Abstract
Some patients with pharmacoresistant epilepsy undergo therapeutic resection of the epileptic focus. At least 12 large-scale microarray studies on brain tissue from epilepsy surgery have been published over the last 10 years, but they have failed to make a significant impact upon our understanding of pharmacoresistance, because (1) doubts have been raised about their reproducibility, (2) only a small number of the gene expression changes found in each microarray study have been independently validated and (3) the results of different studies have not been integrated to give a coherent picture of the genetic changes involved in epilepsy pharmacoresistance. To overcome these limitations, we (1) assessed the reproducibility of the microarray studies by calculating the overlap between lists of differentially regulated genes from pairs of microarray studies and determining if this was greater than would be expected by chance alone, (2) used an inter-study cross-validation technique to simultaneously verify the expression changes of large numbers of genes and (3) used the combined results of the different microarray studies to perform an integrative analysis based on enriched gene ontology terms, networks and pathways. Using this approach, we respectively (1) demonstrate that there are statistically significant overlaps between the gene expression changes in different publications, (2) verify the differential expression of 233 genes and (3) identify the biological processes, networks and genes likely to be most important in the development of pharmacoresistant epilepsy. Our analysis provides novel biologically plausible candidate genes and pathways which warrant further investigation to assess their causal relevance.
INTRODUCTION
Epilepsies are among the most common neurological disorders, affecting up to 1% of the population (1). Most patients with epilepsy become seizure-free with antiepileptic drug (AED) therapy. However, ∼30% of epilepsy patients have a medically intractable condition even if treated with various AEDs at maximal dosages either alone or in combination (2). A subset of patients with intractable epilepsy has the potential for a surgical cure, most commonly those with hippocampal sclerosis (HS) (3). Surgical samples from patients with intractable epilepsy provide a unique opportunity to directly analyse the human epileptic focus in order to determine the causes of pharmacoresistance. Many different causes of pharmacoresistance have been postulated, e.g. dysregulation of multidrug transporters (MDTs) (4), reduced drug-target sensitivity (4), increased neuronal apoptosis (5), cytoskeletal alterations (6) and reorganization of neuronal networks (7). The exact molecular mechanisms underlying pharmacoresistance, however, are still poorly understood. Alterations in the expression of a large number of genes are thought to be responsible, but most of the numerous genes that participate in the development of pharmacoresistance in epilepsy remain unidentified.
To date, the vast majority of studies on samples from epilepsy surgery have focused primarily on a number of selected candidate genes. However, these techniques are not suitable for dissection of multiple interacting molecular pathways or screening potential molecular abnormalities when the list of candidate genes is extensive. In contrast, large-scale microarray studies offer the advantage of assaying gene expression in a comprehensive, unbiased and genome-wide fashion. Analysing the expression profile of many genes simultaneously in large-scale expression studies, without making prior assumptions about candidate genes, allows the identification of new genes and molecular pathways associated with the condition. There have been at least 12 published large-scale gene expression studies on tissue from epilepsy surgery in the last 10 years (see below). However, they have failed to make a significant impact on our understanding of the causes of pharmacoresistance. In our view, this failure is due to several reasons.
First, authors of most microarray studies have focused on selected genes or pathways in their published results. Large-scale gene profiling studies suffer from large numbers of false-positive results. Before drawing conclusions about the differential expression of a specific gene, it is thought necessary to demonstrate independent experimental validation (8) using techniques such as reverse transcription–polymerase chain reaction. Therefore, it is commonplace to use the microarray as a screening tool, then to validate a few chosen genes for additional investigation. However, this methodology under uses the ‘depth’ inherent in the original microarray data set and is prone to missing potentially important genes and networks. Hence, experts (9) have proposed using multiple microarray data sets that address similar hypotheses to simultaneously cross-validate all of the positive results. This inter-study cross-validation approach has been adopted in the present work.
Second, doubts have been raised about the reproducibility of microarray studies in epilepsy (10–12). It has been suggested that the majority of changes in gene expression are specific to laboratory or experimental conditions with very few genes demonstrating changes in more than two publications (12).
