Abstract
Autism is a neurodevelopmental disorder with increasing evidence of heterogeneous genetic etiology including de novo and inherited copy number variants (CNVs). We performed array comparative genomic hybridization using a custom Agilent 1 M oligonucleotide array intended to cover 197 332 unique exons in RefSeq genes; 98% were covered by at least one probe and 95% were covered by three or more probes with the focus on detecting relatively small CNVs that would implicate a single protein-coding gene. The study group included 99 trios from the Simons Simplex Collection. The analysis identified and validated 55 potentially pathogenic CNVs, categorized as de novo autosomal heterozygous, inherited homozygous autosomal, complex autosomal and hemizygous deletions on the X chromosome of probands. Twenty percent (11 of 55) of these CNV calls were rare when compared with the Database of Genomic Variants. Thirty-six percent (20 of 55) of the CNVs were also detected in the same samples in an independent analysis using the 1 M Illumina single-nucleotide polymorphism array. Findings of note included a common and sometimes homozygous 61 bp exonic deletion in SLC38A10, three CNVs found in lymphoblast-derived DNA but not present in whole-blood derived DNA and, most importantly, in a male proband, an exonic deletion of the TMLHE (trimethyllysine hydroxylase epsilon) that encodes the first enzyme in the biosynthesis of carnitine. Data for CNVs present in lymphoblasts but absent in fresh blood DNA suggest that these represent clonal outgrowth of individual B cells with pre-existing somatic mutations rather than artifacts arising in cell culture. GEO accession number GSE23765 (http://www.ncbi.nlm.nih.gov/geo/, date last accessed on 30 August 2011). Genboree accession: http://genboree.org/java-bin/gbrowser.jsp?refSeqId=1868&entryPointId=chr17&from=53496072&to=53694382&isPublic=yes, date last accessed on 30 August 2011.
INTRODUCTION
Autism spectrum disorders (ASDs) represent a heterogeneous group of patients characterized by impaired social interaction and communication and by restricted and repetitive behaviors. The clinical spectrum extends from two extremes. At the severe end of the spectrum are children with intellectual disability, congenital malformations, dysmorphic features and impairments so severe as to make reproduction unlikely. The frequency of cytogenetic abnormalities and pathologic copy number variants (CNVs) in this population is 25–30% (1). The genetic abnormalities in this most severe group are typically de novo, and the sex (male-to-female) ratio is ∼3.2:1 with some mutations occurring on the X chromosome (2). A middle group of patients covers a wide spectrum, and pathological CNVs occur in 5–10% of cases using current methods (3,4). These CNVs may be inherited or de novo, and penetrance is frequently incomplete. At the mild end of the spectrum are patients who have IQs in or near the normal range and are not dysmorphic. This mild end of the spectrum includes the Asperger syndrome, and the sex (male-to-female) ratio can be as high as 8:1 (5,6). The etiology for these milder patients is largely unknown.
Use of exon-focused arrays to analyze 3743 samples in a clinical laboratory setting detected many small, disease-causing CNVs that are not detected by most arrays currently used for research and for clinical diagnosis (7). We reasoned that the exon-focused arrays used here should detect many CNVs that would be below the resolution of most clinical arrays. This study was performed concurrently with analysis of these same cases using Illumina 1 M single-nucleotide polymorphism (SNP) arrays (8). We hoped to detect disease-causing mutations that would not be detected by the Illumina arrays, although the small size of this pilot study would be a limitation.
RESULTS
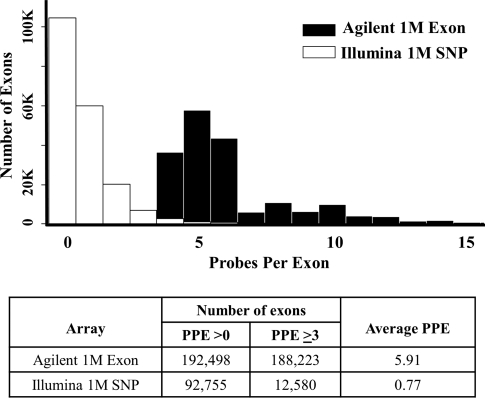
We designed a 1 million (1 M) Agilent comparative genomic hybridization (CGH) whole-genome exon-focused array with probes selected to give six probes per exon (PPE) for the majority of exons in the genome as described in Materials and methods. For each exon, three oligonucleotides were selected and both strands were utilized with identical coordinates. Certain genes were omitted on the assumption that they were unlikely to be mutated as a cause of neurobehavioral abnormalities as specified in Materials and methods. We analyzed 297 samples from 99 trios (probands with ASD, mother, father) from the Simons Simplex Collection (SSC) (9) and used a single male reference on the exon array (Supplementary Material, Table S1). Ninety of these trios were also analyzed using an SNP array (Illumina Human 1 M—single BeadChip array) by collaborators (8). CNV prediction focused on the CNVs most likely to be deleterious, specifically de novo heterozygous gains or losses, inherited homozygous deletions and hemizygous gains or losses of the X chromosome in males. All CNVs were validated by comparison with Illumina CNVs, further Agilent CGH arrays or polymerase chain reaction (PCR) amplification and sequencing of junction sites. We hypothesized that this design should provide substantially increased sensitivity for detecting small exon-containing deletions and duplications when compared with the Illumina data. The exon-focused nature of the 1 M Agilent array gave far greater probe density over exons than the more evenly distributed whole-genome Illumina 1 M SNP array. From the 197 332 unique exons targeted, 98% had ≥1 PPE for the exon array compared with 47% of exons on the SNP array. The mean number of PPE was 5.91 for the custom exon array versus a mean of 0.77 PPE for the SNP array (Fig. 1).
Figure 1.
Oligonucleotide exonic coverage in Agilent whole-genome custom exon array compared with Illumina 1 M SNP array. The coverage of human exons in the Agilent custom exon and Illumina SNP array was calculated based on the RefSeq database (June 2008, hg18). The 197 332 exons described in Materials and methods were used for these calculations; coverage allowed 300 bp flanking both sides of each exon location. PPE, probes per exon.
CNV findings
Using the whole-genome custom exon-focused array, a total of 267 CNVs of potential interest were found in the 99 SSC trios studied (Table 1, Supplementary Material, Table S2). Because the criteria for calling CNVs were intentionally set to minimize false negatives and accept false positives, 193 (72%) of the 267 potential CNVs (mostly rare) were not confirmed leaving 74 CNVs for further analysis. These 74 were broken down as shown in Table 1 with 16 being rare and 58 being common as defined in Materials and methods. The common calls were most likely benign and of less interest being frequent in the population, although some common CNVs could be pathogenic. These included 26 de novo autosomal heterozygous, 15 complex (to be defined and discussed below), 13 autosomal heterozygous first called de novo but later proven inherited, 2 homozygous autosomal, 1 hemizygous (chrX) and 1 autosomal heterozygous in cell line DNA but not blood DNA. The rare group was of greater interest and included six de novo autosomal heterozygous, two complex, three autosomal heterozygous first called de novo but later proven inherited, one homozygous autosomal, two hemizygous (chrX) and two autosomal heterozygous in cell line DNA but not blood DNA.
