Abstract
Despite the fact that 45% of all human gene promoters do not contain CpG islands, the role of DNA methylation in control of non-CpG island promoters is controversial and its relevance in normal and pathological processes is poorly understood. Among the few studies which investigate the correlation between DNA methylation and expression of genes with non-CpG island promoters, the majority do not support the view that DNA methylation directly leads to transcription silencing of these genes. Our reporter assays and gene reactivation by 5-aza-2′-deoxycytidine, a DNA demethylating agent, show that DNA methylation occurring at CpG poor LAMB3 promoter and RUNX3 promoter 1(RUNX3 P1) can directly lead to transcriptional silencing in cells competent to express these genes in vitro. Using Nucleosome Occupancy Methylome- Sequencing, NOMe-Seq, a single-molecule, high-resolution nucleosome positioning assay, we demonstrate that active, but not inactive, non-CpG island promoters display a nucleosome-depleted region (NDR) immediately upstream of the transcription start site (TSS). Furthermore, using NOMe-Seq and clonal analysis, we show that in RUNX3 expressing 623 melanoma cells, RUNX3 P1 has two distinct chromatin configurations: one is unmethylated with an NDR upstream of the TSS; another is methylated and nucleosome occupied, indicating that RUNX3 P1 is monoallelically methylated. Together, these results demonstrate that the epigenetic signatures comprising DNA methylation, histone marks and nucleosome occupancy of non-CpG island promoters are almost identical to CpG island promoters, suggesting that aberrant methylation patterns of non-CpG island promoters may also contribute to tumorigenesis and should therefore be included in analyses of cancer epigenetics.
INTRODUCTION
Epigenetic processes play important roles in many key biological processes, including but not limited to embryonic development, genomic imprinting and X-chromosome inactivation (1). Gene expression can be regulated epigenetically by DNA methylation, histone variants and modifications and nucleosome positioning. These modifications work together to create an epigenetic landscape, which determines the potential for gene expression (2–4). DNA methylation, occurring in the context of the CpG dinucleotide, has profound effects on gene expression by modifying the accessibility of DNA to transcription factors (5). It is well accepted that DNA methylation occurring on CpG islands acts as a silencing mechanism during normal development and tumorigenesis (1).
Histone modifications, variants and nucleosome positioning work coordinately with DNA methylation to regulate gene expression by modifying the accessibility of promoter regions to the transcription machinery (6,7). The N-terminal tails of histones undergo a variety of post-translational modifications to generate permissive or refractory chromatin conformations depending upon the type and location of the modifications (3,8). For instance, trimethylation of lysine 4 on Histone 3 (H3K4me3) is enriched at the promoters of transcriptionally active genes (9,10), whereas trimethylation of H3K9 and H3K27 is associated with transcriptionally inactive gene promoters (10,11). The presence of a nucleosome-depleted region (NDR) immediately upstream of transcription start sites (TSSs) is critical for active gene transcription by providing sites for transcription machinery binding (12). Transcriptionally active promoters display a NDR, which can accommodate at least one nucleosome immediately upstream of the TSS. In contrast, transcriptionally inactive promoters lack this NDR (7,13–15).
The role of CpG methylation in long-term silencing of genes, whose promoters contain CpG islands is well understood (7,16). Despite the fact that 45% of all human gene promoters, particularly those controlling the expression of tissue-specific genes, do not lie within CpG islands (17), little is known about their regulation and the potential role of methylation as a transcriptional control mechanism. A number of genes with non-CpG island promoters have been reported to be methylated in normal tissues, displaying a tissue-specific methylation pattern, suggesting DNA methylation may play a role in the establishment and maintenance of tissue-specific expression patterns (18). Some studies, including those from our own laboratory, have shown that there is an inverse correlation between DNA methylation and gene expression, as it has been demonstrated for CpG island promoters (18,19), whereas other studies have reported that CpG-poor promoters could be still expressed when they are methylated (20). However, when we re-examined this in more detail by categorizing promoters into low, intermediate and high CpG density and found that the inverse correlation between methylation and expression still holds in promoters with low CpG density (19).
In this study, using two non-CpG island promoters as models, we show that the promoter of Laminin Beta 3 (LAMB3) and Runt related transcription factor 3 promoter 1(RUNX3 P1) are methylated in a tissue-specific manner, suggesting that methylation at non-CpG island promoters has a functional significance in the establishment and maintenance of cell identity. In addition, the LAMB3 promoter is aberrantly hypomethylated in primary bladder tumors, indicating a potential role of aberrant methylation occurring at non-CpG islands in tumorigenesis. We further show that methylation of the promoter of LAMB3 and RUNX3 P1 directly leads to transcriptional silencing in cells that are capable of expressing their gene products. Demethylation is necessary but not sufficient to induce gene expression. Other transcription machineries, such as necessary transcription factors are required. Finally, using Nucleosome Occupancy Methylome-Sequencing (NOMe-Seq) in combination with chromatin immunoprecipitation (ChIP), we demonstrate that nucleosome occupancy and histone tail modifications associated with the TSS of LAMB3 and RUNX3 P1 active promoters are remarkably similar to those found in CpG island promoters. In summary, these observations show that genes with non-CpG island promoters share many epigenetic features that are associated with CpG island promoter genes, despite their low CpG density. We propose that the regulation of non-CpG island promoters by DNA methylation plays an important role in the establishment and/or maintenance of cell identity. Furthermore, it has a potential contribution to tumorigenesis, and therefore, should be taken into consideration in the study of cancer epigenetics.
