Abstract
We present the BPIFAn/BPIFBn systematic nomenclature for the PLUNC (palate lung and nasal epithelium clone)/PSP (parotid secretory protein)/BSP30 (bovine salivary protein 30)/SMGB (submandibular gland protein B) family of proteins, based on an adaptation of the SPLUNCn (short PLUNCn)/LPLUNCn (large PLUNCn) nomenclature. The nomenclature is applied to a set of 102 sequences which we believe represent the current reliable data for BPIFA/BPIFB proteins across all species, including marsupials and birds. The nomenclature will be implemented by the HGNC (HUGO Gene Nomenclature Committee).
Introduction
The PLUNC (palate lung and nasal epithelium clone) family has been introduced elsewhere in this issue [1]. The purpose of the present article is to introduce a unified nomenclature for these genes/proteins which arose out of discussions at the recent focussed meeting ‘Proteins with a BPI/LBP/Plunc-like Domain: Revisiting the Old and Characterizing the New’ held in Nottingham, U.K. As discussed below, various confusing and duplicate names exist for these proteins, which hinders communication between workers in the field, and makes it difficult for those from outside the field to understand the relationships between the proteins. Our aim in proposing this nomenclature is to make the relationships between the different proteins as clear as possible.
A feature of the PLUNC proteins is that they are rapidly evolving: orthologues show high sequence dissimilarity (compared to interspecies comparisons for other proteins), and there are significantly different numbers of paralogues in different species [1–4]. It was therefore felt necessary to define a nomenclature that could most distinctly indicate 1:1 orthology between proteins, but also indicate the more subtle relationships where species or lineage-specific paralogues have arisen.
The meeting was not simply focussed on the PLUNC family, but also on the wider BPI (bactericidal/permeability-increasing protein) fold-containing superfamily, which we define as all proteins that can be strongly predicted to contain either one or both domains of the BPI fold. This superfamily can be robustly segregated by simple phylogenetic analysis into PLUNC and non-PLUNC branches [2]. The non-PLUNC branch includes BPI, LBP (lipopolysaccharide-binding protein), C E T P (cholersteryl ester-transfer protein), PLTP (phospholipid-transfer protein), and also other proteins such as BPIL2 [5] or LBPBPI1 [6]. Suitable names already exist for BPI, LBP, CETP and PLTP, and very little is characterized about many of the other proteins in the non-PLUNC branch. Nomenclature discussions were therefore initially restricted to the PLUNC branch. However, as discussed below, this restriction was later relaxed to allow for future inclusion of the non-PLUNC proteins.
Our proposals, which maintain a strong correspondence with a nomenclature that has become common in much of the literature and in database annotations, have been approved by the HGNC (HUGO Gene Nomenclature Committee).
Historical perspective
Over 25 years ago, rodent PSP (parotid secretory protein) was the first member of the PLUNC family that was identified and cloned [7]. Studies on PSP have been largely focused on expression analysis and it is well recognised as a highly abundant protein in rodent saliva. A second related gene, SMGB (submandibular gland protein B) was subsequently cloned from rat salivary glands [8]. The cloning of mouse PLUNC in 1999 [9] added another related protein to this family, and analysis contained within this paper recognized the similarity that these three proteins shared with the database sequences for mouse VEMSGP (von Ebner minor salivary gland protein) (GenBank® accession number U46068) and cow BSP30 (bovine salivary protein) A (accession number U79413) [10]. It was following the analysis of the human and mouse PLUNC genes that, in 2002, we showed that humans contain at least seven expressed genes in the PLUNC locus [11]. Subsequently, this number has undergone a number of revisions and has now been refined to eight authentic genes and three pseudogenes within the human locus. As has been highlighted elsewhere in this issue, this number varies across mammalian species [1,12]. At that time, we used a nomenclature which distinguished between the two possible types of proteins, based on length: SPLUNCn (short PLUNCn; e.g. SPLUNC2) for the one domain (‘short’) proteins comprising approximately 250 amino acids, and LPLUNCn (long PLUNC) for the two domain (‘long’) proteins comprising approximately 450 amino acids [11].