Table 1.
Classification and validation of complex, de novo heterozygous, inherited homozygous and hemizygous preliminary calls from 99 SSC probands using the whole-genome custom exon array
| Type | CNV calls | CNV failed to confirm | CNV confirmed | Confirmed category | Confirmed inheritance | Identified by SNP array and exon array | Unique to exon array | Total |
|---|---|---|---|---|---|---|---|---|
| Rare | 196 | 180 | 16 | Complex | Unclear | 0 | 2 | 2 |
| Autosomal heterozygous | Inheriteda | 0 | 3 | 3 | ||||
| Autosomal homozygous | Inherited | 0 | 1 | 1 | ||||
| Hemizygous (chrX) | Inherited | 0 | 2 | 2 | ||||
| Autosomal heterozygous in cell line DNA but not blood DNA | De novo | 0 | 2 | 2 | ||||
| Autosomal heterozygous in blood DNA | De novo | 5 | 1 | 6 | ||||
| Common | 71 | 13 | 58 | Complex | Unclear | 2 | 13 | 15 |
| Autosomal heterozygous | Inheriteda | 4 | 9 | 13 | ||||
| Autosomal homozygous | Inherited | 1 | 1 | 2 | ||||
| Hemizygous (chrX) | Inherited | 1 | 0 | 1 | ||||
| Autosomal heterozygous in cell line DNA but not blood DNA | De novo | 1 | 0 | 1 | ||||
| Autosomal heterozygous in blood DNA | De novo | 11 | 15 | 26 | ||||
| Total | 267 | 193 | 74 | 25 | 49 | 74 |
aInitial call de novo but proved inherited.
Specific information for the 55 calls of greatest interest is provided in Table 2, which does not include 16 CNVs initially called de novo but proven to be inherited and 3 CNVs present in cell line DNA but absent in blood DNA.
Table 2.
Description of whole-genome custom exon array confirmed CNV calls of interest
| SSC ID | Sex | Chromosome | Min. start | Min. size | Type | RefSeq gene (hg19) | Probes exon | Validation testa | DGV |
|---|---|---|---|---|---|---|---|---|---|
| De novo autosomal heterozygous: 26 common and 6 rare | |||||||||
| 11458 | M | 1q21.3 | 152573248 | 13 045 | Loss | LCE3C, LCE3B | 5 | D/− | Common |
| 11076 | M | 1q31.3 | 196711807 | 111 947 | Gain | CFH, CFHR3, CFHR1 | 35 | B/− | Common |
| 11291 | M | 1q31.3 | 196712510 | 144 771 | Loss | CFH, CFHR3, CFHR1, CFHR4 | 80 | D/− | Common |
| 11443 | M | 1q31.3 | 196718380 | 104 459 | Gain | CFHR3, CFHR1 | 88 | B/− | Common |
| 11489 | M | 2q13 | 110852828 | 408 305 | Loss | NPHP1+ 5 genes | 104 | B/+ | Rare |
| 11046 | M | 3p26.1 | 4344934 | 533 721 | Loss | SETMAR, SUMF1, ITPR1, EGOT | 352 | D/+ | Rare |
| 11378 | M | 4q13.2 | 69402972 | 130 793 | Loss | UGT2B17, UGT2B15 | 45 | D/− | Common |
| 11412 | M | 4q13.2 | 69402972 | 126 404 | Loss | UGT2B17, UGT2B15 | 42 | D/− | Common |
| 11338 | M | 5q13.2 | 70193462 | 72 211 | Loss | 5 genes | 16 | D/− | Common |
| 11415 | M | 7q22.1 | 100328032 | 9797 | Gain | ZAN | 34 | B/− | Common |
| 11524 | M | 7q22.1 | 100328032 | 12 551 | Gain | ZAN | 32 | B/− | Common |
| 11345 | M | 11q12.2 | 60965139 | 55 347 | Loss | PGA3, PGA4, PGA5 | 49 | B/− | Common |
| 11499 | M | 11q12.2 | 60965716 | 55 365 | Gain | PGA3, PGA4, PGA5 | 63 | B/− | Common |
| 11197 | M | 12p11.21 | 31267642 | 85 948 | Gain | 0 | 136 | B/− | Common |
| 11303 | M | 12p11.21 | 31277416 | 76 688 | Loss | 0 | 124 | A,B/− | Common |
| 11469 | M | 12p11.21 | 31277416 | 76 368 | Loss | 0 | 142 | B/− | Common |
| 11550 | M | 12p11.21 | 31277416 | 76 368 | Loss | 0 | 120 | B/− | Common |
| 11152 | M | 13q21.1 | 57722436 | 25 162 | Gain | 5 genes | 4 | B/− | Common |
| 11149 | M | 15q11.1 | 20588546 | 45 025 | Loss | 0 | 7 | D/− | Common |
| 11443 | M | 15q11.1-q11.2 | 20588546 | 1 442 028 | Loss | 13 genes | 130 | D/− | Common |
| 11265 | M | 15q11.2 | 22835869 | 600 861 | Loss | 8 genes | 339 | D/+ | Common |
| 11178 | M | 15q13.2 | 30653646 | 35 270 | Loss | CHRFAM7A | 41 | D/− | Common |
| 11178 | M | 15q13.3 | 32445752 | 16 891 | Loss | CHRNA7 | 27 | D/− | Common |
| 11168 | M | 16p13.2 | 8895680 | 317 315 | Gain | PMM2, CARHSP1, USP7, C16orf72 | 240 | D/+ | Rare |
| 11378 | M | 16p11.2 | 32676757 | 618 367 | Gain | TP53TG3, TP53TG3B, LOC653550, SLC6A10P | 21 | B/− | Common |
| 11006 | M | 16q22.1 | 70174866 | 20 901 | Loss | PDPR | 105 | D/− | Common |
| 11327 | M | 16q23.2-q24.1 | 81183435 | 4 772 529 | Loss | 37 genes | 2009 | D/+ | Rare |
| 11353 | F | 17q12 | 34482071 | 2 047 404 | Loss | 27 genes | 1146 | D/+ | Rare |
| 11406 | M | 17q21.31 | 44403033 | 70 700 | Gain | LRRC37A, ARL17B | 6 | B/− | Common |
| 11186 | M | 17q25.3 | 79027376 | 32 869 | Loss | BAIAP2 | 25 | D/+ | Rare |
| 11075 | M | 22q11.1 | 16226253 | 225 102 | Loss | POTEH, OR11H1 | 56 | B/− | Common |
| 11554 | M | 22q11.1 | 16345359 | 89 835 | Loss | 0 | 56 | B/− | Common |
| Inherited autosomal homozygous: 2 common and 1 rare | |||||||||
| 11418 | M | 5q35.3 | 180412358 | 17 430 | Loss | BTNL3 | 23 | A/− | Common |
| 11089 | M | 17q25.3 | 79225034 | 61 | Loss | SLC38A10 | 4 | B,C/− | Rare |
| 11550 | M | 22q13.1 | 39359112 | 26 373 | Loss | APOBEC3A, APOBEC3B | 60 | A,D/− | Common |
| Autosomal complex: 15 common and 2 rare | |||||||||
| 11417 | M | 1q21.3 | 152555599 | 30 626 | Loss | LCE3C | 5 | B/− | Common |