RESULTS
The LAMB3 promoter and RUNX3 P1 show tissue-specific and lineage-specific methylation patterns
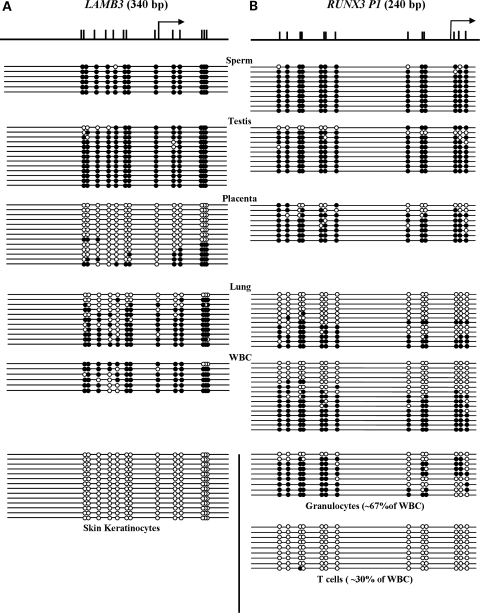
LAMB3 encodes the β subunit of laminin 5, which facilitates the attachment of epidermal cells to the basement membrane (21). RUNX3, a member of the highly conserved DNA-binding RUNT domain family of transcription factors, is highly expressed in the hematopoietic system (22). RUNX3 is regulated by two distantly located promoters, P1 and P2 (22), and most studies of RUNX3 focus on the transcript from P2, a CpG island promoter. It has been shown that the transcripts generated by RUNX3 P2 have tumor suppressor activity (23). RUNX3 P2 has been reported repeatedly to be hypermethylated in various cancers (24–27), whereas little is known about RUNX3 P1. The LAMB3 promoter and RUNX3 P1 are not located within CpG islands according to the criteria established by Takai and Jones (17,28), which defines a CpG island as regions of DNA greater than 500 bp with a G + C content ≥55% and an observed CpG/expected CpG ratio that is >65%. To determine the DNA methylation status of the LAMB3 promoter and RUNX3 P1, we carried out bisulfite genomic sequencing in a panel of normal human tissues. Thirteen CpG sites within the 340 bp promoter region of LAMB3 were analyzed. It is heavily methylated in the sperm, testis, lung and white blood cells (WBC) (Fig. 1A). In contrast, it is minimally methylated in placenta lining and almost completely devoid of methylation in skin keratinocytes (Fig. 1A), where LAMB3 is endogenously expressed (data not shown). These data suggest that DNA methylation of the LAMB3 promoter may play a role in controlling its tissue-specific expression.
Figure 1.
LAMB3 and RUNX3 P1 display tissue-specific methylation patterns. The DNA methylation pattern of the LAMB3 promoter (A) and RUNX3 P1 (B) in normal tissues and cells was analyzed by genomic bisulfite sequencing. Filled and open circles represent methylated and unmethylated CpG sites, respectively. Schematic diagrams of the LAMB3 promoter and RUNX3 P1 are shown. Upper tick marks represent individual CpG sites and black arrows represent TSSs.
We also analyzed 13 CpG sites within a span of 240 bp in RUNX3 P1. It is heavily methylated in the sperm, testis and placenta, and is ∼50% methylated in the lung and WBC, with a majority of the strands being either completely methylated or completely unmethylated (Fig. 1B). This suggests the possibility of cell type-specific methylation pattern. WBC are a mixture of cell types, so we fractionated them using a Ficoll-Paque PLUS density gradient to isolate granulocytes and lymphocytes. T-cells were further purified from the lymphocytes. The granulocytes had a mixed pattern of methylation, as previously seen in unfractionated WBCs; however, the T-cells were almost completely devoid of methylation. Importantly, lack of methylation across RUNX3 P1 was not observed in any other tissues analyzed (Fig. 1B), suggesting that the transcript generated from RUNX3 P1 is crucial to T-cells function, in agreement with its role in blood cell development (29,30). Our data show that RUNX3 P1 displays cell type-specific methylation pattern.
LAMB3 is hypomethylated in primary bladder tumors
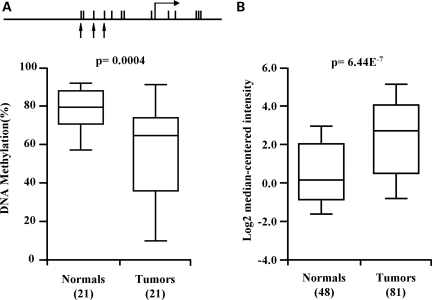
The importance of abnormal methylation of CpG island promoters in carcinogenesis is well established (1,31). In contrast, the potential role of aberrant methylation of non-CpG island promoters during tumorigenesis is poorly understood. To evaluate the methylation patterns of the LAMB3 promoter and RUNX3 P1 in cancer, we investigated the DNA methylation level of the LAMB3 promoter in 21 matched sets of invasive primary bladder tumors and their paired ‘normal’ urothelium using methylation-sensitive single-nucleotide primer extension (Ms-SNuPE). The results show a significant reduction (P-value = 0.004) in the levels of DNA methylation of the LAMB3 promoter in the tumors (56%) compared with ‘normal’ urothelium (79%) (Fig. 2A). These results are in agreement with our in silico analysis of LAMB3 expression using the cancer microarray database ONCOMINE (32), which shows significant up-regulation of LAMB3 (P-value = 6.44E−7) in primary bladder tumors compared with normal urothelium (Fig. 2B). Together, these data show that in primary bladder tumors, hypomethylation of the LAMB3 promoter is associated with its over-expression, suggesting that LAMB3 may play a functional role in tumorigenesis by loss of methylation. Furthermore, our results also suggest that the inverse relationship between promoter methylation and gene expression still holds true in CpG-poor regions. In contrast to the LAMB3 promoter, RUNX3 P1 showed no significant methylation changes in primary bladder tumors compared with normal urothelium (data not shown).