Community discussion¶
At the meeting, a parallel discussion session considered the various issues surrounding defining a satisfactory nomenclature. These discussions were then continued in a plenary session. One issue that was discussed was whether PLUNC was suitable as a basis for a ‘root’ name for the family. The term PLUNC was originally coined as an acronym for palate lung and nasal epithelium clone [9]. This acronym does not accurately convey the true variety of localizations of PLUNC, and furthermore the word ‘carcinoma’ has been substituted for ‘clone’ in many instances in the databases: a rewording that seems to have little basis in science. There is also a widespread concern that the SPLUNC1 and LPLUNC1 style of nomenclature can lead to confusion, whereby SPLUNC1 is misinterpreted as a short form of a longer protein LPLUNC1.
Continuation of the discussions following the meeting led to the proposal of a nomenclature that can encompass the whole BPI fold-containing superfamily with the introduction of a BPIF root. New gene symbols will be allocated by the HGNC for the PLUNC branch proteins, whereas the symbols will have the status of aliases for the well-established BPI, LBP, CETP and PLTP proteins.
The BPIF superfamily is divided into BPIFA, BPIFB, BPIFC etc., subfamilies. BPIFA will replace the SPLUNC root, BPIFB will replace the LPLUNC root, BPIFC will replace the symbol for BPIL2 [5] and BPIFD, BPIFE and so on, will be used by the HGNC as aliases for BPI, LBP etc., in a manner yet to be finalized. The allocation of these latter aliases will not be discussed further here.
Nomenclature for the PLUNC branch proteins
-
The nomenclature system will be a modification of the existing SPLUNCn/LPLUNCn style names:
The SPLUNC root will be replaced by BPIFA
The LPLUNC root will be replaced by BPIFB
Wherever possible the existing numbering of proteins will be retained
Assignment to families will be on the basis of inspection of sequence-based phylogenetic trees and pairwise identity patterns as described below.
Small amendments will be made to the nomenclature of a limited number of proteins. A number of proteins for which no SPLUNCn/LPLUNCn style name existed will have a name allocated.
The main amendment to the current usage is where expansion of the number of paralogues has occurred in a lineage, and where none of the resulting proteins can be identified at the sequence level to have retained significantly the most similarity to the presumptive orthologue. In these cases ‘A’, ‘B’, ‘C’ etc. will be appended to the gene name. Where expansions have occurred in two lineages separately, different letters will then be used in the two lineages.
Application of the nomenclature proposals
For the purpose of refining and testing the nomenclature proposals above, a collection of 102 sequences was established, which is a combination of well-established PLUNC branch sequences, along with sequences in which we have moderately high confidence from the current NCBI nr database. Initially, BLAST searches against the nr database were performed using all human and/or mouse LPLUNC and SPLUNC proteins as queries until a converged set of proteins was obtained. The sequences were reduced to maximum 90% pairwise identity using cd-hit and sequences from the set in Chiang et al. [13] were then added. A number of manually collected sequences were added, especially to populate more fully the BASE branch. An iterative process of constructing phylogenetic trees and inspecting apparent anomalies against EST (expressed sequence tag) databases was then followed. In this process, a number of sequences were either discarded as faulty predictions, or replaced by more secure predictions. Phylogenetic trees were prepared using ClustalW [14] and visualized using ITOL [15]. Sequence analysis was facilitated by using the Jalview resource [16] and in-house Python Scripts. A phylogenetic tree of the resulting dataset is shown in Figure 1. We believe that this represents a good snapshot of the current state of knowledge of the species distribution of PLUNC proteins, and this is our basis for assessing the effectiveness of the proposed nomenclature.
Figure 1. Phylogenetic tree of the 102 proteins in the collection described in the text.