| 11376 | M | 4q13.2 | 69373903 | 117 903 | Gain | UGT2B17 | 15 | B/− | Common |
| 11146 | M | 5q13.2 | 68830681 | 72 261 | Loss | 7 genes | 39 | B/− | Rare |
| 11469 | M | 5q13.2 | 68851539 | 1 518 420 | Loss | 15 genes | 34 | B/− | Rare |
| 11146 | M | 5q13.2 | 70305524 | 83 320 | Loss | 7 genes | 58 | B,D/− | Common |
| 11271 | M | 7q22.1 | 100327877 | 12 750 | Loss | ZAN | 30 | A,B/− | Common |
| 11178 | M | 7q22.1 | 100327877 | 12 750 | Loss | ZAN | 32 | B/− | Common |
| 11076 | M | 7q22.1 | 100328193 | 12 434 | Loss | ZAN | 7 | A,B/− | Common |
| 11152 | M | 14q24.3 | 73994722 | 54 336 | Loss | HEATR4, ACOT1, ACOT2 | 34 | B/− | Common |
| 11156 | M | 14q24.3 | 73994722 | 54 336 | Loss | HEATR4, ACOT1, ACOT2 | 34 | B/− | Common |
| 11197 | M | 14q24.3 | 73994722 | 55 467 | Loss | HEATR4, ACOT1, ACOT2 | 36 | B/− | Common |
| 11353 | F | 14q24.3 | 73994722 | 54 336 | Loss | HEATR4, ACOT1, ACOT2 | 42 | B,D/− | Common |
| 11443 | M | 14q24.3 | 73994722 | 55 467 | Loss | HEATR4, ACOT1, ACOT2 | 34 | B/− | Common |
| 11479 | M | 14q24.3 | 73994722 | 54 336 | Loss | HEATR4, ACOT1, ACOT2 | 34 | B/− | Common |
| 11399 | M | 14q24.3 | 73995172 | 53 886 | Loss | HEATR4, ACOT1, ACOT2 | 38 | A,B/− | Common |
| 11523 | M | 17q21.32 | 45616195 | 54 400 | Gain | NPEPPS | 58 | B/− | Common |
| 11442 | M | 19q13.41 | 52134771 | 13 818 | Loss | SIGLEC14 | 6 | B/− | Common |
| Hemizygous chromosome X: 1 common and 2 rare (all maternal) | |||||||||
| 11411 | M | Xq12 | 67412653 | 21 207 | Loss | OPHN1b | 44 | A/− | Rare |
| 11443 | M | Xq28 | 153857308 | 23 928 | Loss | NCRNA00204B, NCRNA00204, CTAG2 | 13 | D/− | Common |
| 11000 | M | Xq28 | 154770740 | 13 835 | Loss | TMLHE | 5 | B,C/− | Rare |
aValidation test(s) used. A, 1 M Agilent catalog; B, design ID 027305, C, PCR; D, Illumina SNP array. Although not available at the time of submission, we can now report that another analysis of these same samples (17) identified some and not others of the CNVs reported here; /+, reported by Levy et al.; /−, not reported by these authors.
bThe OPHN1 proband call was among the Illumina 1 M SNP array high-confidence parameters; however, the proband's mother DNA did not pass the QC tests.
De novo autosomal heterozygous CNVs
The exon array successfully identified six rare de novo autosomal heterozygous CNVs present in blood out of 99 probands giving a de novo CNV burden of 6.1%; five of these rare de novo events were also detected by the Illumina array and have been described previously (8). Two of these CNVs are likely disease-causing and include a 4.8 Mb deletion at 16q23.2-q24.1 and a 2.0 Mb deletion at 17q12. The other three rare events of uncertain significance detected by both arrays included deletions of 534 kb at 3p26.2 and 33 kb at 17q25.3 and a duplication of 317 kb at 16p13.2 (Table 2). The single de novo autosomal heterozygous rare call not found by the Illumina array proved to be of low interest. This CNV was at 2q13 involving six genes, including the nephronophthisis 1 (NPHP1) gene. Homozygous deletion of this region is found in a large percentage of patients with familial juvenile nephronophthisis [NPHP1 (MIM 256100)]. The heterozygous deletion is seen frequently in clinical diagnostic labs and is usually considered to be a benign heterozygous carrier state. Although not rare, we observed a duplication of a region that occurs on both chromosomes X and 16. This CNV region was previously described (10) and includes SLC6A8 and BCAP31 at Xq28 and pseudogenes at 16p11.2. Because the duplicated region in the SSC case includes segments unique to 16p11.2, the duplication likely involves the pseudogenes on chromosome 16 and not the functional genes on the X chromosome. This duplication does not overlap with the deletions and duplications on 16p11.2 that are associated with cognitive or psychiatric abnormalities.
‘Complex’ CNVs
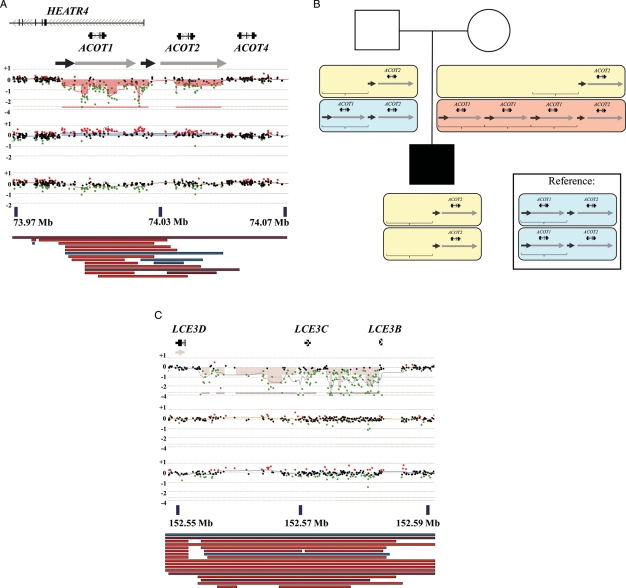
The CNVs described here as complex appeared de novo on first analysis, but closer evaluation revealed that these calls had variation in copy number within the CNV and almost always involved numerous related genes or unprocessed pseudogenes in highly repetitive and complex genomic regions densely represented in the Database of Genomic Variants (DGV). Two examples of complex calls are presented in Figure 2. Fifteen out of 17 of the complex calls were located in common CNV regions, and the same region was often involved in more than one family. For example, 7 of 17 complex calls were apparent deletions involving the ACOT1 and other genes at 14q24.3, while the second most common complex calls were three apparent deletions involving the ZAN gene at 7q22.1. In addition, proband SSC 11146.p1 had two adjacent deletions at 5q13.2, which may be the product of a complex rearrangement (data not shown). We propose that these ‘complex’ findings are most likely not truly de novo but are explained by low copy repeat sequences with a variable number of repeats on individual chromosomes (see Discussion).
