Abstract
Bone morphogenetic proteins (BMPs) play essential roles in the control of skin development, postnatal tissue remodelling and tumorigenesis. To explore whether some of the effects of BMP signalling are mediated by microRNAs, we performed genome-wide microRNA (miRNA) screening in primary mouse keratinocytes after BMP4 treatment. Microarray analysis revealed substantial BMP4-dependent changes in the expression of distinct miRNAs, including miR-21. Real-time PCR confirmed that BMP4 dramatically inhibits miR-21 expression in the keratinocytes. Consistently, significantly increased levels of miR-21 were observed in transgenic mice overexpressing the BMP antagonist noggin under control of the K14 promoter (K14-noggin). By in situ hybridization, miR-21 expression was observed in the epidermis and hair follicle epithelium in normal mouse skin. In K14-noggin skin, miR-21 was prominently expressed in the epidermis, as well as in the peripheral portion of trichofolliculoma-like hair follicle-derived tumours that contain proliferating and poorly differentiated cells. By transfecting keratinocytes with a miR-21 mimic, we identified the existence of two groups of the BMP target genes, which are differentially regulated by miR-21. These included selected BMP-dependent tumour-suppressor genes (Pten, Pdcd4, Timp3 and Tpm1) negatively regulated by miR-21, as well as miR-21-independent Id1, Id2, Id3 and Msx2 that predominantly mediate the effects of BMPs on cell differentiation. In primary keratinocytes and HaCaT cells, miR-21 prevented the inhibitory effects of BMP4 on cell proliferation and migration. Thus, our study establishes a novel mechanism for the regulation of BMP-induced effects in the skin and suggests miRNAs are important modulators of the effects of growth factor signalling pathways on skin development and tumorigenesis.
Key words: BMP, miR-21, skin
Introduction
Bone morphogenetic proteins (BMPs) belong to a large group of secreted polypeptide growth factors that form the transforming growth factor-β (TGF-β) superfamily. BMP signalling plays essential roles in the control of organ development, postnatal remodelling and regeneration by regulating proliferation, differentiation and apoptosis in many cell types (Miyazono et al., 2010; Walsh et al., 2010). During skin development, BMP signalling controls the initiation phase of hair follicle (HF) morphogenesis, as well as being required for a proper control of keratinocyte differentiation in the HF and epidermis (Botchkarev et al., 1999; Kobielak et al., 2003; Sharov et al., 2003; Andl et al., 2004; Guha et al., 2004). In adult skin, BMP signalling is also involved in the control of HF cycling, HF size and hair pigmentation (Sharov et al., 2006; Sharov et al., 2009). Furthermore, BMP signalling operates as a potent tumour suppressor in the skin and inhibits formation of epidermal- and HF-derived tumours (Blessing et al., 1995; Sharov et al., 2009).
MicroRNAs (miRNA) are a large family of regulatory molecules found in all multicellular organisms. MiRNAs are small non-coding RNAs of approximately 22 nucleotides in length that are created by the enzymatic cleavage of the endogenous primary transcript that contains a local hairpin structure (Ambros, 2001; Bartel, 2004; Farajollahi and Maas, 2010). Currently, it is estimated that miRNA genes make up approximately 3% of the known genes, and up to 30% of genes might be regulated by miRNAs in eukaryotes. MiRNAs regulate gene expression at the post-transcriptional level by forming the Watson–Crick base pairs between its seed region and miRNA recognition elements in the target mRNA 3′ UTRs (Brennecke et al., 2005).
The expression pattern of miRNAs is tissue- and developmental stage-specific. Recently, miR-203 was identified as a skin-specific miRNA that targets the p63 transcription factor in the developing epidermis, restricts keratinocyte proliferation and induces differentiation (Lena et al., 2008; Yi et al., 2008). We demonstrated that in postnatal skin, miRNAs are differentially expressed during distinct stages of the hair cycle (Mardaryev et al., 2010). In particular, miR-31 is highly expressed in actively growing HFs, and, by targeting a number of growth regulatory molecules and cytoskeletal proteins, is involved in establishing an optimal balance of gene expression required for proper HF growth and hair fibre formation (Mardaryev et al., 2010).