Third, the results of different microarray studies have not been integrated to give a coherent picture of the genomic changes involved in epilepsy pharmacoresistance.
We believe that the above problems can be overcome by testing the validity and reproducibility of the individual microarray studies and by performing an integrative analysis of all available microarray results. We are aware of only two previous published reviews which have included microarray studies on non-cancerous brain tissue from epilepsy surgery (10,12). However, neither of these reviews included all currently available microarray studies on brain tissue from epilepsy surgery; in fact, the study by Lukasiuk and Pitkanen (10) includes only one human microarray study, and the study by Wang et al. (12) includes only three human large-scale gene expression studies. These reviews found a very small number of genes that showed similar expression profiles and, hence, found it difficult to draw robust conclusions about the possible mechanism(s) of pharmacoresistance in epilepsy. In the present work, we have performed an integrative analysis of all available microarray studies on non-cancerous brain tissue from epilepsy surgery with the aim of identifying novel and important genes and networks that play a role in pharmacoresistance.
RESULTS
Gene lists
We found 12 published large-scale genome-wide gene expression profiling studies on non-neoplastic tissue from epilepsy surgery (11,13–20). Gene lists were available for nine genome-wide gene expression studies, and these were included in the present review. Seven studies published one differentially regulated gene list each. However, Lee et al. (13) and van Gassen et al. (11) used more than one type of control tissue and published more than one list of differentially regulated genes each. Lee et al. (13) used two types of controls: (1) histologically normal CA1 from patients with ‘paradoxical temporal lobe epilepsy (PTLE)’, citing the evidence that the poor seizure-free outcome (44%) in PTLE following hippocampectomy suggests that the hippocampus is unlikely to be epileptogenic in this group, and (2) histologically normal CA1 tissue from ‘mass-associated temporal lobe epilepsy (MaTLE)’ where the mass lesion was outside the hippocampus, citing the evidence that mesial temporal lobe epilepsy (MTLE) granule cells are hyperexcitable, while those in MaTLE are not. The authors presented two lists of differentially regulated genes: (1) genes regulated by 1.5-fold or more for both MTLE versus PTLE and MTLE versus MaTLE, and (2) genes regulated 1.5-fold or more when MaTLE and PTLE were considered replicates of non-sclerotic hippocampi. Regulated genes from both lists were integrated into the current systematic review, giving a list of 758 unique genes that could be mapped to Entrez gene numbers. Similarly, van Gassen et al. (11) compared slices of sclerosed hippocampus with histologically normal hippocampal slices from patients with MTLE but no HS. However, van Gassen et al. (11) also compared normal hippocampal autopsy samples with histologically normal hippocampal slices from patients with MTLE but no HS. The non-HS MTLE and HS groups were found to share a large group of differentially expressed genes when compared with the autopsy group, and there was a significant overlap between functional gene classes affected in non-HS MTLE and HS groups when compared with autopsy samples. Hence, the autopsy versus non-HS MTLE gene list was also deemed appropriate for inclusion in the current systematic review. In total, van Gassen et al. (11) presented three differentially regulated gene lists: (1) autopsy versus HS, (2) autopsy versus non-HS and (3) non-HS versus HS. Regulated genes from all three lists were integrated into the current systematic review. Twenty-four genes had conflicting changes in expression (down-regulated in one list but up-regulated in another) and were excluded, leaving 521 unique genes.
Study characteristics
The nine included studies had sample sizes ranging from 6 to 60. All studies used test tissue from the temporal lobe, and all but three studies (14,16,20) used test tissue from the hippocampus. Gene expression profiling platforms used included one-channel microarrays, two-channel microarrays and serial analysis of gene expression (Table 1). The smallest microarray chip used was able to assay up to 588 unique genes [Clontech's atlas human neurobiology array used by Becker et al. (19)], while the largest was able to assay up to 21,329 unique genes [Operon's Human Array-Ready oligo set version 2.0 used by van Gassen et al. (11)].