Figure 2.
Validation of complex deletions. (A) Chromosome 14q24.3: relative position of UCSC genome browser RefSeq (UCSC genome browser GRCh37/hg19 assembly) genes (http://genome.ucsc.edu, date last accessed on 30 August 2011) is aligned above the CGH probe plot of proband SSC 11443 (top), mother (middle) and father (bottom). x-axis, chromosome position; y-axis, log 2 ratio. Arrowheads indicate orientation of transcription. Semi-transparent filled boxes on CGH plots highlight the region of aberration. Large gray arrows below genes represent segmental duplications from the UCSC web browser (data last updated: 19 February 2010). Arrows of same color represent regions that share more than 90% similarity. Colored bars below the CGH plot display represent relative position of CNVs from the DGV obtained through the UCSC web browser (data version: v10; data last updated: 22 February 2011). Blue bars represent a gain in copy number compared with the reference; red bars represent a loss in copy number compared with the reference; brown bars represent both a loss and a gain in copy number compared with the reference. (B) A diagram explaining the possible origin of the complex character of deletion at 14q24.3 found in proband SSC 11443 (A) is depicted (see text). Brackets represent a region that is deleted or duplicated in different individuals. Yellow, blue and pink rectangles represent chromosomes that contain a different number of copies of the genomic locus within brackets. Depiction is similar to (A). (C) Chromosome 1q21.3: relative position of UCSC genome browser RefSeq (hg19) genes (http://genome.ucsc.edu, date last accessed on 30 August 2011) is aligned above the CGH probe plot of proband SSC 11417 (top), mother (middle) and father (bottom). Depiction is similar to (A).
Homozygous deletions
The de novo detection algorithm for the exon array identified changes in copy number present in the proband but not the parents; this included the presence of homozygous deletions inherited from two parents with heterozygous deletions; one rare and two common CNVs were found in the homozygous form. One apparently homozygous common CNV was that of a deletion involving the first four exons of the BTNL3 or butyrophilin-like-3 gene inherited from both heterozygous parents. The BTNL3 gene is flanked by BTNL8 and BTNL9, and complex rearrangements are possible. Very little is known about this gene other than the fact that other butyrophilins are thought to be involved in inflammation and immune response (11). Although the deletion is in a common region, it is not clear that a homozygous deletion of this locus is benign, although we favor the interpretation that it is not the cause of autism in this case. The other homozygous common CNV was that of a deletion at 22q13.1 inherited from both heterozygous parents. The genes involved are APOBEC3A and APOBEC3B; they belong to the cytidine deaminase family and are reported to be the inhibitors of LTR retrotransposon and to affect certain viral infections (12,13). This deletion is of uncertain significance relative to the autism phenotype.
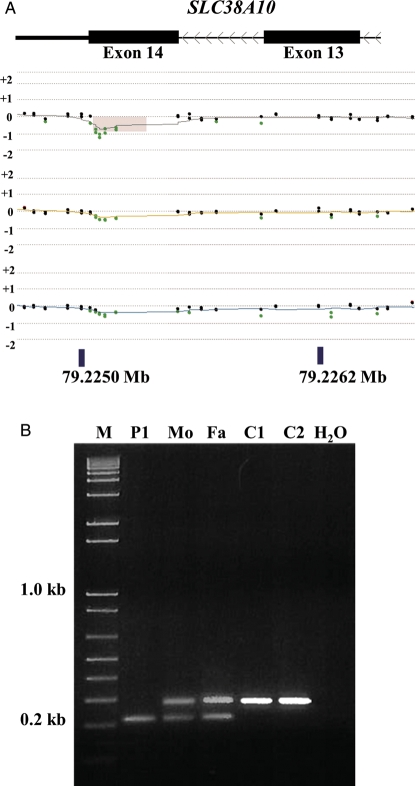
The sole autosomal homozygous rare variant was a 61 bp homozygous deletion of the last exon (exon 14) of one of the isoforms of the SLC38A10 gene; the deletion was inherited from both of the proband's heterozygous parents (Fig. 3). We used PCR and sequencing to define the breakpoints of this deletion and found that both alleles had the identical sequence with 8 bp of microhomology at the breakpoints and that the deletion was located within a 61 bp simple tandem repeat (Supplementary Material, Table S3). This deletion would cause frame-shift and result in a premature stop codon. SLC38A10, or solute carrier family 38 member 10, belongs to a family of solute carriers that transport neutral amino acids. One possibility is that this is a common benign CNV. We performed PCR across the deletion to detect two products in heterozygotes differing in size by 61 bp to test all remaining 98 probands of our collection, and an additional 485 SSC probands, 72 autism males from male affected sib pairs from the Autism Genetic Resource Exchange (AGRE) collection and 341 National Institute of Mental Health (NIMH) controls for the presence of the same 61 bp deletion. Based on the PCR studies, all the deletion products were indistinguishable in size. We found that 45 (or 7.7%) of the SSC probands were heterozygous and 2 (or 0.3%) were homozygous (counting SSC 11089.p1). We found that three parents of heterozygous SSC probands were homozygous for the deletion. This would be consistent with Hardy–Weinberg equilibrium. Additionally, we found that 8 (11.1%) AGRE probands and 32 (9.4%) NIMH controls were heterozygous. There were no homozygotes in the AGRE and NIMH samples tested (Supplementary Material, Tables S4–S6). These results indicate that heterozygosity for this SLC38A10 deletion is common. We suspect that both heterozygous and homozygous deletion genotypes are benign, but we cannot rule out the possibility that this deletion is a risk factor for autism.
Figure 3.
Validation of homozygous deletion in SLC38A10. (A) The browser display and labels are as in Fig. 2 showing exons 13 and 14 of one isoform of SLC38A10 (UCSC genome browser GRCh37/hg19 assembly). Analysis of family proband SSC 11089 (top), mother (middle) and father (bottom) is shown. (B) PCR for the SSC 11089 family showing the homozygous deletion in the patient (P1) and heterozygous deletion in the mother (Mo) and the father (Fa) but not in the unaffected controls (C1 and C2).