Figure 2.
LAMB3 is hypomethylated in primary bladder tumors. (A) Box plots indicate DNA methylation of the LAMB3 promoter in primary bladder tumors and paired ‘normal’ tissues. The P-value was calculated using t-test. A schematic diagram of the LAMB3 promoter is shown. Black arrows indicate the 3 CpG sites, which were interrogated by Ms-SNuPE. (B) The expression levels of LAMB3 in normal urothelium and bladder tumors were obtained from ONCOMINE. Numbers in the parentheses indicate number of samples used to generate box plots.
5-Aza-CdR can induce expression of non-CpG island promoter genes
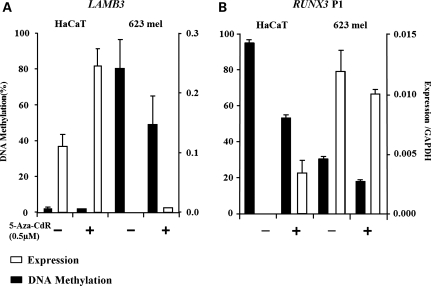
Our results presented thus far suggest that DNA methylation regulates non-CpG island promoters and leads to gene silencing. To elucidate whether this effect is direct or indirect, we examined whether expression of endogenous LAMB3 and the transcript generated by RUNX3 P1 can be induced by treating cells that do not express these genes with a DNA methylatransferase inhibitor, 5-aza-2′-deoxycytidine (5-Aza-CdR). We first screened a panel of cell lines for endogenous expression of LAMB3 and the transcript produced from RUNX3 P1 (data not shown) and selected a suitable pair of cell lines that expressed just one of the two transcripts. A dose-response experiment was performed to determine the optimal concentration of 5-Aza-CdR for each cell line. The maximum demethylation level was reached for both cell lines when 0.5 μm of 5-Aza-CdR was used (Supplementary Material, Fig. S1). HaCaT cells express LAMB3, and 623 melanoma cells express RUNX3 P1. Both cell lines were treated with 0.5 µm of 5-Aza-CdR for 24 h. DNA and RNA were harvested 8 days after drug treatment to evaluate methylation and gene expression levels, respectively. The LAMB3 promoter showed an 80% methylation and undetectable expression level in 623 melanoma cells prior to 5-Aza-CdR treatment. However, its expression remained low after 5-Aza-CdR treatment, even though DNA demethylation was observed (Fig. 3A), suggesting that LAMB3 expression requires other transcription regulators that are absent in 623 melanoma cells. Prior to 5-Aza-CdR treatment, LAMB3 was expressed and its promoter was unmethylated in HaCaT cells. Interestingly, the expression level increased following 5-Aza-CdR treatment, suggesting that 5-Aza-CdR-induced genome-wide demethylation may lead to up-regulation of other factors required for LAMB3 expression.
Figure 3.
5-Aza-CdR treatment induces expression of genes with non-CpG island promoters. (A) The expression levels (white bars) of LAMB3 (A) and RUNX3 P1 (B) were determined by RT–PCR in HaCaT and 623 melanoma cells. Expression is normalized to GAPDH. The DNA methylation (black bars) levels were determined by Ms-SNuPE. Error bars represent the range between two biological duplicates.
RUNX3 P1 displayed a 95% methylation and its transcript was undetectable in HaCaT cells (Fig. 3B). Treatment with 5-Aza-CdR induced both DNA demethylation and an increase in transcription from RUNX3 P1, suggesting that expression from this non-CpG island promoter is regulated by DNA methylation (Fig. 3A, left panel). Surprisingly, our results showed a 35% methylation on RUNX3 P1 in 623 melanoma cells, which endogenously express the transcript from this promoter (Fig. 3B, right panel). Overall, our data illustrate that the expression of genes controlled by non-CpG island promoters can be induced after 5-Aza-CdR treatment only in cells that contain necessary transcription machinery for such expression.
DNA methylation directly silences non-CpG island gene promoters
Results from the 5-Aza-CdR treatment suggested that DNA methylation can regulate gene expression from non-CpG island promoters. However, this agent induces genome-wide demethylation, the observed increase in expression after treatment could be indirect. To eliminate this possibility, we investigated whether DNA methylation plays a direct role in regulating non-CpG island promoters using a luciferase reporter assay based on a CpG-less luciferase vector-pCpGL (33).