Increasing evidence suggests that the misexpression of miRNAs is associated with alterations in the cellular signalling networks and gene expression profiles, and leads to the development of different pathological conditions including abnormal hair growth, inflammatory skin diseases and tumorigenesis (Andl et al., 2006; Sonkoly and Pivarcsi, 2009; Yi and Fuchs, 2010). However, the mechanisms underlying the cross-talk between individual miRNAs and the activity of signalling pathways that control keratinocyte proliferation and differentiation in normal skin and during tumorigenesis remain largely unknown. In this study, we identify the profile of miRNAs the expression of which is modulated by BMP signalling in primary epidermal keratinocytes. Specifically, we show that miR-21 expression in keratinocytes is negatively regulated by BMP4. In turn, miR-21 might inhibit the anti-tumour activity of BMPs in the skin by targeting a number of the BMP-regulated tumour-suppressor genes.
Results and Discussion
BMP4 induces changes in the expression of distinct miRNAs in primary mouse keratinocytes
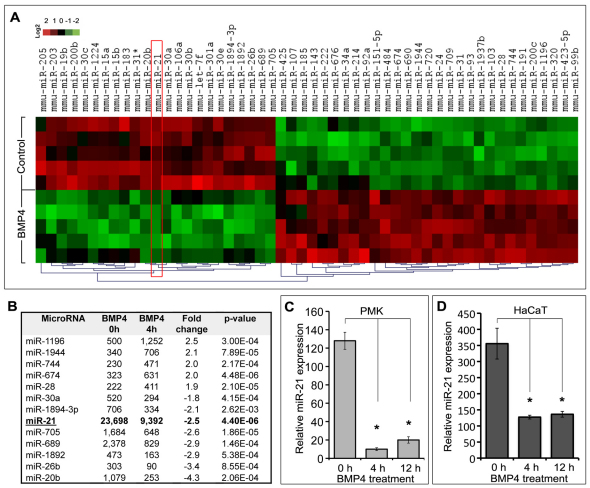
To define whether BMP4 treatment of primary mouse epidermal keratinocytes leads to changes in their global miRNA signature, cells isolated from 2- to 3-day-old mice were treated with BMP4 for 4 hours and miRNA expression profiling was performed using microarray analysis, as described previously (Mardaryev et al., 2010). Global miRNA microarray analysis revealed that 13 out of 226 miRNAs analyzed showed significant (P<0.01) twofold and higher changes in their expression in response to BMP4 treatment (Fig. 1A,B).
Fig. 1.
miRNA expression profile of mouse primary keratinocytes treated with BMP4. (A,B) Microarray analysis. (A) A heat map depicting the miRNAs for which the expression significantly (P<0.01) changed after BMP4 treatment; red and green colours represent elevated and decreased expression of miRNAs, respectively. (B) Table of miRNAs showing ≥2-fold or higher changes in expression in the keratinocytes treated with BMP4 (levels of expression are shown in arbitrary units). (C,D) Detection of miR-21 by qRT-PCR. BMP4 treatment significantly (*P<0.01) reduced expression of the mature form of miR-21 in the primary mouse epidermal keratinocytes (PMK; C) and HaCaT cells (D).
The function of the majority of miRNAs, whose expression in keratinocytes was either positively or negatively regulated by BMP4, has not been identified yet. Among those miRNAs, miR-21 showed very high basal expression levels in keratinocytes (i.e. 10- to 20-fold higher than other miRNAs), which was markedly decreased in response to BMP4 treatment (Fig. 1B). Quantitative RT-PCR (qRT-PCR) analysis confirmed that BMP4 significantly (P<0.01) inhibited miR-21 expression in mouse primary keratinocytes, as well as in HaCaT cells (Fig. 1C,D). However, the levels of primary miR-21 and precursor miR-21 (pri-miR-21 and pre-miR-21, respectively) transcripts in the keratinocytes remained unchanged (data not shown). It was previously shown that BMP signalling can differentially influence miR-21 biogenesis in distinct cell types (Davis et al., 2008; Du et al., 2009; Sahni et al., 2010). Our data suggest that BMP4 negatively regulates the expression of miR-21 post-transcriptionally, affecting the cytoplasmic processing of pre-miR-21 to mature miR-21 in the keratinocytes, which is consistent with data obtained from astrocytes (Sahni et al., 2010).