Hemizygous (chromosome X)
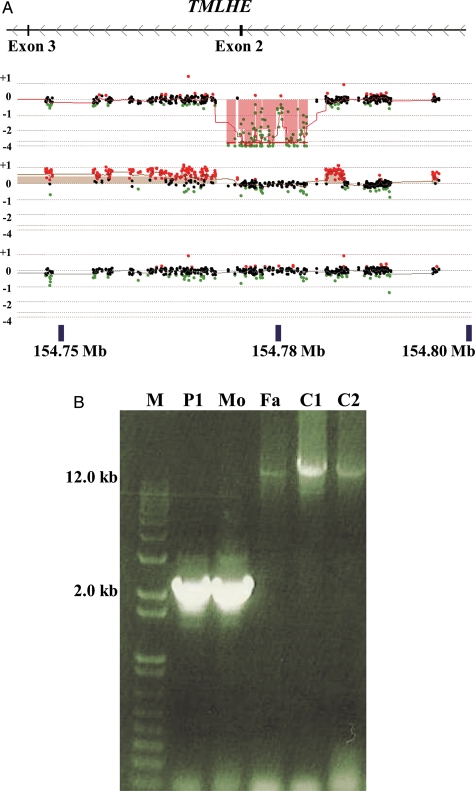
There were two rare hemizygous deletions on the X chromosome. One was a deletion of exons 7–15 of OPHN1 at Xq12 in a male. Loss-of-function mutations of OPHN1 in males are known to cause intellectual disability, seizures and cerebellar hypoplasia [OPHN1 (MIM 300127)] (14). The rare variant of potentially greatest interest in this study was a hemizygous loss of exon 2 of the X-linked trimethyllysine hydroxylase epsilon (TMLHE) gene in a male proband (SSC 11000.p1), whose mother was heterozygous for the deletion based on array analysis (Fig. 4A). PCR amplification identified a deleted fragment which was present in the proband and heterozygous mother (Fig. 4B). Sequencing of the junction fragment confirmed a deletion of 13 835 bp with a 7 bp repeat sequence at the junction (Supplementary Material, Table S3). The TMLD enzyme functions in the first step of l-carnitine biosynthesis, and there are no reports of human loss-of-function mutations, although one deletion of exon 2 in a healthy CEPH male (NA12003) is found in publicly available data (15).
Figure 4.
Validation of hemizygous deletion of exon 2 of TMLHE. (A) The browser display and labels are as in Fig. 2 showing exons 2 and 3 of TMLHE (UCSC genome browser GRCh37/hg19 assembly). Analysis of family proband SSC 11000 (top), mother (middle) and father (bottom) is shown. (B) PCR for the SSC 11000 family showing the deletion in the patient (P1) and mother (Mo), but not in the father (Fa) or unaffected controls (C1 and C2). There is bias of amplification of the smaller band in the mother so that the normal band is faint.
There was one common hemizygous deletion on Xq28. This 24 kb deletion involved three genes and was validated by the Illumina SNP array. Two of the genes transcribe non-coding RNAs (NCRNA00204B and NCRNA00204), while the third gene, CTAG2, encodes a cancer antigen [CTAG2 (MIM 300396)].
High-confidence de novo calls unique to the SNP arrays
In addition to obtaining calls that were in common between the exon array and the SNP array and calls that were unique to the exon array, there were two rare de novo CNVs with 20 SNP probes or more that were identified in 90 SSC probands using the Illumina SNP array (8). The two de novo CNVs were both large duplications (1 and 0.7 Mb at 16p13.11 and 16p12.2, respectively) spanning hundreds of probes on the exon array. The CNVs were detected by the exon array but were erroneous scored as inherited and were not followed up properly. In retrospect, we believe that the algorithm can be improved to avoid this problem in the future, although the experience points out the need not only to score the presence or the absence of a CNV, but also to correctly identify it as de novo or inherited, since it is usually the case that de novo heterozygous CNVs deserve much greater attention than inherited CNVs in this setting.
Cell culture artifact versus clonal enrichment of pre-existing cells?
Deletions associated with immunoglobulin VDJ recombination at 22q11.22 were observed in 55% of lymphoblast samples (data not shown), but these were not detected by the Illumina arrays which analyzed blood DNA. These events have been reported (16), but they are rarely presented in CNV tabulations. These VDJ recombination events typically appeared to be heterozygous or homozygous and not to be mosaic. These finding suggest that many or most of these lymphoblast cultures are monoclonal or oligoclonal in origin. These VDJ deletions were also detected in many parents (Supplementary Material, Fig. S1). Apart from the immunoglobulin rearrangements, there were three additional validated calls (two rare and one common) that were present in lymphoblast DNA but were not detected in DNA from whole blood (Supplementary Material, Table S7). These may represent the clonal expansion of individual B cells with pre-existing somatic mutations rather than artifacts arising in culture. If so, data on CNVs (and perhaps point mutations as well) present in lymphoblasts but undetected in DNA from fresh blood can provide information on somatic mutation in B cells.
DISCUSSION
This was a small pilot study designed to address the question of whether similar analysis of a larger series of other research cases would be worthwhile. We undertook the study with the expectation that genome-wide exon-focused copy number arrays would detect mutations that would not be detected by some of the arrays now widely used for the analysis of CNVs. Detection of events missed by other arrays might be infrequent, but their small size could identify specific genes of disease relevance. We continue to believe that this is the case, and there is evidence that arrays with exon-by-exon coverage detect disease-causing mutations missed by other array designs (7). Preferential positioning of oligonucleotides in and near exons has the potential to improve the utility of all genomic copy number analysis, whether performed for research or diagnostic purposes. Since the study was initiated, the feasibility of whole-exome or whole-genome sequencing of research samples has increased, but whole-genome exon copy number analysis still offers a more economical method to discover new potential disease gene relationships as occurred in this study for the TMLHE locus. The exon array method has value in detecting inherited CNVs, but analysis of this group was not carried out for this report. Improved designs of exon-focused arrays should continue to be useful for research and for clinical diagnosis. The array used in this study identified a large number of false-positive calls, and this is a draw back. We believe that the number of false-positive calls can be reduced by improved oligonucleotide selection and/or using more oligonucleotides per exon on a single array as is possible with a 4.2 million oligonucleotides per array design that is now available from Nimblegen (Madison, WI, USA). In addition, better algorithms for reducing false-positive calls are likely to be possible with increased experience. With a large number of inherited CNVs, the difficulty in determining if a CNV was inherited or de novo proved surprisingly challenging, and some CNVs initially thought to be de novo were proven to be inherited.
We identified a number of CNVs that most likely are benign, but could have some relevance to autism. Analysis of numerous additional autism and control samples suggests that the 61 bp deletion in SLC38A10 is not associated with autism. Although we doubt that they are relevant to the etiology of autism, similar studies of additional autism and control samples would be needed to assure that the 14q24.3 (ACOT cluster), ZAN, APOBEC3A/APOBEC3B and BTNL3 CNVs are not associated with autism.