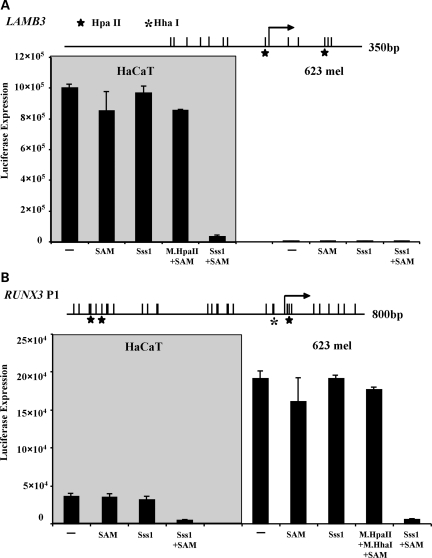
Methylated and unmethylated pCpGL-LAMB3 and pCpGL-RUNX3P1 were transfected into HaCaT cells and 623 melanoma cells. In agreement with their endogenous expression status, high luciferase activities were observed when unmethylated promoters, including mock-treated control plasmids, were transfected into cells which are competent to express each gene. In contrast, low luciferase activities were observed when the promoters were methylated, indicating methylation can directly suppress promoter activity (Fig. 4A, left panel, and B, right panel). To rule out the possibility that plasmid methylation could affect transfection efficiency, we took advantage of HpaII sites present in the promoter of LAMB3 and HpaII and HhaI sites in RUNX3 P1. Cells transfected with plasmids that were only methylated by HpaII (Fig. 4A) or HpaII and HhaI methyltransferases (Fig. 4B) showed a level of luciferase activity comparable with that of unmethylated controls arguing against the possibility that the low level of luciferase activity detected in our study was caused by low transfection efficiency of methylated plasmids. In addition, this result suggests that partial or sporadic methylation may not be sufficient to silence these genes and that extensive methylation may be required to achieve gene silencing. Luciferase activity remained low in 623 melanoma cells transfected with methylated or unmethylated pCpGL-LAMB3 (Fig. 4A, right panel), suggesting that expression also requires the presence of additional factors to initiate transcription in host cells. These results are consistent with our previous findings that 5-Aza-CdR treatment fails to induce LAMB3 expression in 623 melanoma cells (Fig. 3A). Consistent with our expression data of the RUNX3 P1 in HaCaT cells after 5-Aza-CdR treatment, we observed moderate luciferase activity when transfecting unmethylated RUNX3 P1 into HaCaT cells, which do not express this gene endogenously. As expected, no luciferase activity was observed when methylated RUNX3 P1 was transfected into HaCaT cells (Fig. 4B, left panel). Taken together, these data show that DNA methylation can directly lead to transcriptional silencing. Furthermore, the results also show that promoter demethylation is not sufficient for re-expression of genes controlled by non-CpG island promoters.
Figure 4.
DNA methylation directly silences LAMB3 and RUNX3 P1. (A) Luciferase activities of methylated and unmethylated pCpGL-LAMB3 in HaCaT and 623 melanoma cells. M.SssI with the presence of SAM methylates all CpG sites. (B) Luciferase activities of methylated and unmethylated pCpGL-RUNX3 P1 in HaCaT and 623 melanoma cells. Bars are the average of technical triplicates + the standard deviation. Same trend was observed in three biological experiments. Stars represent HpaII sites and asterisks represent HhaI sites.
Distinct histone modifications at LAMB3 and RUNX3 promoters correlate with their expression statuses
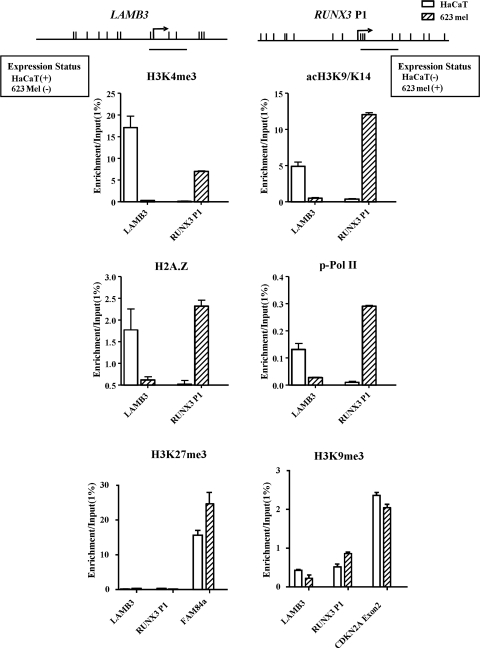
Specific histone tail modifications have been previously shown to be associated with active and inactive CpG island promoters (34,35). For instance, methylation of histone H3 lysine 4 (H3K4), acetylation of histone H3 lysine 9 (acH3K9) and H2A.Z have been shown to be important for transcriptional activation of CpG island promoters, whereas the methylation of histone H3 lysine 9(H3K9) is associated with transcriptionally silenced genes (34,36). To characterize the chromatin architecture associated with the LAMB3 promoter and RUNX3 P1, we performed ChIP-qPCR analysis of histone tail modifications. An enrichment of H3K4me3 and acetylation of H3, H2A.Z and p-Pol II were observed at the LAMB3 promoter in HaCaT cells, whereas minimal or no enrichment of such active marks was observed in LAMB3 non-expressing 623 melanoma cells (Fig. 5). Similarly, active marks were associated with RUNX3 P1 in its expressing 623 melanoma cells but not in the non-expressing HaCaT cells (Fig. 5). Surprisingly, enrichment of inactive histone marks H3K9me3 and H3K27me3 was not observed at transcriptionally inactive non-CpG island promoters and was not correlated with expression status. Our positive controls, FAM84a, a known polycomb silenced gene, and p16 exon 2, a region that is silenced by DNA methylation in these two cell lines (data not shown), showed an enrichment of H3K27me3 and H3K9me3, respectively (Fig. 5). Our results demonstrate that non-CpG island promoters may have limited similarity in histone modifications to CpG island promoters. Only active non-CpG island promoters display a histone tail modification repertoire similar to that of CpG island promoters, thereby showing a link between DNA methylation status and chromatin configuration at such non-CpG promoters.
Figure 5.
Active histone modifications mark active non-CpG promoters; inactive non-CpG island promoters lack inactive histone marks. ChIP was performed using antibodies for H3K4me3, acetylated H3, H2A.Z, phosphorylated Pol II, H3K27me3 and H3K9me3. Y-axis represents IP-IgG/Input-IgG of two biological experiments + the range between the two experiments. FAM84a and CDKN2A Exon 2 are used as positive controls for H3K27me3 and H3K9me3, respectively.