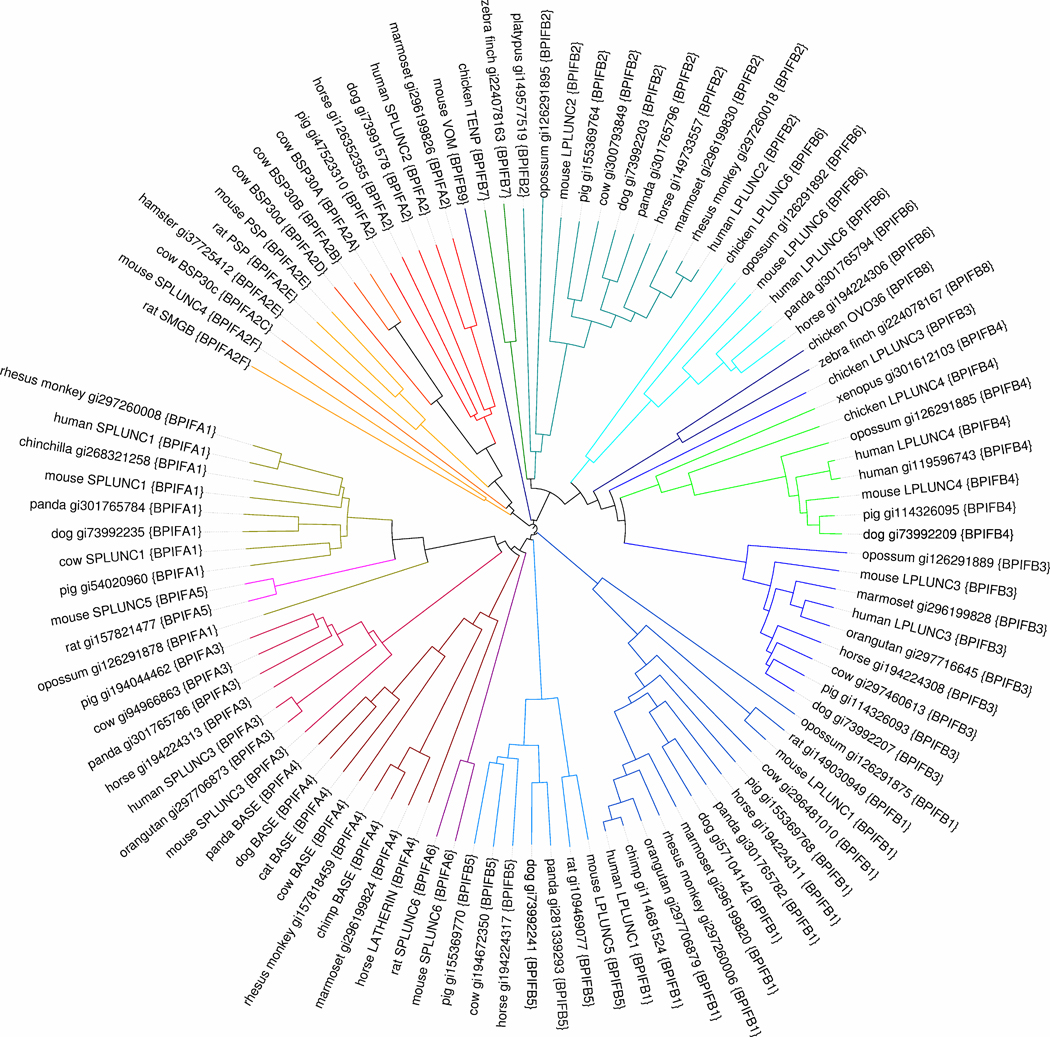
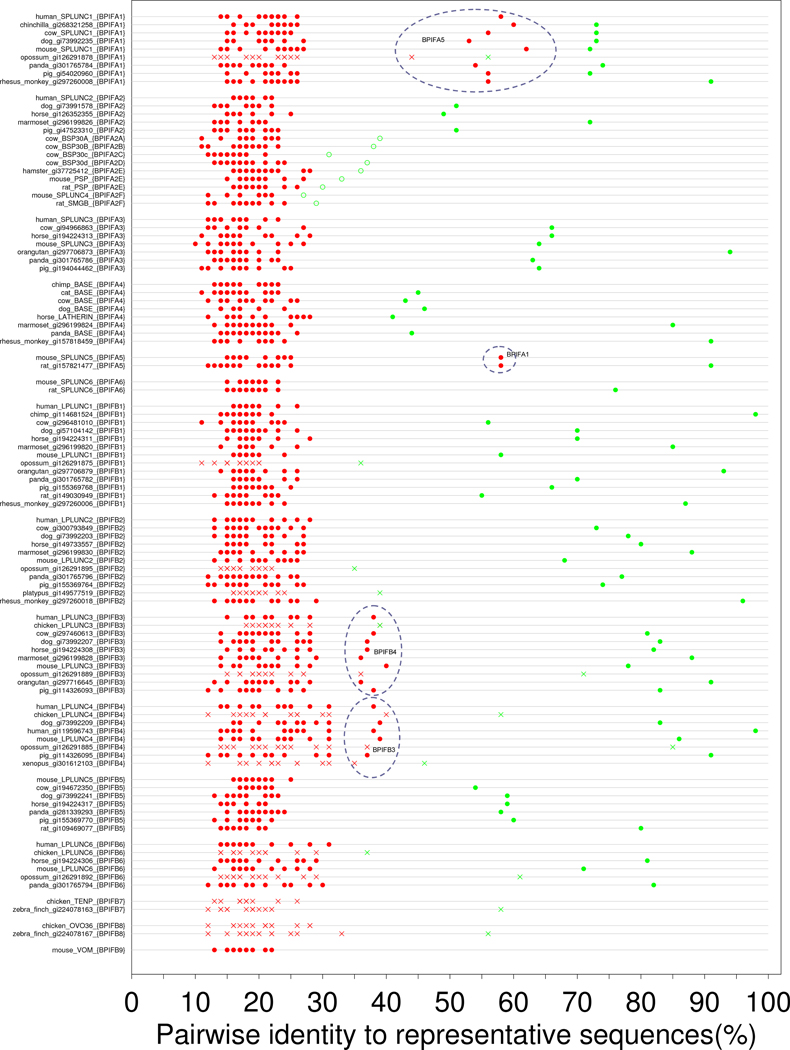
The tree was constructed using clustalw and displayed using ITOL. The names of proteins are of the style X {Y}. For proteins that are described in the literature, X is the common name generally used there. For proteins derived from BLAST searches for this work the proteins are identified by NCBI gi accession number. Y is the systematic name established by inspection of the tree, and further justified by the analysis in Figure 2. Branches joining proteins within the same family are in the same colour. A range of similar hues is used for the subfamilies of BPIFA2. For one domain BPIFA proteins, colours at the red end of the spectrum are used, whereas the blue end is used for the two domain BPIFB proteins. Species included in this figure: chicken (Gallus gallus), chimp (Pan troglodytes), chinchilla (Chinchilla lanigera), cow (Bos taurus), dog (Canis familiaris), hamster (Mesocricetus auratus), horse (Equus caballus), human (Homo sapiens), marmoset (Callithrix jacchus), mouse (Mus musculus), opossum (Monodelphis domestica), orangutan (Pongo abelii), panda (Ailuropoda melanoleuca), pig (Sus scrofa), platypus (Ornithorhynchus anatinus), rabbit (Oryctolagus cuniculus), rat (Rattus norvegicus), rhesus monkey (Macaca mulatta), xenopus (Xenopus silurana), zebra finch (Taeniopygia guttata). Note that in order to maintain a reasonable size to this collection of sequences, a 90% maximum pairwise identity was imposed during its assembly (see main text for details). Therefore if a protein is not shown for a particular species it does not necessarily imply its absence from that species. This is particularly the case for the well conserved BPIFB3 and BPIF4 families.