miR-21 is expressed in the epidermis and hair follicle and is increased in the skin of K14-noggin transgenic mice
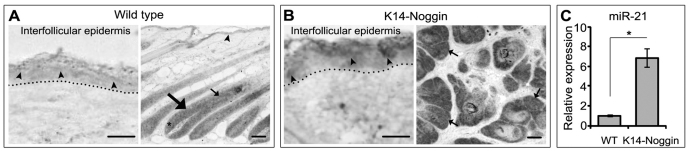
Recent reports have identified miR-21 as a unique miRNA that is consistently overexpressed in a number of tumours, including melanoma and squamous cell carcinoma (Krichevsky and Gabriely, 2009; Selcuklu et al., 2009; Dziunycz et al., 2010). However, the spatiotemporal expression of miR-21 in normal skin has not yet been elucidated. By in situ hybridisation, miR-21 expression was seen in the basal and suprabasal layers of the epidermis of 12-week-old wild-type mice, but was not detected in the dermis (Fig. 2A). In addition, prominent miR-21 expression was seen in the HF outer and inner root sheaths and in the hair matrix, but not in the follicular papilla (Fig. 2A).
Fig. 2.
Spatiotemporal expression of miR-21 in the skin. (A,B) Representative photomicrographs of in situ hybridization for miR-21. (A) miR-21 expression in the basal and suprabasal layers of the epidermis (arrowheads), in the HF outer and inner root sheaths (large and small arrows, respectively) and in the hair matrix (asterisk) in the skin of a 12-week-old wild-type mouse. (B) Prominent miR-21 expression in the epidermis (arrowheads) and in the periphery of the tumours (arrows) in K14-noggin skin. (C) Detection of miR-21 by qRT-PCR: there were significantly (*P<0.01) increased levels of miR-21 in the total skin of K14-noggin mice than in wild-type mice.
BMP signalling suppresses the development of epithelial tumours in the chemical skin carcinogenesis model (Blessing et al., 1995), whereas BMP anti-tumour activity is compromised by extracellular BMP antagonists such as noggin and gremlin (Sneddon et al., 2006; Sharov et al., 2009). To define whether deficiency of BMP signalling affects miR-21 expression, we analyzed its expression in the skin of transgenic mice in which noggin was expressed under the K14 promoter (K14-noggin), which are characterized by spontaneous development of HF-derived trichofolliculoma-like tumours (Sharov et al., 2009). In K14-noggin skin, prominent miR-21 expression was seen in the epidermis, whereas in the HF-derived tumours miR-21 was expressed predominantly in their peripheral portion that contains proliferating and partially differentiated cells (Fig. 2B) (Sharov et al., 2009). The in situ hybridization data (Fig. 2B) were confirmed by qRT-PCR analysis that showed significantly (P<0.01) higher levels of miR-21 in the total skin of K14-noggin mice compared with wild-type mice (Fig. 2C).
These results are consistent with data demonstrating inhibitory effects of BMP4 on miR-21 expression in the keratinocytes (Fig. 1), as well as with recent findings showing that BMP6 causes post-transcriptional repression of miR-21 in breast cancer cells (Du et al., 2009). Because miR-21 functions are generally considered as pro-carcinogenic and associated with tumour progression and metastasis (Krichevsky and Gabriely, 2009), negative regulation of miR-21 by the BMP signalling identified here suggests a novel mechanism, which might mediate anti-tumour activity of BMP signalling in the skin.
miR-21 differentially regulates expression of the BMP target genes and modulates effects of BMP-4 on cell proliferation and migration
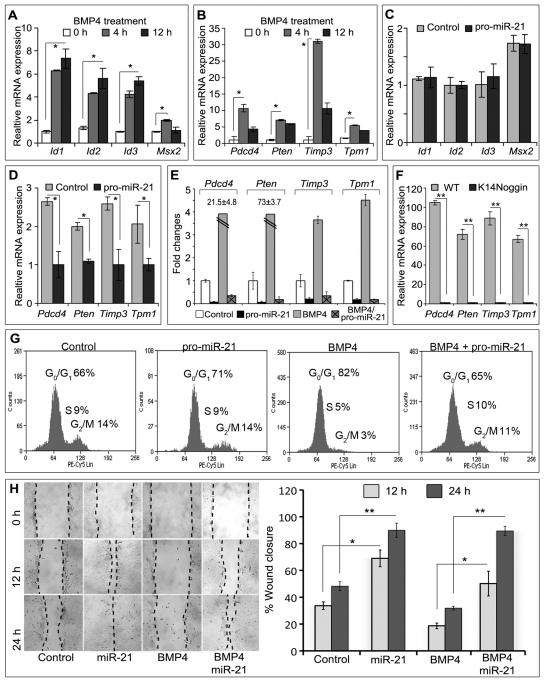
BMP signalling regulates expression of a large number of genes in keratinocytes (Fessing et al., 2010), and some of them (such as Pdcd4, Pten, Timp3 and Tpm1) also serve as direct targets of miR-21 in other cell types (Zhu et al., 2007; Asangani et al., 2008; Song et al., 2010; Zhang et al., 2010). To explore whether miR-21 is indeed involved in the regulation of the BMP-dependent gene expression program in keratinocytes, primary epidermal keratinocytes were treated with BMP4 or an miR-21 mimic (pro-miR-21) and the corresponding controls. The expression of the selected BMP target genes (Id1, Id2 Id3, Msx2, Pten, Pdcd4, Timp3 and Tpm1) known to mediate the effects of BMP in different cell types including keratinocytes (Ogata et al., 1993; Hussein et al., 2003; Fessing et al., 2010) was assessed by qRT-PCR.