Distinct nucleosome occupancies at LAMB3 and RUNX3 promoters are associated with their methylation status
Nucleosome positioning participates in transcription regulation by modifying the accessibility of DNA to transcription machinery (14,37). It is well-known that active CpG island promoters contain a NDR upstream of their TSSs, whereas inactive CpG island promoters lack this NDR (7,15,37). To characterize the chromatin architecture of non-CpG island promoters, we used a single-molecule, high-resolution nucleosome positioning assay, NOMe-Seq (37,38), which is based on the ability of the M.CviPI enzyme to methylate GpC sites (39) that are not bound by nucleosomes providing a digital footprint of nucleosome positioning. This enzyme has an advantage over M.SssI due to the lack of endogenous GpC methylation in humans, except that occurring in the context of GpCpG. M.CviPI enables us to analyze nucleosome position at CpG poor promoters irrespective of their endogenous methylation status.
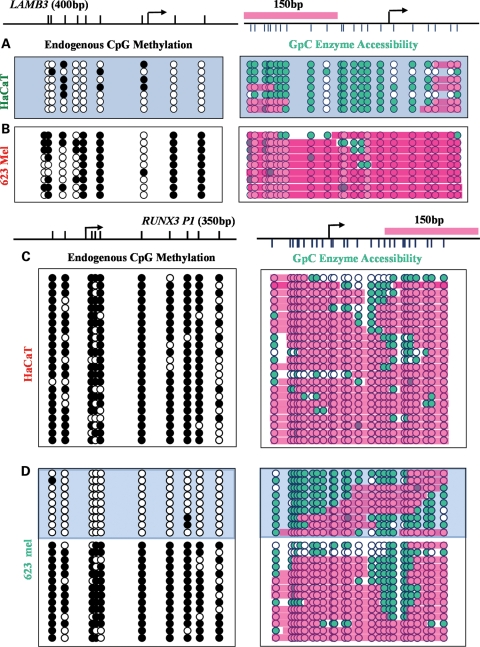
The region upstream of the TSS of the LAMB3 promoter is accessible to M.CviPI in HaCaT cells, indicating a NDR, which might allow the binding of appropriate transcription factors (Fig. 6A). The presence of the NDR correlated with high LAMB3 expression and the unmethylated status of the LAMB3 promoter in these cells (Fig. 3A). In contrast, 623 melanoma cells, which exhibit extensive methylation at the LAMB3 promoter, show almost no accessibility to M.CviPI at the region immediately upstream of the TSS, indicating the presence of nucleosomes (Fig. 6B).
Figure 6.
Distinct chromatin structures at the LAMB3 promoter and RUNX3 P1 correlate with expression status. Endogenous methylation level and M.CviPI, accessibility of LAMB3 in HaCaT cells (A) and 623 melanoma cells (B). Endogenous methylation level and M.CviPI, accessibility of RUNX3 in HaCaT cells (C) and 623 melanoma cells (D). Filled and unfilled circles represent methylated and unmethylated CpG sites, respectively. Green circles indicate accessible GpC sites. Pink boxes represent inaccessibility, indicating nucleosome occupancy.
Despite the fact that RUNX3 P1 showed a 35% methylation, a transcript produced from this promoter is endogenously expressed in 623 melanoma cells (Fig. 3B). To our surprise, NOMe-Seq revealed that RUNX3 P1 exists in two distinct nucleosome configurations, which are associated with two different methylation patterns in these cells. A major strength of NOMe-Seq is the ability to correlate nucleosome positioning and DNA methylation within the same DNA strand. We found that ∼40% of the strands were either completely unmethylated or sporadically methylated, displaying an NDR upstream of the TSS. On the other hand, methylated strands lacked this NDR (Fig. 6C). Conversely, in non-expressing HaCaT cells, RUNX3 P1 was heavily methylated (Fig. 4B), and had minimal accessibility to M.CviPI, indicating nucleosome occupancy at the TSS (Fig. 6D). Taken together, these data indicate that in non-CpG island promoters, the pattern of nucleosome occupancy at the TSS correlates with their methylation and expression status, similar to that of CpG islands.
Monoallelic methylation of RUNX3 P1 in 623 melanoma cells
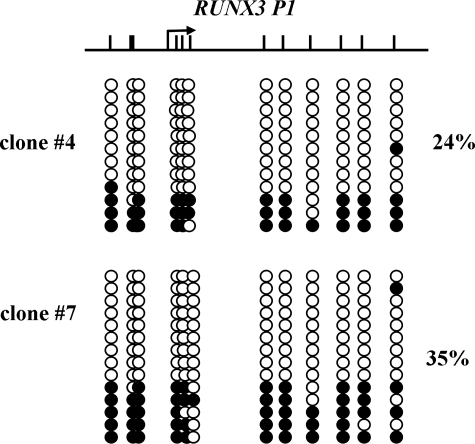
The methylation pattern of RUNX3 P1 we found in 623 melanoma cells was reminiscent of that observed in monoallelically methylated genes (40,41). To investigate the possibility that RUNX3 P1 is monallelically methylated, we cloned single 623 melanoma cells and the methylation level of RUNX3 P1 and expression level of the transcript generated by RUNX3 P1 assessed in each clone as well as the parental cells. Parental cells and all the clones showed comparable DNA methylation and expression levels (data not shown). Subsequently, we selected two clones, one with the highest and one with the lowest DNA methylation level and performed bisulfite genomic sequencing to determine the methylation pattern at a single-molecule resolution. The results show that two clones displayed a 24% and a 35% methylation and exhibited a monoallelic methylation pattern with majority of the DNA strands being either completely methylated or unmethylated (Fig. 7).
Figure 7.
Monoallelic methylation of RUNX3 P1. The DNA methylation pattern of RUNX3 P1 was analyzed in clone #4 and #7 by bisulfite genomic sequencing. Filled and unfilled circles represent methylated and unmethylated CpG sites, respectively. Schematic diagram of RUNX3 P1 is shown. The black arrow represents the TSS.