Table 1.
Included large-scale gene expression profiling studies on brain tissue from epilepsy surgery
| Name | Test tissue | Control tissue | Chip | Number of regulated genes |
|---|---|---|---|---|
| Lee et al. (13) | Sclerosed CA1 from MTLE (n = 8) | Histologically normal CA1 from MaTLE (n = 6) or PTLE (n = 6) | Affymetrix U133A | 674 (MTLE versus PTLE and MTLE versus MaTLE); 947 (MTLE versus others) |
| van Gassen et al. (11) | Sclerosed hippocampus from MTLE patients (n = 4) | Hippocampus from MTLE patients without HS (n = 4) and autopsy (n = 4) | Human Array-Ready oligo set version 2.0, Operon Biotechnologies | Autopsy versus non-HS: 322; Autopsy versus HS: 322; non-HS versus HS: 206 |
| Xi et al. (14) | Anterior temporal neocortex epileptogenic zone (n = 40) | Anterior temporal neocortex removed for raised ICP (n = 20) | n.s. | 143 |
| Ozbas-Gerceker et al. (15) | Anterior hippocampus (n = 6) | Anterior hippocampus tissue from autopsy (n = 1) | SAGE | 143 |
| Xiao et al. (16) | Temporal lobe (n = 40): HS, FCD etc. | Temporal neocortex, hippocampus, parietal cortex or frontal cortex from brain trauma patients (n = 20) | Biostar H-40s | 142 |
| Arion et al. (17) | ‘Spiking areas’ from anterolateral temporal cortical samples (n = 6) | ‘Non-spiking’ from anterolateral temporal cortical samples (n = 6) | Affymetrix U133A | 76 |
| Becker et al. (19) | Sclerosed CA1 from MTLE (n = 5) | Dentate gyrus from same patients (n = 5) | Affymetrix U133A | 25 |
| Becker et al. (19) | Sclerosed hippocampus (n = 3) | Normal hippocampus from tumour patients (n = 2) or epilepsy patients (n = 1) | Atlas, Clontech | 21 |
| Jamali et al. (20) | Entorhinal cortex (n = 5) | Lateral temporal neocortex (n = 5) | Micromax | 16 |
FCD, focal cortical dysplasia; HS, hippocampal sclerosis; ICP, intracranial pressure; IE, intractable epilepsy; MaTLE, mass-associated temporal lobe epilepsy; MTLE, mesial temporal lobe epilepsy; n.s., not specified; PTLE, paradoxical temporal lobe epilepsy; SAGE, serial analysis of gene expression.
We assessed the methodological quality of the included studies. One limitation applies to all the studies: no power calculations were performed to determine the appropriate sample size. In addition, each study had specific methodological strengths and weaknesses. For example, van Gassen et al. (11) performed a like-for-like tissue comparison, detailed probe- and array-level quality-control procedures, corrected P-values for multiple testing and deposited Minimum Information About a Microarray Experiment (MIAME)-compliant data in a publically accessible repository, but used a modest sample size and applied no fold-change criterion to differentially regulated gene lists. On the other hand, Xiao et al. (16) used a large sample size and applied appropriate fold-change criterion, but did not perform a like-for-like tissue comparison, or detail probe- and array-level quality-control procedures, or perform correction for multiple testing, or deposit their data in a public database. Table 2 lists the criteria used to assess methodological quality and the quality assessment outcomes for the individual studies.
Table 2.