In agreement with data published previously (Fessing et al., 2010), treatment of the keratinocytes with BMP4 resulted in a marked increase in the mRNA levels of all BMP target genes selected for analyses, compared with the controls (Fig. 3A,B). However, miR-21 treatment alone differentially modulated the expression of those genes that were positively regulated by BMP4 in keratinocytes. The levels of transcripts of some BMP target genes (Id1, Id2 Id3 and Msx2) were the same in the keratinocytes transfected with pro-miR-21 as in the controls (Fig. 3C). However, pro-miR-21 caused significant (P<0.01) downregulation of the expression of Pten, Pdcd4, Timp3 and Tpm1 in keratinocytes versus the controls (Fig. 3D). To determine whether miR-21 could prevent the BMP4-induced increased expression of these genes, primary mouse keratinocytes and HaCaT cells were first transfected with the miR-21 mimic and then treated with BMP4. Pretreatment of the keratinocytes with pro-miR-21 resulted in the failure of BMP4 to increase the mRNA levels of Pten, Pdcd4, Timp3 and Tpm1, compared with the BMP4 treatment alone (Fig. 3E). Similar changes in gene expression were seen in the HaCaT keratinocytes (data not shown). Consistent with these results, significantly (P<0.01) decreased levels of Pten, Pdcd4, Timp3 and Tpm1 transcripts were found in skin of K14-noggin mice compared with wild-type skin (Fig. 3F).
Fig. 3.
miR-21 modulates the effects of BMP4 on gene expression, cell proliferation and migration in keratinocytes. (A–E) qRT-PCR analysis of gene expression in the primary mouse keratinocytes relatively to Gapdh levels. (A) BMP4 treatment causes a significant increase in the expression of Id1, Id2, Id3 and Msx2 compared with untreated cells. (B) Upregulation in the expression of Pdcd4, Pten, Timp3 and Tpm1 transcripts by BMP4 treatment. (C) Expression of Id1, Id2, Id3 and Msx2 is not affected by transfection with miR-21 mimic (pro-miR-21). (D) Expression of Pdcd4, Pten, Timp3 and Tpm1 is decreased after treatment with miR-21 mimic. (E) miR-21 mimic prevents BMP4-induced expression of Pdcd4, Pten, Timp3 and Tpm1 in keratinocytes. (F) Expression of Pten, Pdcd4, Timp3 and Tpm1 in the skin of K14-noggin transgenic mice is decreased compared with wild-type mice. (G) Flow cytometry analysis of the cell cycle in primary keratinocytes: a significant decrease in number of proliferating cells caused by BMP-4 treatment was prevented by the miR-21 mimic. (H) Cell migration (scratch) assay. miR-21 mimic significantly increases HaCaT cell migration compared with the control, and interferes with the suppression of cell migration induced by BMP4; *P<0.01, **P<0.001.
These data therefore suggest that miR-21 is indeed involved in mediating the effects of BMP signalling in keratinocytes by modulating the expression of selected BMP target genes that also serve as miR-21 targets. Although the full profile of the miR-21 target mRNAs in keratinocytes remains to be defined, some of the genes that in keratinocytes are regulated by both BMP4 and miR-21 (Pten, Pdcd4, Timp3 and Tpm1) serve as tumour suppressors in many organs and cell types (Zhu et al., 2007; Asangani et al., 2008; Song et al., 2010; Zhang et al., 2010). Loss of PTEN expression in the skin contributes to tumour development (reviewed by Ming and He, 2009), whereas transgenic overexpression of Pdcd4 in the epidermis causes an inhibition of tumour growth in chemical skin carcinogenesis tests (Jansen et al., 2005). TIMP3 and TPM1 also suppress tumour growth by regulating epithelial–mesenchymal interactions and inhibiting cell motility and tumour invasion (Zheng et al., 2008; Rodgers et al., 2009).