From a positive perspective, the concordance for the exon and SNP arrays detecting larger CNVs was excellent. Even a single observation can identify a new disease gene, as was the case for TMLHE in this study. This deletion was not detected as a high-confidence call in one proband (SSC 11000.p1) in common between our exon array approach and the larger study using SNP arrays. Furthermore, the exon array certainly can detect both de novo and inherited small exonic events that are far beyond the resolution of conventional arrays. This is exemplified by the 61 bp deletion in SLC38A10. Another strength of the exon-focused array is the detection of small inherited exonic CNVs. These inherited heterozygous exonic CNVs are not yet fully analyzed and not included in this report. Based on a manual review of five trios, there were ∼74 such inherited autosomal heterozygous CNVs per trio detected by the exon arrays and not detected by the high-confidence SNP array calls. Because of the inherited detection, it is likely that the majority of these heterozygous inherited CNVs are true positive rather than false-positive calls.
We detected numerous CNVs that we refer to as complex. In chromosome region 14q24.3, the ACOT1 and ACOT2 genes are embedded in two sets of paralogous, very Alu-rich segmental duplications, 20 and 22, and 7.5 and 5 kb in size of 95 and 96% overall DNA sequence identity, respectively (Fig. 2A). The log ratios of the oligonucleotide probes in the proximal ACOT1 segmental duplication copies indicate their homozygous deletion together with the unique DNA segment between two segmental duplications, leaving the distal ACOT2 segmental duplication copies intact. The mother's plot with a copy number gain (when compared with the same reference DNA) suggests an apparently non-Mendelian inheritance with de novo deletion(s). However, the 0/0 homozygous deletion pattern in the patient can be simply explained in a Mendelian fashion with, for example, three copy number alleles of ACOT1, 0/3 in the mother, 0/1 in the father and 1/1 in the reference DNA (Fig. 2B).
In the second complex CNV involving the LCE3 gene family in chromosome region 1q21.3 (Fig. 2C), the homozygous deletion pattern (0/0 copy number) in the patient may also suggest an apparently non-Mendelian de novo deletion events on the normal copy number parental chromosomes. However, this pattern can be simply explained by a Mendelian segregation of two alleles, 0/1 in the mother and 0/1 in the father (0/1 in the reference DNA). In this case, the lower ratios of the oligonucleotide probes in the proximal portion of the CNV likely results from the presence of a large set of LINE elements that ‘diluted’ their dynamic range. Similar interpretations of these complex CNVs have been suggested by others using the term ‘Mendelian violator’ (17).
We observed a number of findings that were present in lymphoblasts but absent in blood DNA (Supplementary Material, Table S7). Most of these were related to known immunoglobulin rearrangement, but three of these were novel. These three CNVs were much smaller than those that were previously reported as ‘cell culture artifacts’ (18). For example, we detected two CNVs that encompassed only one gene and were <60 kb in size. In a previous study (18), the smallest CNV that originated from ‘cell culture artifact’ was 6.5 Mb. We saw very prominent examples of this arising through VDJ recombination as reported previously (16). The patterns observed for VDJ deletions had sharp but somewhat variable boundaries and appeared to be homozygous or heterozygous (Supplementary Material, Fig. S1) as opposed to appearing mosaic as would be expected for a polyclonal culture of lymphoblasts. The appearance of the VDJ deletions suggests that cultures showing prominent and homogeneous deletions are monoclonal or oligoclonal at the time of DNA harvest. The majority of samples do not show VDJ deletions which could be due to the cultures being polyclonal or more likely because they are derived from B cells that have not undergone VDJ recombination. These interpretations suggest that some fraction of mutations assumed to have arisen as tissue culture artifacts may represent somatic mutation pre-existing in a B cell that was the origin for a monoclonal or oligoclonal lymphoblast culture. The report that somatic aneuploidy is common in hepatocytes (19) might have some parallel within B cells that ultimately result in monoclonal or oligoclonal lymphoblast cultures. Both gross chromosomal mutations and point mutations might exist as pre-existing somatic events in B cells. Whatever the mechanisms, it is common to see mutations in lymphoblast DNA that are not detected in DNA from mixed leukocytes. Recently, one study found single amino acid changes due to ‘cell culture artifacts’ (20).
In conclusion, this work demonstrates that array CGH using exon-focused arrays can detect small exonic CNVs that are not detected by most arrays in widespread use for diagnostic and research purposes. These arrays detect variation that we describe as ‘complex’ typically involving groups of related genes whose architecture is highly polymorphic. Analysis of VDJ rearrangements indicated that many lymphoblast cultures are monoclonal or oligoclonal, and it is possible that mutations often described as tissue culture artifacts represent clonal expansions of mutations that pre-exist in B cells. Finally, the most important finding in this pilot study was the detection of deletions of exon 2 of the TMLHE gene that led to the discovery of a novel inborn error of metabolism.
MATERIALS AND METHODS
Families
One hundred trios from the SSC were randomly selected from among families enrolled in the first year. The SSC enrolls young simplex cases of autism as described elsewhere (9). A list of all families studied is present in Supplementary Material, Table S1. After these samples were analyzed, 19 families (identified in Supplementary Material, Table S1) were removed from the SSC because they did not meet the full criteria for simplex autism; since this paper focuses on the methods of CNV detection, these cases were not removed. Family 11154 failed our quality control (QC) and was eliminated.
To follow-up on a deletion identified in SLC38A10 by this study, we also analyzed an additional 485 SSC simplex probands, 72 males with multiplex autism and at least one affected brother from the AGRE (http://research.agre.org/, date last accessed on 30 August 2011) and 341 controls from the NIMH Human Genetics Initiative (NIMH-HGI, https://www.nimhgenetics.org/nimh_human_genetics_initiative/, date last accessed on 30 August 2011) (identified in Supplementary Material, Tables S4–S6).
DNA samples
Blood from each individual was used to establish lymphoblastoid cell lines and extract DNA at the Rutgers University Cell and DNA Repository through the SSC. DNA derived from lymphoblast cell culture was used for array CGH; if the lymphoblast DNA failed the DNA digestion step or the array data failed QC, we used DNA from blood. All CNVs described in this paper were validated using whole-blood derived DNA by either array CGH or PCR.
Array design
Array targets included 273 832 exons from 18 579 genes in the RefSeq database (June 2008, hg18). In order to conserve space on the array, 642 genes were removed on the basis that the genes were unlikely to be implicated in a neurodevelopmental disease; these included HLA genes, immunoglobulin genes, T-cell receptor genes, olfactory receptors and collagen genes. After intentionally removing these genes, 197 332 exons that were unique and mapped to annotated regions remained. All 3′- and 5′-UTRs were included in the list of exons. Six probes were selected to locate partially or completely within the exon. If less than three pairs of probes were not available, the target area was extended into the adjacent intron; three of these probes were on the positive strand and three on the negative strand resulting in three pairs of identical coordinates. The three probe pairs were selected to ensure that they were distributed as evenly as possible across each target.
Probes were selected from the Agilent Technologies e-array high density (HD) CGH database. To avoid cross-hybridization, each probe was aligned to the hg18 genome using BLAST; any probe that did not map uniquely was removed except for those in the pseudoautosomal regions on chromosomes X and Y for which two locations were tolerated. Although no probes were used if there was a perfect match elsewhere in the genome, some probes were near-perfect matches and would not distinguish pseudogenes and the parent genes, particularly for unprocessed pseudogenes. Where multiple suitable probes were available, the probes with the highest Agilent HD CGH database probe score where chosen. If the exon length differed between isoforms, the longest possible exon from all isoforms was used to design the six probes.