By inspection of the phylogenetic tree, all proteins in the collection were assigned to the appropriate BPIFAn or BPIFBn families, giving rise to the annotations in Figure 1. A table of correspondences between the new nomenclature and existing names is given in Table 1. The delineation of families was largely guided by analysis of proteins from eutherian mammals (i.e. excluding avian and marsupial sequences). As discussed by Chiang et al [13], a number of chicken proteins can be assigned to the families defined by the eutherian mammals. For the more divergent proteins chicken TENP [17] and chicken OVO36 [18] we preferred to create new family designations (BPIFB7 and BPIFB8 respectively). The mouse protein vomeromodulin [19,20] is only weakly placed in the PLUNC branch by phylogenetic methods, however, its genomic location confirms this analysis. It is assigned to its own family (BPIFB9).
Table1. BPIFA and BPIFB families.
For each family, the new systematic name is given, along with previous names that have had common usage.
| Protein family | Previous names | Species specific notes |
|---|---|---|
| BPIFA | ||
| BPIFA1 | SPLUNC1, PLUNC*, LUNX, SPURT | |
| BPIFA2 | SPLUNC2, PSP (human), C20orf70* | |
| BPIFA2A | BSP30A | Cow only |
| BPIFA2B | BSP30B | Cow only |
| BPIFA2C | BSP30C | Cow only |
| BPIFA2D | BSP30D | Cow only |
| BPIFA2E | SPLUNC2, PSP (mouse and rat) | Rodent only |
| BPIFA2F | SMGB (rat), SPLUNC4 (mouse) | Rat; pseudogene in mouse (mouse gene symbol = Bpifa2f-ps) |
| BPIFA3 | SPLUNC3, C20orf71* | |
| BPIFA4 | RP11-49G10.8, BASE, latherin | Pseudogene in human (human gene symbol = BPIFA4P); authentic gene in chimpanzee. Absent from rodents. Latherin sequence is rather divergent, which may reflect a change of function in horse. |
| BPIFA5 | SPLUNC5 | Rodent only |
| BPIFA6 | SPLUNC6, XM | Rodent only. May be highly divergent form of BPIFA2 from genomic position |
| BPIFB | ||
| BPIFB1 | LPLUNC1, C20orf114*, VEMSGP | |
| BPIFB2 | LPLUNC2, BPIL1*, C20orf184 | |
| BPIFB3 | LPLUNC3, RYA3(rat), C20orf185* | |
| BPIFB4 | LPLUNC4, RY2G5(rat), C20orf186* | |
| BPIFB5 | LPLUNC5 | Pseudogene in primates (human gene symbol = BPIFB5P) |
| BPIFB6 | LPLUNC6, BPIL3* | |
| BPIFB7 | TENP | Avian |
| BPIFB8 | OVO36 | Avian |
| BPIFB9 | vomeromodulin | Highly divergent; pseudogene in primates (human gene symbol = BPIFB9P) |
HGNC names (that will be replaced by this new nomenclature) are marked with an asterisk.
The bold font used in the Table is for ease of reading.
In order to test the robustness of this procedure of naming of proteins we analysed the pairwise identities of all the collected sequences to a single representative from each family. This method is more objective than a method based on inspecting a phylogenetic tree, since it is less affected by the details of a large multiple-sequence alignment. We selected the human protein (where it exists as an expressed protein) as the prime representative sequence for each family. Where no human proteins exists, we chose the mouse protein (for BPIFA5, BPIFA6 and BPIFB5), and the chimpanzee protein (for BPIFA4[BASE]).
We then used ClustalW to pairwise align each sequence against the representative sequence from each family. The resulting pairwise identities are plotted in Figure 2. With the exception of the BPIFA2 proteins, in all the eutherian proteins the identity to the cognate representative (i.e. to the representative sequence for the family to which the protein is assigned) is significantly higher than to any of the non cognate representatives. In most cases the pairwise identity to the cognate representative protein exceeds 50%, and the pairwise identity to non-cognate representatives is less than 30%. There are, however, a number of cases that complicate the analysis, so that a ‘>50%-cognate vs <30%-non-cognate’ rule does not universally apply. The most simple cases are for BPIFB3/4, BPIFA5, and BPIFA4.