To determine whether miR-21 is indeed capable of interfering with the inhibitory effects of BMP on keratinocyte proliferation and motility, cell cycle and in vitro ‘scratch’ migration assays were performed using primary keratinocytes and HaCaT cells, respectively. Consistent with data published previously (Sharov et al., 2006), BMP4 significantly (P<0.05) inhibited cell proliferation in primary keratinocytes (Fig. 3G). Pro-miR-21 treatment alone did not alter keratinocyte proliferation, but significantly (P<0.05) diminished the anti-proliferative effects of BMP4 on these cells (Fig. 3G). The scratch assay revealed that miR-21 mimic significantly (P<0.01) accelerated migration of HaCaT cell compared with the control, as well as preventing the inhibitory effects of BMP4 on the motility of HaCaT cells (Fig. 3H).
Thus, BMP signalling might have an anti-tumour functions in the skin in part by downregulating miR-21 expression and thereby removing its negative effects on the expression of Pten, Pdcd4, Timp3 and Tpm1 as tumour suppressor genes. Our data also demonstrate that miR-21 does not interfere with BMP4-induced expression of Id1, Id2, Id3 and Msx2 in the keratinocytes (Fig. 3C). Id1–3 are established targets for BMP signalling, and they mediate its effect on cell differentiation (Langlands et al., 2000; Sharov et al., 2003). Msx2 also mediates the effects of BMP signalling during skin development and in postnatal hair follicles, as well as contributing to re-epithelialisation in the epidermis (Yeh et al., 2009). This suggests that the effects of BMP on cell differentiation are likely to occur in an miR-21-independent manner.
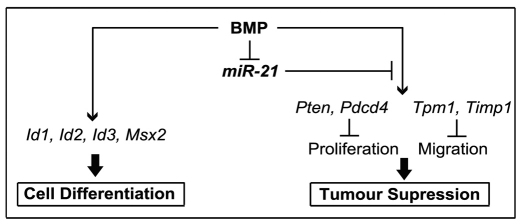
Taken together, we identified the existence of two groups of BMP target genes, the expressions of which are differentially regulated by miR-21. BMP-dependent tumour-suppressor genes such as Pten, Pdcd4, Timp3 and Tpm1 are negatively regulated by miR-21, whereas Id1, Id2, Id3 and Msx2, which predominantly mediate the effects of BMPs on cell differentiation, are miR-21 independent (Fig. 4). Although additional studies are required to fully elucidate how BMP signalling controls miR-21 expression in keratinocytes, our data reveal miR-21 as an important downstream component that modulates the BMP-regulated gene expression program and its effects on the distinct groups of target genes. In conclusion, our study establishes a novel mechanism for the regulation of BMP-induced effects in the skin and suggests that, together with the BMP target genes described previously (Fessing et al., 2010), BMP signalling also exerts its effects on keratinocytes through regulation of the expression of the selected miRNAs, including miR-21.
Fig. 4.
Schematic illustrating an involvement of miR-21 in the modulation of BMP4-mediated effects on gene expression in keratinocytes. miR-21 modulates the BMP anti-tumour activity by negative regulation of the expression of BMP-dependent tumour-suppressor genes (Pten, Pdcd4, Timp3 and Tpm1) in keratinocytes. However, the effects of BMP on keratinocyte differentiation mediated through its targets Id1, Id2, Id3 and Msx2 are miR-21 independent.
Materials and Methods
Cell culture
Primary mouse epidermal keratinocytes (PMEKs) were prepared from newborn mice at postnatal days 2–3, as described previously (Lichti et al., 2008). HaCaT keratinocytes were grown in Dulbecco's modified Eagle's medium (Invitrogen, UK) supplemented with heat-inactivated 10% foetal bovine serum. PMEKs and HaCaT were treated with 200 ng/ml of recombinant human BMP4 (98% identity with mouse Bmp4 protein; R&D system, UK) for 4 and 12 hours as described previously (Fessing et al., 2010). Cells were transfected with 200 nM miR-21 mimic (pro-miR-21) or miRNA negative controls (Dharmacon, USA), using Lipofectamine RNAiMax (Invitrogen, UK), and harvested 24 hours later. To examine the regulatory effects of miR-21 on BMP-induced gene expression, keratinocytes were transfected with 200 nM synthetic pro-miR-21 or miRNA negative controls for 4 hours, followed by quick washing in PBS and treatment with 200 ng/ml of BMP4 for 4 hours.
