubMed] [Google Scholar]
- 70.Anestal K, Arner ES. Rapid induction of cell death by selenium-compromised thioredoxin reductase 1 but not by the fully active enzyme containing selenocysteine. J Biol Chem. 2003;278:15966–15972. doi: 10.1074/jbc.M210733200. [DOI] [PubMed] [Google Scholar]
- 71.Anestal K, et al. Cell death by SecTRAPs: thioredoxin reductase as a prooxidant killer of cells. PLoS ONE. 2008;3(4):e1846. doi: 10.1371/journal.pone.0001846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cassidy PB, et al. Thioredoxin reductase is required for the inactivation of tumor suppressor p53 and for apoptosis induced by endogenous electrophiles. Carcinogenesis. 2006;27(12):2538–2549. doi: 10.1093/carcin/bgl111. [DOI] [PubMed] [Google Scholar]
- 73.Huber RE, Criddle RS. Comparison of the chemical properties of selenocysteine and selenocystine with their sulfur analogs. Arch Biochem Biophys. 1967;122(1):164–73. doi: 10.1016/0003-9861(67)90136-1. [DOI] [PubMed] [Google Scholar]
- 74.Danehy JP, Noel CJ. The relative nucleophilic character of several mercaptans toward ethylene oxide. J Am Chem Soc. 1960;82:2511–2515. [Google Scholar]
- 75.Mugesh G, Sing HB. Synthetic organoselenium compounds as antioxidants: glutathione peroxidase activity. Chem Soc Rev. 2000;29:347–357. [Google Scholar]
- 76.Lee SR, et al. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc Natl Acad Sci U S A. 2000;97(6):2521–6. doi: 10.1073/pnas.050579797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.