DISCUSSION
Gene expression is regulated epigenetically by DNA methylation, histone modifications and nucleosome positioning. Although genome-wide studies have revealed the pattern of methylation across the whole genome both at CpG island and at non-CpG island promoters (35), the role of DNA methylation at non-CpG island promoters has often been overlooked. It has been previously speculated that CpG methylation at non-CpG island promoters is involved in the establishment and maintenance of tissue-specific gene expression based on a few genome-wide DNA methylation analyses (18,35). However, no clear examples of such genes have been identified in humans. We found that the promoter of LAMB3 and RUNX3 P1 shows a tissue-specific methylation pattern; in addition, the promoter of LAMB3 is hypomethylated in primary bladder tumors, suggesting that DNA methylation at non-CpG island promoters is involved in regulating both normal cell functions and pathological processes. It has been shown that aberrant methylation occurring at non-CpG island promoters can also result in ectopic gene activation or silencing, thereby contributing to tumorigenesis (42). LAMB3 hypermethylation and gene silencing have been reported to occur in various types of cancers, including bladder cancer, causing its silencing (43–45). However, our results clearly show that hypomethylation of the LAMB3 promoter correlates with its upregulation in primary bladder tumors, thus potentially contributing to tumorigenesis, as it has been previously shown to increase cell motility (46). In addition, our laboratory has demonstrated that both aberrant hypermethylation and hypomethylation occur in bladder tumors when compared with cancer-free urothelium (47) by using Illumina GoldenGate methylation assay, which interrogates 1370 CpG loci. Most of the hypomethylated loci were located in non-CpG island promoters. Hypomethylation of non-CpG island promoters was correlated with gene reactivation in both invasive and superficial bladder tumors, supporting that the view of hypomethylation occurring at non-CpG island promoters may play an important role in carcinogenesis.
To assess whether DNA methylation can directly regulate non-CpG island promoters, we utilized a CpG-less luciferase gene containing vector to perform the luciferase reporter assay. This CpG-less vector overcomes the problems previously associated with testing methylation-sensitive promoter activity in vitro. For example, the traditional way of testing methylation-sensitive promoter activity involves methylating an entire reporter vector (promoter and backbone) in vitro. This can be problematic due to the existence of CpG sites in the coding region of the luciferase gene and vector backbone, thus potentially contributing to the repression of the promoter (33). Another traditional way is patch methylation, which involves in vitro methylation of the promoter before ligating it to a vector to avoid methylating the vector, which is technically difficult and labor intensive. Our luciferase assay results demonstrate that DNA methylation can directly silence genes with non-CpG island promoters in cells competent to express these genes, arguing against a proposal that DNA methylation is unlikely to play a functional role in regulating non-CpG island promoters (20). Our results also addressed some earlier concerns regarding a lack of examples of methylation of a tissue-specific gene that can prevent its transcription in a cell type which is capable of expressing the gene (48). Our results also show, as expected, that promoter demethylation is not sufficient for a gene to induce gene re-expression, which requires the assembly of the transcriptional machinery, including proper specific transcription factors. The 623 melanoma cells presumably lack the necessary transcription factors for LAMB3 expression, thus resulting in both the failure of 5-Aza-CdR to induce re-expression as well as the inability of unmethylated LAMB3 constructs to be expressed in these cells.
We also showed that transcriptionally active non-CpG island promoters possess a chromatin structure that is similar to that in CpG island promoters, with an enrichment of active histone marks. However, enrichments of inactive histone marks were not observed and were not correlated with expression status at transcriptionally inactive promoters. It is possible that inactive histone marks are not shared by non-CpG island promoters (49). The acquisition of inactive marks may require a higher concentration of methylated CpG sites. In the future, genome-wide analyses will be necessary to elucidate histone tail modifications that are associated with non-CpG island promoters. NOMe-Seq allowed us to map nucleosome occupancy at non-CpG island promoters at a single-molecule resolution, revealing that the existence of a NDR upstream of TSSs is not restricted to CpG island promoters. Using this powerful method, which allows for the correlation of DNA methylation status and nucleosome positioning within the same DNA strand, we also found that RUNX3 P1 is monoallelically methylated in 623 melanoma cells. To our knowledge, this is the first report showing that monoallelic methylation corresponds to distinct nucleosome occupancy.
In summary, our detailed study shows the underlying mechanism of epigenetic regulation at non-CpG island promoters. We demonstrate that DNA methylation can directly silence genes with non-CpG island promoters and contribute to the establishment of tissue-specific methylation patterns. The distinct chromatin architectures associated with active non-CpG island promoters are associated with an NDR immediately upstream of the TSS and the acquisition of active histone marks, such as H3K4me3, H3ac H2A.Z and p-Pol II, behaving similarly to active CpG island promoters. Lack of acquisition of inactive histone marks at inactive non-CpG island promoters may make them more ‘plastic’ and therefore more changeable during development.
MATERIALS AND METHODS
Isolation of human lymphocytes from whole blood
Blood (40 ml) was drawn (in accordance with the IRB protocol) from a healthy donor into a BD Vacutainer containing sodium heparin to prevent coagulation. Blood was layered on top of Ficoll-Paque PLUS (GE Healthcare, Piscataway, NJ, USA) in 15 ml centrifuge tubes. Tubes were spun in a clinical centrifuge (440g) for 30 min. The lymphocytes which were found at the interface between the plasma and the Ficoll-Paque PLUS were recovered and subjected to short washing steps with Hank's balanced salt solution and phosphate buffered solution (Mediatech Inc., Herndon, VA, USA) to remove any platelets. CD8+ and CD4+ lymphocytes were further purified using Microbeads (Miltenyi, Auburn, CA, USA) conjugated to monoclonal anti-human CD8+ or CD4+ antibodies. Remaining granulocytes located on the layer immediately above the erythrocytes were also recovered separately and washed.