Animals and tissue collection
All animal work was performed under local research ethics committee approval. K14-noggin-overexpressing mice were generated in an FVB background as described previously (Sharov et al., 2009). Skin samples were collected from adult mice (12-weeks old, n=5 for each mouse strain) and processed for further analyses as described elsewhere (Mardaryev et al., 2010).
Microarray and real-time PCR
Total RNA was isolated using the miRNeasy kit (Qiagen, UK). Microarray analysis was performed by LC Sciences (Houston, TX). qRT-PCR for miR-21 was performed using TaqMan® Real Time PCR Assay (Applied Biosystems, USA). Differences between samples and controls were calculated based on the Ct (ΔΔCt) method and normalized to the small nucleolar RNA 202 values (SnoRNA). Data were pooled, the means ± s.e.m. were calculated, and statistical analysis was performed using an unpaired Student's t-test. qRT-PCR for mRNAs was performed as described previously using Gapdh as an internal control (Mardaryev et al., 2010). PCR primers were designed on Beacon Designer software (Premier Biosoft, Palo Alto, CA; supplementary material Table S1).
In situ hybridization
Skin cryosections (10 μm) were fixed in 4% paraformaldehyde for 10 minutes at room temperature (RT). After acetylation in triethanolamine buffer (4.5 mM triethanolamine, 6 M HCl, 3 mM acetic anhydride) for 10 minutes and pre-mobilisation (1% Triton X-100 in 1× PBS) for 30 minutes, slides were hybridized with 2.5 pmol digoxigenin-labelled miR-21 probe (Exiqon, Vedbaek, Denmark) diluted in hybridization buffer (50% formamide DI, 2× SCC, 1% dextran sulphate, 0.4 mg/ml tRNA) for 16–18 hours at 55°C. Slides were then washed in 2× SCC (10 minutes, four times, 67°C), 0.1× SCC (60 minutes, 65°C) and 0.2× SCC (10 minutes, RT). Immunodetection of miR-21 was performed with sheep alkaline-phosphatase-conjugated anti-digoxigenin antibody (1:5000; Roche, Germany) followed by a staining reaction with nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolyphosphate (NBT/BCIP) solution (Roche, Germany) for 16–18 hours at RT.
Flow cytometry
To assess proliferation rate, cells were washed with PBS and fixed in 70% ethanol followed by incubation with 20 μg/ml of 7-aminoactinomycin D (VWR, Lutterworth, UK) for 15 minutes at RT. The percentage of cells at distinct phases of the cell cycle was analysed by flow cytometry with a Beckman Coulter – CyAn™ ADP analyser (Beckman Coulter, UK) equipped with Summit™ Software, version 4.2. For each sample, 10,000 events were collected.
Cell migration assay
HaCaT cells were treated as described above. Using a P10 pipette tip, a scratch was made in the middle of the plate. Mitomycin C (2 μg/ml; VWR, UK) was included in the migration assay to block cell proliferation. The distance between the leading edges of the migrating keratinocytes was measured using ImageJ software (http://rsbweb.nih.gov/ij/), as described previously (Chmielowiec et al., 2007). Data from triplicate samples were pooled, the means ± s.d. were calculated, and statistical analysis was performed using an unpaired Student's t-test.
Supplementary Material
Acknowledgements
Prof D. Tobin is acknowledged for his support and critical comments on the manuscript.