Figure 2. Pairwise identities of sequences from Figure 1, aligned against the representative sequence from each BPIF family, as described in the text.
Filled green circles show the pairwise identity with the representative from the family to which the sequence has been assigned (the cognate representative). Filled red circles show the pairwise identity to the representative sequences from the other families. Open green circles show the pairwise identity of a protein from a subfamily (such as mouse BPIFA2E) to the representative of the family of which the subfamily is a part (thus to human BPIFA2 in the case of mouse BPIFA2E). Crosses are used in place of circles for non-eutherian proteins and for mouse vomeromodulin. If an assignment is robust then the green circle should be substantially to the right of the red circles. The dashed ovals identify the non-cognate representative giving rise to pairwise identities above 30%.
BPIFB3 and BPIFB4 have retained a significantly higher similarity to each other, so that the pairwise identities of BPIFB3 proteins with BPIFB4 proteins (and vice versa) is approximately 40%.
BPIFA5, which is specific to rodents [2,3], has clearly arisen from a duplication of the BPIFA1 proteins. The pairwise identities of BPIFA5 proteins to BPIFA1 proteins (and vice versa) are approximately 55%.
The BPIFA4 (BASE) family is quite divergent, with pairwise identities with the representative sequence of between 40–50% [21,22]. The horse BPIFA4 (also known as latherin) has the lowest pairwise identity with chimpanzee BPIFA4, and in the phylogenetic tree its position is somewhat anomalous relative to the BPIFA4 sequences from other species. This suggests that it has been under different evolutionary pressures to the other BPIFA4 proteins and may have developed, at least in part, a different function to the other BPIFA4 proteins [22].
The most complex family is BPIFA2. This is very highly divergent, and is the only case in the current dataset where addition of letters to the protein name is required. In the case of cow, four BPIFA2 related paralogues exist, which are currently referred to as BSP30A, BSP30B, BSP30C, and BSP30D [12]. Three of the proteins have similar pairwise identities with human BPIFA2 of approximately 40%. The fourth protein (BSP30C) has a substantially lower similarity to human BPIFA2; however, it is still clearly a member of this cluster of proteins. As none of these proteins shows strongly greater similarity to human BPIFA2, these proteins are renamed BPIFA2A, BPIFA2B, BPIFA2C and BPIFA2D. Fortunately, it is possible at this stage to retain the correspondence of letters between BSP30A and BPIFA2A, etc. In rats, a similar situation exists, where there are two BPIFA2 paralogues, which have the historical gene symbols Psp and Smgb [23]. There is only a marginal difference in pairwise similarities to human BPIFA2 (and all the values are much lower than 35%), and thus they become BPIFA2E and BPIFA2F respectively. Note that these symbols represent the rat protein; rodent gene symbols are of the format Bpifa2e and Bpifa2f. For other mammalian species mentioned in the present paper, protein and gene symbols are of the same format. The BPIFA2E and BPIFA2F symbols are chosen to use letters of the alphabet distinct from those chosen for the BSP30 proteins. In mouse, Bpifa2e is a protein-coding gene, but the mouse ortholog of rat Bpifa2f is a pseudogene, which previously acquired the name mouse Splunc4 [2], and is now referred to as Bpifa2f-ps in accordance with rules for mouse pseudogene nomenclature. This is the only instance where the numbering in the new system is different to the SPLUNCn/LPLUNCn system. Besides these two lineage specific duplications, the pairwise identities between the remaining non-primate sequences and human BPIFA2 are also low, at approximately 50%, indicating that there is some form of very strong evolutionary pressure acting on all the BPIFA2 proteins.
It is worthwhile briefly returning to the case of BPIFA5, to illustrate the opposite case to BPIFA2. Since mouse and rat BPIFA1 have retained much greater similarity to human BPIFA1 than have the mouse and rat BPIFA5 proteins, a new symbol (BPIFA5) is allocated to it, rather than introducing BPIFA1A and BPIFA1B.