5-Aza-CdR treatment
HaCaT cells and 623 Melanoma cells were plated at 3 × 105 cells/100 mm dish and treated the next day with 0.5 µm of 5-Aza-CdR (Sigma Chemical Co., St Louis, MO, USA). The medium was changed 24 h later. RNA and DNA were extracted 8 days after the treatment.
Tissue collection and DNA isolation
DNA from healthy tissue was purchased from BioChain Institute Inc. (Hayward, CA, USA). DNA from matched sets of adjacent normal bladder and tumor specimens was obtained from bladder cancer patients at University of Southern California/Norris Comprehensive Cancer Center (Los Angeles, CA, USA) with informed consent and IRB approval. DNA was extracted from cells by digestion in lysis buffer [400 mm NaCl, 100 mm Tris–HCl (pH 8.5), 5 mm EDTA, 2% SDS, 20 µg/ml RNase A and 500 µg/ml proteinase K] for 16 h at 55°C. DNA was phenol/chloroform extracted, ethanol precipitated and dissolved in TE buffer.
Real-time RT–PCR
Total RNA was isolated from cells with Trizol reagent (Invitrogen). Two micrograms of RNA were reverse transcribed using random hexamers, deoxynucleotide triphosphate (Boehringer Mannheim, Mannheim, Germany), and superscript II reverse transcriptase (Life Technologies, Inc.) in 50 μl reaction volume. Human Multiple Tissue cDNA was purchased from BD Biosciences Clontech (Mountain View, CA, USA). With each set of PCR, titrations of known amounts of DNA were included as a standard for quantitation. Primer sequences are available upon request.
Methylation-sensitive single-nucleotide primer extension
Genomic DNA was treated with sodium bisulfite as previously described (50). The average cytosine methylation level was determined by the Ms-SNuPE as described previously (51). Briefly, three CpG sites were interrogated for each promoter. The methylation percentage of each sample is the average of the three CpG sites examined by Ms-SNuPE. Primer sequences are available upon request.
M.SssI treatment and luciferase assay
LAMB3 promoter and RUNX3 P1 were cloned into a CpG-less vector, pCpGL using methylation insensitive restriction sites: SpeI and HindIII. The whole plasmid was subjected to in vitro methylation by methyltransferase M.SssI in the presence of methyl group donor S-adenosylmethionine (SAM). Three controls included mock-treated plasmids that were taken through the same steps, but in the absence of either or both of M.SssI and SAM. Methylation was confirmed by digestion of HpaII, a methylation-sensitive restriction enzyme, and MspI, an isoschizomer of HpaII, which is not methylation sensitive. Both the methylated and unmethylated promoters were transfected into HaCaT cells and 623 melanoma cells. After 24 h, cells were assayed for luciferase activities using a single-luciferase reporter assay kit from Promega (Madison, WI, USA).
ChIP assay
ChIP assays were performed as described previously using 4 × 106 cells per IP (7,52). H3, phospharylated Pol II and H3K9me3 antibodies were purchased from Abcam (Cambridge, MA, USA). Acetylated H3, H3K27me3 and IgG antibodies were purchased from Millipore (Billerica, MA, USA). H3K4me3 and H2A.Z antibodies were purchased from Active Motif (Carlsbad, CA, USA).
M.CviPI treatment
Nuclei preparation and GpC Methyltransferase treatment were performed as described previously (37,38). Briefly, freshly extracted nuclei were treated with 200 U of GpC methylatransferase in the presence of SAM for 15 min at 37°C. An equal volume of stopping solution (20 nm Tris–HCl, 600 mm NaCl, 1% SDS, 10 mm EDTA, 400 µg/ml proteinase K) was added to stop the reaction. The whole reaction mixture was incubated at 55°C overnight. DNA was purified by phenol/chloroform extraction and ethanol precipitation, followed by PCR of regions of interest and TA cloning. Primer sequences are available upon request.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by National Institute of Health Grants (R01 CA124518 to G.L., R01 CA83867 to P.A.J.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Michael Rehli for the generous gift of the pCpGL plasmid vector. We also thank Dr Claudia Andreu-Vieyra and Dr Theresa K. Kelly for the helpful discussion and critical reading of the manuscript.