Sixteen percent of probes were entirely within exons, 56% crossed an intron–exon boundary, 26% were intronic and 2% were intergenic; 3.5% of probes overlapped a second probe on the same strand often in the case of alternative splicing and overlapping exons. A excess of candidate probes were printed on five Agilent 244K arrays for empirical selection based on performance (i.e. noisy probes were eliminated), and 960 000 probes were chosen to print an Agilent 1 M array that was used to generate the data presented in this paper. A final list of genes included, genes excluded and the relative distribution of probes to genes and exons in the whole-genome custom exon array is displayed on our Genboree web resource (http://genboree.org/java-bin/gbrowser.jsp?refSeqId=1868&entryPointId=chr17&from=53496072&to=53694382&isPublic=yes, date last accessed on 30 August 2011).
Agilent 1 M array CGH protocol
Two micrograms of DNA derived from lymphoblast cell culture from 100 trios (300 samples) was hybridized to an Agilent 1 M whole-genome custom exon array for CNV discovery. Two micrograms of DNA derived from blood DNA of a single male was used as the reference throughout. The protocol for DNA digestion, labeling, purification and hybridization to the arrays followed the manufacturers' instructions with minor modifications, as described previously (21). Each slide was scanned into an image file using the Agilent G2565 DNA Microarray Scanner at a 3 µm scan resolution.
Data analysis and CNV prediction
Microarray image files were quantified using the Agilent Feature Extraction software (v10.7.3.1). The data were written to a feature extraction file. Original feature extraction files for the 100 SSC probands and their parents can be found under the Gene Expression Omnibus (GEO) accession number GSE23765 (http://www.ncbi.nlm.nih.gov/geo/, date last accessed on 30 August 2011). For CNV predictions for homozygous and de novo heterozygous calls on autosomes, analysis of oligonucleotide CGH microarray data was performed in three steps. First, a filtering procedure was used to flag low-intensity features; the intensity filter is a mixture model analysis on the combined Cy3 and Cy5 intensity data. Features with a combined Cy3 and Cy5 intensity value of more than 3 SD below the mean of the high-intensity mode were flagged and excluded from further analysis. Next, segmentation of the remaining data was performed using a circular binary segmentation method with post-processing to ensure that regions had at least three genomic coordinate consecutive probes with the same sign of deviation in the log 2 ratio as well as the median log 2 ratio which exceeded 0.2 in absolute value on the log scale. Finally, to compute possible de novo events, we repeated the procedure above for the proband, maternal and paternal samples from each trio. Events were considered possible de novo calls when a segmental event was present in the child but not present in either parent. To refine the score discriminating the copy number values in the parents and proband, the difference in the median oligonucleotide log 2 ratio values were also determined and used as an additional filter. Because the trio-segmentation overlap procedure outlined above can generate both false-positive and false-negative results, we also performed an additional segmentation analysis on the oligonucleotide-level differences between the proband and maternal sample and the proband and paternal sample. In this analysis, the direct difference between proband and maternal sample and the proband and paternal sample at the level of individual oligonucleotides were segmented using circular binary segmentation using the procedure defined above. Candidate de novo events were those where a segmental event appeared in both the differences between the proband and maternal sample and the proband and paternal sample. In this case, the median of the direct differences was used as an additional feature that characterized the candidate de novo calls. The combination of these two sets of candidate de novo events was considered in our subsequent analyses of the data.
For CNV predictions for de novo and hemizygous calls on chromosomes X and Y, analysis was done using Agilent's DNA Analytics software (v4.0.76; Agilent Technologies, Santa Clara, CA, USA) with the following settings: aberration algorithm ADM-2, minimum of five consecutive probes per region and a minimum absolute average log 2 ratio of 0.3 for any given region. Since all individuals were hybridized with a male reference, whenever analyzing female probands and the mothers, we expected their X chromosome to normally show an overall gain at a log 2 ratio of +1.
Comparison with DGV
CNVs were defined as ‘rare’ or ‘common’ by comparison with the DGV (variation.hg18.v9.txt, March.2010, http://projects.tcag.ca/variation/, date last accessed on 30 August 2011). Regions in the DGV with CNVs present in ≥1% of samples were identified. A CNV was considered ‘rare’ if less than 50% of its length overlapped with these common DGV regions. All other CNVs were classed as ‘common’. The entire DGV was used to define rare events. The DGV is composed of many different studies and methodologies to identify CNVs, including aCGH, genotyping arrays, computational assessments, FISH and high-throughput sequencing. It is difficult to formulate unbiased criteria for inclusion or exclusion of individual studies or CNVs. Instead of selecting a ‘high-confidence’ subset of the DGV, we chose to use a frequency based approach so that a specific locus would need to have CNVs from multiple samples to be considered common. This approach also has the advantage of defining rare CNVs by a specific frequency (1% of the population of samples making up the DGV) rather than an arbitrary cut-off.
Comparison to Illumina 1 M data
Genotyping data were available for 90 of the 99 trios passing Agilent QC using the Illumina 1 M SNP array (Supplementary Material, Table S1); 8944 high-confidence CNV predictions (≥91% positive predicative value) were generated from the Illumina data as described previously (8). If an Agilent predicted CNV overlapped with an Illumina high-confidence predicted CNV, it was considered validated.
CNVs that were not present in the Illumina high-confidence CNV list were validated by further CGH arrays or by PCR. If the CNVs predicted in the family were covered adequately by the SurePrint G3 Human CGH Microarray 1 × 1 M (Agilent Technologies; design ID 021529) then this array was used; otherwise custom Agilent 4 × 180 and 8 × 60 K arrays were designed for the CNVs predicted in the remaining families. These arrays were designed to cover the specific CNVs with 30 kb of flanking sequence (design IDs: 025211, 027305, 028249, 028812, https://earray.chem.agilent.com/earray/, date last accessed on 30 August 2011).
Validation Agilent array CGH protocol
The Agilent arrays used for validation were run in the same manner as the initial exon-specific array and using the same single male as a reference. The results were analyzed using Agilent's DNA Analytics software (v4.0.76) with the following settings: aberration algorithm ADM-2, minimum of five consecutive probes per region for the 1 × 1 M array (minimum of three for the 4 × 180 and 8 × 60 K arrays) and a minimum absolute average log 2 ratio of 0.3 for any given region for the 1 × 1 M array (minimum of 0.25 for the 4 × 180 and 8 × 60 K arrays).