All sequences used in this study are available as Supplementary Online Data of http://www.biochemsoctrans.org/bst/039/bst0390976add.htm.
The methodology used in Figure 2 should allow the ready checking of the assignment of new protein sequences. The authors are willing to offer assistance in classifying new protein sequences, and would be grateful to be informed about sequences that are in conflict with the system.
Conclusions
We have presented the BPIFAn/BPIFBn nomenclature as a relatively simple modification of the SPLUNCn/LPLUNCn nomenclature to allow systematic treatment of all known PLUNC branch proteins. We have also assembled a set of 102 sequences that we believe represent the current reliable data for BPIFA/BPIFB proteins across all species, including marsupials and birds. There are currently six BPIFA and nine BPIFB families. The BPIFA2 family is, by a significant margin, the most diverse family, with two lineage-specific duplications, and low pairwise identities between the remaining members. The nomenclature will be implemented by the HGNC, who will also finalize proposals for creating BPIF-style aliases for members of the wider BPIF superfamily.
Supplementary Material
Acknowledgements
We are very grateful to Sven Gorr, Tom Wheeler, Peter Di, Klaus Kopec, Mats Lindahl and Joel Gautron who actively contributed to the nomenclature discussions at the meeting and were subsequently able to comment during the development of this framework.
References
- 1.Bingle CD, Bingle L, Craven CJ. Distant cousins: genomic and sequence diversity within the BPI fold-containing (BPIF)/PLUNC protein family. Biochem. Soc. Trans. 2011;39:961–965. doi: 10.1042/BST0390961. [DOI] [PubMed] [Google Scholar]
- 2.Bingle CD, LeClair EE, Havard S, Bingle L, Gillingham P, Craven CJ. Phylogenetic and evolutionary analysis of the PLUNC gene family. Protein Sci. 2004;13:422–430. doi: 10.1110/ps.03332704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeClair EE, Nomellini V, Bahena M, Singleton V, Bingle L, Craven CJ, Bingle CD. Cloning and expression of a mouse member of the PLUNC protein family exclusively expressed in tongue epithelium. Genomics. 2004;83:658–666. doi: 10.1016/j.ygeno.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Wheeler TT, Hood KA, Maqbool NJ, McEwan JC, Bingle CD, Zhao S. Expansion of the bactericidal/permeability increasing-like (BPI-like) protein locus in cattle. BMC Genomics. 2007;8:75. doi: 10.1186/1471-2164-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulero JJ, Boyle BJ, Bradley S, Bright JM, Nelken ST, Ho TT, Mize NK, Childs JD, Ballinger DG, Ford JE, et al. Three new human members of the lipid transfer/lipopolysaccharide binding protein family (LT/LBP) Immunogenetics. 2002;54:293–300. doi: 10.1007/s00251-002-0467-3. [DOI] [PubMed] [Google Scholar]
- 6.Inagawa H, Honda T, Kohchi C, Nishizawa T, Yoshiura Y, Nakanishi T, Yokomizo Y, Soma G. Cloning and characterization of the homolog of mammalian lipopolysaccharide-binding protein and bactericidal permeability-increasing protein in rainbow trout Oncorhynchus mykiss. J. Immunol. 2002;168:5638–5644. doi: 10.4049/jimmunol.168.11.5638. [DOI] [PubMed] [Google Scholar]
- 7.Madsen HO, Hjorth JP. Molecular cloning of mouse PSP mRNA. Nucleic Acids Res. 1985;13:1–138. doi: 10.1093/nar/13.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mirels L, Ball WD. Neonatal rat submandibular gland protein SMG-A and parotid secretory protein are alternatively regulated members of a salivary protein multigene family. J. Biol. Chem. 1992;267:2679–2687. [PubMed] [Google Scholar]