Reference
- 1.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 5.Jones P.A., Liang G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 7.Lin J.C., Jeong S., Liang G., Takai D., Fatemi M., Tsai Y.C., Egger G., Gal-Yam E.N., Jones P.A. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebbes T.R., Thorne A.W., Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang G., Lin J.C., Wei V., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl Acad. Sci. USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 12.Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 13.Mellor J. The dynamics of chromatin remodeling at promoters. Mol. Cell. 2005;19:147–157. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Jiang C., Pugh B.F. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones P.A. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 17.Takai D., Jones P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gal-Yam E.N., Egger G., Iniguez L., Holster H., Einarsson S., Zhang X., Lin J.C., Liang G., Jones P.A., Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc. Natl Acad. Sci. USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 21.Aumailley M., Rousselle P. Laminins of the dermo-epidermal junction. Matrix Biol. 1999;18:19–28. doi: 10.1016/s0945-053x(98)00004-3. [DOI] [PubMed] [Google Scholar]
- 22.Bangsow C., Rubins N., Glusman G., Bernstein Y., Negreanu V., Goldenberg D., Lotem J., Ben-Asher E., Lancet D., Levanon D., et al. The RUNX3 gene—sequence, structure and regulated expression. Gene. 2001;279:221–232. doi: 10.1016/s0378-1119(01)00760-0. [DOI] [PubMed] [Google Scholar]
- 23.Bae S.C., Choi J.K. Tumor suppressor activity of RUNX3. Oncogene. 2004;23:4336–4340. doi: 10.1038/sj.onc.1207286. [DOI] [PubMed] [Google Scholar]
- 24.Kang G.H., Lee S., Lee H.J., Hwang K.S. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J. Pathol. 2004;202:233–240. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- 25.Kim T.Y., Lee H.J., Hwang K.S., Lee M., Kim J.W., Bang Y.J., Kang G.H. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab. Invest. 2004;84:479–484. doi: 10.1038/labinvest.3700060. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y., Tong D., Lou G., Zhang Y., Geng J. Expression of RUNX3 gene, methylation status and clinicopathological significance in breast cancer and breast cancer cell lines. Pathobiology. 2008;75:244–251. doi: 10.1159/000132385. [DOI] [PubMed] [Google Scholar]
- 27.Wolff E.M., Liang G., Cortez C.C., Tsai Y.C., Castelao J.E., Cortessis V.K., Tsao-Wei D.D., Groshen S., Jones P.A. RUNX3 methylation reveals that bladder tumors are older in patients with a history of smoking. Cancer Res. 2008;68:6208–6214. doi: 10.1158/0008-5472.CAN-07-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takai D., Jones P.A. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- 29.Chung D.D., Honda K., Cafuir L., McDuffie M., Wotton D. The Runx3 distal transcript encodes an additional transcriptional activation domain. FEBS J. 2007;274:3429–3439. doi: 10.1111/j.1742-4658.2007.05875.x. [DOI] [PubMed] [Google Scholar]
- 30.Egawa T., Tillman R.E., Naoe Y., Taniuchi I., Littman D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2009;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klug M., Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 34.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Rakyan V.K., Down T.A., Thorne N.P., Flicek P., Kulesha E., Graf S., Tomazou E.M., Backdahl L., Johnson N., Herberth M., et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z., Pugh B.F. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly T.K., Miranda T.B., Liang G., Berman B.P., Lin J.C., Tanay A., Jones P.A. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol. Cell. 2010;39:901–911. doi: 10.1016/j.molcel.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolff E.M., Byun H.M., Han H.F., Sharma S., Nichols P.W., Siegmund K.D., Yang A.S., Jones P.A., Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6:e1000917. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu M., Kladde M.P., Van Etten J.L., Simpson R.T. Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic Acids Res. 1998;26:3961–3966. doi: 10.1093/nar/26.17.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum. Mol. Genet. 2010;19:R210–R220. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Valo Z., Bowers C.W., Smith D.D., Liu Z., Singer-Sam J. Dual DNA methylation patterns in the CNS reveal developmentally poised chromatin and monoallelic expression of critical genes. PLoS ONE. 2010;5:e13843. doi: 10.1371/journal.pone.0013843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calaluce R., Bearss D.J., Barrera J., Zhao Y., Han H., Beck S.K., McDaniel K., Nagle R.B. Laminin-5 beta3A expression in LNCaP human prostate carcinoma cells increases cell migration and tumorigenicity. Neoplasia. 2004;6:468–479. doi: 10.1593/neo.03499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sathyanarayana U.G., Maruyama R., Padar A., Suzuki M., Bondaruk J., Sagalowsky A., Minna J.D., Frenkel E.P., Grossman H.B., Czerniak B., et al. Molecular detection of noninvasive and invasive bladder tumor tissues and exfoliated cells by aberrant promoter methylation of laminin-5 encoding genes. Cancer Res. 2004;64:1425–1430. doi: 10.1158/0008-5472.can-03-0701. [DOI] [PubMed] [Google Scholar]
- 44.Sathyanarayana U.G., Padar A., Huang C.X., Suzuki M., Shigematsu H., Bekele B.N., Gazdar A.F. Aberrant promoter methylation and silencing of laminin-5-encoding genes in breast carcinoma. Clin. Cancer Res. 2003;9:6389–6394. [PubMed] [Google Scholar]
- 45.Sathyanarayana U.G., Padar A., Suzuki M., Maruyama R., Shigematsu H., Hsieh J.T., Frenkel E.P., Gazdar A.F. Aberrant promoter methylation of laminin-5-encoding genes in prostate cancers and its relationship to clinicopathological features. Clin. Cancer Res. 2003;9:6395–6400. [PubMed] [Google Scholar]
- 46.Aumailley M., El Khal A., Knoss N., Tunggal L. Laminin 5 processing and its integration into the ECM. Matrix Biol. 2003;22:49–54. doi: 10.1016/s0945-053x(03)00013-1. [DOI] [PubMed] [Google Scholar]
- 47.Wolff E.M., Chihara Y., Pan F., Weisenberger D.J., Siegmund K.D., Sugano K., Kawashima K., Laird P.W., Jones P.A., Liang G. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010;70:8169–8178. doi: 10.1158/0008-5472.CAN-10-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh C.P., Bestor T.H. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou V.W., Goren A., Bernstein B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2010;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 50.Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., Molloy P.L., Paul C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalgo M.L., Liang G. Methylation-sensitive single-nucleotide primer extension (Ms-SNuPE) for quantitative measurement of DNA methylation. Nat. Protoc. 2007;2:1931–1936. doi: 10.1038/nprot.2007.271. [DOI] [PubMed] [Google Scholar]
- 52.Gal-Yam E.N., Jeong S., Tanay A., Egger G., Lee A.S., Jones P.A. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.