Footnotes
Funding
This work was supported by a Royal Society Research Grant to N.V.B.; and the National Institutes of Health [KO1 award number AR056771 to A.A.S.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.086710/-/DC1
References
- Ambros V. (2001). microRNAs: tiny regulators with great potential. Cell 107, 823-826 [DOI] [PubMed] [Google Scholar]
- Andl T., Ahn K., Kairo A., Chu E. Y., Wine-Lee L., Reddy S. T., Croft N. J., Cebra-Thomas J. A., Metzger D., Chambon P., et al. (2004). Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development 131, 2257-2268 [DOI] [PubMed] [Google Scholar]
- Andl T., Murchison E. P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J. W., Andl C. D., Seykora J. T., Hannon G. J., Millar S. E. (2006). The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 16, 1041-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani I. A., Rasheed S. A., Nikolova D. A., Leupold J. H., Colburn N. H., Post S., Allgayer H. (2008). MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128-2136 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281-297 [DOI] [PubMed] [Google Scholar]
- Blessing M., Nanney L. B., King L. E., Hogan B. L. (1995). Chemical skin carcinogenesis is prevented in mice by the induced expression of a TGF-beta related transgene. Teratog. Carcinog. Mutagen 15, 11-21 [DOI] [PubMed] [Google Scholar]
- Botchkarev V. A., Botchkareva N. V., Roth W., Nakamura M., Chen L. H., Herzog W., Lindner G., McMahon J. A., Peters C., Lauster R., et al. (1999). Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat. Cell Biol. 1, 158-164 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R. B., Cohen S. M. (2005). Principles of microRNA-target recognition. PLoS Biol. 3, e85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielowiec J., Borowiak M., Morkel M., Stradal T., Munz B., Werner S., Wehland J., Birchmeier C., Birchmeier W. (2007). c-Met is essential for wound healing in the skin. J. Cell Biol. 177, 151-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008) SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454, 56-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Yang S., An D., Hu F., Yuan W., Zhai C., Zhu T. (2009). BMP-6 inhibits microRNA-21 expression in breast cancer through repressing deltaEF1 and AP-1. Cell Res. 19, 487-496 [DOI] [PubMed] [Google Scholar]
- Dziunycz P., Iotzova-Weiss G., Eloranta J. J., Lauchli S., Hafner J., French L. E., Hofbauer G. F. (2010). Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. J. Invest. Dermatol. 130, 2686-2689 [DOI] [PubMed] [Google Scholar]
- Farajollahi S., Maas S. (2010). Molecular diversity through RNA editing: a balancing act. Trends Genet. 26, 221-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessing M. Y., Atoyan R., Shander B., Mardaryev A. N., Botchkarev V. V., Jr., Poterlowicz K., Peng Y., Efimova T., Botchkarev V. A. (2010). BMP signaling induces cell-type-specific changes in gene expression programs of human keratinocytes and fibroblasts. J. Invest. Dermatol. 130, 398-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha U., Gomes W. A., Samanta J., Gupta M., Rice F. L., Kessler J. A. (2004). Target-derived BMP signaling limits sensory neuron number and the extent of peripheral innervation in vivo. Development 131, 1175-1186 [DOI] [PubMed] [Google Scholar]
- Hussein S. M., Duff E. K., Sirard C. (2003). Smad4 and beta-catenin co-activators functionally interact with lymphoid-enhancing factor to regulate graded expression of Msx2. J. Biol. Chem. 278, 48805-48814 [DOI] [PubMed] [Google Scholar]
- Jansen A. P., Camalier C. E., Colburn N. H. (2005). Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 65, 6034-6041 [DOI] [PubMed] [Google Scholar]
- Kobielak K., Pasolli H. A., Alonso L., Polak L., Fuchs E. (2003). Defining BMP functions in the hair follicle by conditional ablation of BMP receptor IA. J. Cell Biol. 163, 609-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A. M., Gabriely G. (2009). miR-21: a small multi-faceted RNA. J. Cell Mol. Med. 13, 39-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlands K., Down G. A., Kealey T. (2000). Id proteins are dynamically expressed in normal epidermis and dysregulated in squamous cell carcinoma. Cancer Res. 60, 5929-5933 [PubMed] [Google Scholar]
- Lena A. M., Shalom-Feuerstein R., Rivetti di Val Cervo P., Aberdam D., Knight R. A., Melino G., Candi E. (2008). miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 15, 1187-1195 [DOI] [PubMed] [Google Scholar]