- 8.Mirels L, Ball WD. Neonatal rat submandibular gland protein SMG-A and parotid secretory protein are alternatively regulated members of a salivary protein multigene family. J. Biol. Chem. 1992;267:2679–2687. [PubMed] [Google Scholar]
- 9.Weston WM, LeClair EE, Trzyna W, McHugh KM, Nugent P, Lafferty CM, Ma L, Tuan RS, Greene RM. Differential display identification of plunc, a novel gene expressed in embryonic palate, nasal epithelium, and adult lung. J. Biol. Chem. 1999;274:13698–13703. doi: 10.1074/jbc.274.19.13698. [DOI] [PubMed] [Google Scholar]
- 10.Rajan GH, Morris CA, Carruthers VR, Wilkins RJ, Wheeler TT. The relative abundance of a salivary protein, BSP30, is correlated with susceptibility to bloat in cattle herds selected for high or low bloat susceptibility. Anim. Genet. 1996;27:407–414. doi: 10.1111/j.1365-2052.1996.tb00507.x. [DOI] [PubMed] [Google Scholar]
- 11.Bingle CD, Craven CJ. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Hum. Mol. Genet. 2002;11:937–943. doi: 10.1093/hmg/11.8.937. [DOI] [PubMed] [Google Scholar]
- 12.Wheeler TT, Haigh BJ, Broadhurst MK, Hood KA, Maqbool NJ. The BPI-like/PLUNC family proteins in cattle. Biochem. Soc. Trans. 2011;39:1006–1011. doi: 10.1042/BST0391006. [DOI] [PubMed] [Google Scholar]
- 13.Chiang SC, Veldhuizen EJ, Barnes FA, Craven CJ, Haagsman HP, Bingle CD. Identification and characterisation of the BPI/LBP/PLUNC-like gene repertoire in chickens reveals the absence of a LBP gene. Dev. Comp. Immunol. 2011;35:285–295. doi: 10.1016/j.dci.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 15.Letunic I, Bork P. Interactive Tree Of Life (ITOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 16.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan RT, Wang SZ. Identification and characterization of tenp, a gene transiently expressed before overt cell differentiation during neurogenesis. J. Neurobiol. 1998;34:319–328. [PubMed] [Google Scholar]
- 18.Gautron J, Murayama E, Vignal A, Morisson M, McKee MD, R´ehault S, Labas V, Belghazi M, Vidal ML, Nys Y, et al. Cloning of ovocalyxin-36, a novel chicken eggshell protein related to lipopolysaccharide-binding proteins, bactericidal permeabilityincreasing proteins, and plunc family proteins. J. Biol. Chem. 2007;282:5273–5286. doi: 10.1074/jbc.M610294200. [DOI] [PubMed] [Google Scholar]
- 19.Khew-Goodall Y, Grillo M, Getchell ML, Danho W, Getchell TV, Margolis FL. Vomeromodulin, a putative pheromone transporter: cloning, characterization, and cellular localization of a novel glycoprotein of lateral nasal gland. FASEB J. 1991;5:2976–2982. doi: 10.1096/fasebj.5.14.1752363. [DOI] [PubMed] [Google Scholar]
- 20.Dear TN, Boehm T, Keverne EB, Rabbitts TH. Novel genes for potential ligand-binding proteins in subregions of the olfactory mucosa. EMBO J. 1991;10:2813–2819. doi: 10.1002/j.1460-2075.1991.tb07830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egland KA, Vincent JJ, Strausberg R, Lee B, Pastan I. Discovery of the breast cancer gene BASE using a molecular approach to enrich for genes encoding membrane and secreted proteins. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1099–1104. doi: 10.1073/pnas.0337425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald RE, Fleming RI, Beeley JG, Bovell DL, Lu JR, Zhao X, Cooper A, Kennedy MW. Latherin: a surfactant protein of horse sweat and saliva. PLoS ONE. 2009;4:e5726. doi: 10.1371/journal.pone.0005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ball WD, Mirels L, Hand AR. Psp and Smgb: a model for developmental and functional regulation in the rat major salivary glands. Biochem. Soc. Trans. 2003;31:777–780. doi: 10.1042/bst0310777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.