PCR and DNA sequencing
PCR was used to validate the TMLHE deletion, followed by sequencing to identify the deletion junction in proband SSC 11000.p1 and in CEPH sample NA12003. PCR was performed using Takara's LA PCR kit (Takara Bio, Inc., Shiga, Japan). Briefly, 50–100 ng of genomic DNA was used in a 25 µl reaction that also contained 0.5 µm primers (Integrated DNA Technologies, Inc., Coralville, IA, USA; Supplementary Material, Table S8), 400 µm of each dNTP, 1.25 units of Takara LA Taq and 10× LA PCR buffer. The PCR was performed with the following reaction conditions: 94°C for 1 min; 35 cycles of 94°C for 30 s and 68°C for 30–60 s/expected kilo-bases of extended DNA; 72°C for 10 min. PCR products were analyzed using agarose gel electrophoresis.
PCR was also used to confirm the SLC38A10 deletion in proband SSC 11089.p1 and to analyze for the presence of the identical deletion in 996 samples from SSC, AGRE and NIMH controls. PCR was performed using Roche's FastStart Taq-DNA Polymerase, dNTPack (Roche Diagnostics, Mannheim, Germany). Briefly, 50–100 ng of genomic DNA was used in a 25 µl reaction that also contained 0.25 µm primers (Integrated DNA Technologies, Inc.; Supplementary Material, Table S8), 200 µm of each dNTP, 1 unit of FastStart Taq-DNA Polymerase and 10× PCR buffer. The PCR was performed with the following reaction conditions: 95°C for 6 min; 40 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 1 min; and final extension at 72°C for 7 min. PCR products were analyzed using agarose gel electrophoresis.
PCR products were sent for nucleotide sequencing by Sanger di-deoxynucleotide sequencing (Macrogen USA, Rockville, MD, USA, and Genewiz, Inc, South Plainfield, NJ, USA) with or without prior purification from agarose gel using the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA).
SUPPLEMENTARY MATERIAL
Conflict of Interest statement. A.L.B. is Chair of the Department of Molecular and Human Genetics at Baylor College of Medicine which offers extensive genetic laboratory testing including the use of arrays for genomic copy number analysis, and the department derives revenue from this activity.
FUNDING
This work was supported by a grant from the Simons Foundation (SFARI 124827 to the investigators of the SSC Genetic Consortium); and grants from the National Institutes of Health (HD-37283 to A.L.B., HG-4009 to A.M.).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all of the families and the clinicians at the participating SFARI Simplex Collection (SSC) sites; the principal investigators of the SSC Genetic Consortium are as follows: A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, E. Wijsman. We appreciate obtaining access to phenotypic data on SFARI Base. Approved researchers can obtain further information about the SSC data presented here by applying at https://base.sfari.org, date last accessed on 30 August 2011. We also thank Robin Goin-Kochel and April Hall for assistance and helpful discussions.
REFERENCES
- 1.Jacquemont M.-L., Sanlaville D., Redon R., Raoul O., Cormier-Daire V., Lyonnet S., Amiel J., Le M.M., Heron D., De Blois M.-C., et al. Array-based comparative genomic hybridization identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J. Med. Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miles J.H., Takahashi T.N., Hong J., Munden N., Flournoy N., Braddock S.R., Martin R.A., Bocian M.E., Spence M.A., Hillman R.E., Farmer J.E. Development and validation of a measure of dysmorphology: useful for autism subgroup classification. Am. J. Med. Genet. A. 2008;146A:1101–1116. doi: 10.1002/ajmg.a.32244. [DOI] [PubMed] [Google Scholar]
- 3.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yamrom B., Yoon S., Krasnitz A., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y., et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott F.J., Baron-Cohen S., Bolton P., Brayne C. Brief report: prevalence of autism spectrum conditions in children aged 5–11 years in Cambridgeshire, UK. Autism. 2002;6:231–237. doi: 10.1177/1362361302006003002. [DOI] [PubMed] [Google Scholar]
- 6.Kalra V., Seth R., Sapra S. Autism—experiences in a tertiary care hospital. Indian J. Pediatr. 2005;72:227–230. [PubMed] [Google Scholar]
- 7.Boone P.M., Bacino C.A., Shaw C.A., Eng P.A., Hixson P.M., Pursley A.N., Kang S.H., Yang Y., Wiszniewska J., Nowakowska B.A., et al. Detection of clinically relevant exonic copy-number changes by array CGH. Hum. Mutat. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A., et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischbach G.D., Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Eichler E.E., Lu F., Shen Y., Antonacci R., Jurecic V., Doggett N.A., Moyzis R.K., Baldini A., Gibbs R.A., Nelson D.L. Duplication of a gene-rich cluster between 16p11.1 and Xq28: a novel pericentromeric-directed mechanism for paralogous genome evolution. Hum. Mol. Genet. 1996;5:899–912. doi: 10.1093/hmg/5.7.899. [DOI] [PubMed] [Google Scholar]
- 11.Arnett H.A., Escobar S.S., Viney J.L. Regulation of costimulation in the era of butyrophilins. Cytokine. 2009;46:370–375. doi: 10.1016/j.cyto.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Bogerd H.P., Wiegand H.L., Doehle B.P., Lueders K.K., Cullen B.R. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abe H., Ochi H., Maekawa T., Hatakeyama T., Tsuge M., Kitamura S., Kimura T., Miki D., Mitsui F., Hiraga N., et al. Effects of structural variations of APOBEC3A and APOBEC3B genes in chronic hepatitis B virus infection. Hepatol. Res. 2009;39:1159–1168. doi: 10.1111/j.1872-034X.2009.00566.x. [DOI] [PubMed] [Google Scholar]
- 14.Zanni G., Saillour Y., Nagara M., Billuart P., Castelnau L., Moraine C., Faivre L., Bertini E., Durr A., Guichet A., et al. Oligophrenin 1 mutations frequently cause X-linked mental retardation with cerebellar hypoplasia. Neurology. 2005;65:1364–1369. doi: 10.1212/01.wnl.0000182813.94713.ee. [DOI] [PubMed] [Google Scholar]
- 15.McCarroll S.A., Hadnott T.N., Perry G.H., Sabeti P.C., Zody M.C., Barrett J.C., Dallaire S., Gabriel S.B., Lee C., Daly M.J., Altshuler D.M. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 16.Craddock N., Hurles M.E., Cardin N., Pearson R.D., Plagnol V., Robson S., Vukcevic D., Barnes C., Conrad D.F., Giannoulatou E., et al. Genome-wide association study of CNVs in 16000 cases of eight common diseases and 3000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy D., Ronemus M., Yamrom B., Lee Y., Leotta A., Kendall J., Marks S., Lashmi B., Ye K., Buja A., et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan A.W., Taylor M.H., Hickey R.D., Hanlon Newell A.E., Lenzi M.L., Olson S.B., Finegold M.J., Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Awadalla P., Gauthier J., Myers R.A., Casals F., Hamdan F.F., Griffing A.R., Cote M., Henrion E., Spiegelman D., Tarabeux J., et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am. J. Hum. Genet. 2010;87:316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou Z., Kang S.H., Shaw C.A., Carmack C.E., White L.D., Patel A., Beaudet A.L., Cheung S.W., Chinault A.C. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet. Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.