- Lichti U., Anders J., Yuspa S. H. (2008). Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 3, 799-810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev A. N., Ahmed M. I., Vlahov N. V., Fessing M. Y., Gill J. H., Sharov A. A., Botchkareva N. V. (2010). Micro-RNA-31 controls hair cycle-associated changes in gene expression programs of the skin and hair follicle. FASEB J. 24, 3869-3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming M., He Y. Y. (2009). PTEN: new insights into its regulation and function in skin cancer. J. Invest. Dermatol. 129, 2109-2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K., Kamiya Y., Morikawa M. (2010). Bone morphogenetic protein receptors and signal transduction. J. Biochem. 147, 35-51 [DOI] [PubMed] [Google Scholar]
- Ogata T., Wozney J. M., Benezra R., Noda M. (1993). Bone morphogenetic protein 2 transiently enhances expression of a gene, Id (inhibitor of differentiation), encoding a helix-loop-helix molecule in osteoblast-like cells. Proc. Natl. Acad. Sci. USA 90, 9219-9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers U. R., Kevorkian L., Surridge A. K., Waters J. G., Swingler T. E., Culley K., Illman S., Lohi J., Parker A. E., Clark I. M. (2009). Expression and function of matrix metalloproteinase (MMP)-28. Matrix Biol. 28, 263-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni V., Mukhopadhyay A., Tysseling V., Hebert A., Birch D., McGuire T. L., Stupp S. I., Kessler J. A., (2010). BMPR1a and BMPR1b signaling exert opposing effects on gliosis after spinal cord injury. J. Neurosci. 30, 1839-1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcuklu S. D., Donoghue M. T., Spillane C. (2009). miR-21 as a key regulator of oncogenic processes. Biochem. Soc. Trans. 37, 918-925 [DOI] [PubMed] [Google Scholar]
- Sharov A. A., Weiner L., Sharova T. Y., Siebenhaar F., Atoyan R., Reginato A. M., McNamara C. A., Funa K., Gilchrest B. A., Brissette J. L., et al. (2003). Noggin overexpression inhibits eyelid opening by altering epidermal apoptosis and differentiation. EMBO J. 22, 2992-3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A. A., Sharova T. Y., Mardaryev A. N., Tommasi di Vignano A., Atoyan R., Weiner L., Yang S., Brissette J. L., Dotto G. P., Botchkarev V. A. (2006). Bone morphogenetic protein signaling regulates the size of hair follicles and modulates the expression of cell cycle-associated genes. Proc. Natl. Acad. Sci. USA 103, 18166-18171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A. A., Mardaryev A. N., Sharova T. Y., Grachtchouk M., Atoyan R., Byers H. R., Seykora J. T., Overbeek P., Dlugosz A., Botchkarev V. A. (2009). Bone morphogenetic protein antagonist noggin promotes skin tumorigenesis via stimulation of the Wnt and Shh signaling pathways. Am. J. Pathol. 175, 1303-1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon J. B., Zhen H. H., Montgomery K., van de Rijn M., Tward A. D., West R., Gladstone H., Chang H. Y., Morganroth G. S., Oro A. E., et al. (2006). Bone morphogenetic protein antagonist gremlin 1 is widely expressed by cancer-associated stromal cells and can promote tumor cell proliferation. Proc. Natl. Acad. Sci. USA 103, 14842-14847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Wang C., Liu J., Wang X., Lv L., Wei L., Xie L., Zheng Y., Song X. (2010). MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J. Exp. Clin. Cancer Res. 29, 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E., Pivarcsi A. (2009). Advances in microRNAs: implications for immunity and inflammatory diseases. J. Cell Mol. Med. 13, 24-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D. W., Godson C., Brazil D. P., Martin F. (2010). Extracellular BMP-antagonist regulation in development and disease: tied up in knots. Trends Cell Biol. 20, 244-256 [DOI] [PubMed] [Google Scholar]
- Yeh J., Green L. M., Jiang T. X., Plikus M., Huang E., Chang R. N., Hughes M. W., Chuong C. M., Tuan T. L. (2009). Accelerated closure of skin wounds in mice deficient in the homeobox gene Msx2. Wound Repair Regen. 17, 639-648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Fuchs E. (2010). MicroRNA-mediated control in the skin. Cell Death Differ. 17, 229-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Poy M. N., Stoffel M., Fuchs E. (2008). A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 452, 225-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. G., Wang J. J., Zhao F., Liu Q., Jiang K., Yang G. H. (2010). MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin. Chim. Acta 411, 846-852 [DOI] [PubMed] [Google Scholar]
- Zheng Q., Safina A., Bakin A. V. (2008). Role of high-molecular weight tropomyosins in TGF-beta-mediated control of cell motility. Int. J. Cancer 122, 78-90 [DOI] [PubMed] [Google Scholar]
- Zhu S., Si M. L., Wu H., Mo Y. Y. (2007). MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 282, 14328-14336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.