Critical appraisal of included studies
| Study | Sample size | Like-for-like test and control tissues used | Data QC procedures detailed | Correction for multiple testing | Effect size criterion applied | Raw data provided |
|---|---|---|---|---|---|---|
| Lee et al. (13) | 20 | ✓ | ✗ | FDR < 0.05 | Fold change ≥ 1.5 | ✗ |
| van Gassen et al. (11) | 12 | ✓ | ✓ | Family-wise error correction < 0.05 | ✗ | ✓ |
| Arion et al. (17) | 12 | ✓ | ✓ | FDR = 0.057 | Fold change ≥ 1.2 | ✗ |
| Jamali et al. (20) | 10 | ✗a | ✓ | n.a.a | Fold change ≥ 2.56 | ✗ |
| Xi et al. (14) | 60 | ✓ | ✗ | ✗b | Fold change ≥ 2 | ✗ |
| Ozbas-Gerceker et al. (15) | 7 | ✓c | ✗ | ✗ | ✗ | ✓ |
| Xiao et al. (16) | 60 | ✗ | ✗ | ✗ | Fold change ≥ 2 | ✗ |
| Becker et al. (18) | 10 | ✗ | ✓ | ✗ | Fold change ≥ 1.5 | ✗ |
| Becker et al. (19) | 6 | ✓ | ✗ | ✗b | ✗b | ✗ |
aIn the study by Jamali et al. (20), tissue from the entorhinal cortex (test) was compared with tissue from the lateral temporal neocortex (control). However, genes were only considered regulated in the epileptic focus if quantitative reverse transcription–polymerase chain reaction confirmed no significant difference in expression between the entorhinal cortex and the lateral temporal neocortex of non-epileptic autopsy brain samples. The statistical test employed was the ‘z-score’.
bDid not specify what, if any, statistical test used.
cAutopsy control used.
Study heterogeneity
There was heterogeneity within the studies and the number of genes shown to be regulated in the individual studies varied widely (Table 1). The largest parts of this variation can be explained by:
the number of genes that can be assayed by the different arrays (see above) and
the widely differing criteria used to define statistically significant change in gene expression (Table 2).
Other sources of heterogeneity were:
differences in sample handling and RNA extraction techniques;
different gene expression profiling platforms used;
varying microarray data normalization procedures;
test tissue used: sclerosed hippocampus was used by most studies, but other anatomical regions and other pathologies were also employed by some studies (Table 1) and
the use of at least four different types of control tissue: (1) non-epileptic brain tissue removed from the same patients as part of the surgical procedure, (2) normal hippocampal tissue from autopsy, (3) normal brain tissue removed from other patients for various indications and (4) histologically normal hippocampal tissue from patients with MTLE.
While acknowledging the heterogeneity within the studies, it is important to note that there is also an underlying commonality in the design of the studies: comparing gene expression between pharmacoresistant epileptic brain samples and non-epileptic or pharmacoresponsive brain samples. Although some of the differentially expressed genes identified in individual studies will be specific to the histological characteristics of the tissues being compared and to laboratory conditions, a significant subset of genes in each list represents the universal genetic changes linked with pharmacoresistance. As validation of this concept, we determined the size of the overlap between pairs of gene lists and calculated if this size was greater than would be expected by chance alone (see the Data validation section below).
Data validation
The overlapping probabilities of differentially regulated gene lists were calculated—all the gene lists from the included studies were tested in pairs. The size and the statistical significance of the overlaps are shown in Supplementary Material, Table S1. Our results show that there was a statistically significant overlap between gene lists from studies done on different types of brain tissue using different array platforms (elaborated further below).
To test the validity of the list of genes differentially regulated in only one study, we compared this with a list of all human genes and human homologues of mouse and rat genes from CarpeDB (www.carpedb.ua.edu), a dynamic continuously updated epilepsy genetics database. Of the 278 genes thus extracted from CarpeDB, 54 overlapped with our list of genes differentially regulated in only one study. This overlap was statistically significant [false discovery rate (FDR) = 1.2 × 10−24].
Overlapping gene lists
Genes differentially expressed in three or four studies are listed in Table 3. Prominently represented in these two lists are genes involved in neuroinflammation, in the control of synaptic transmission and in the restructuring of neuronal networks—specific examples are discussed below (see Discussion section). Genes differentially regulated in one study or two studies are shown in Supplementary Material, Table S2.
Table 3.
Genes differentially expressed in three or four studies
| Symbol | Description | Molecular function | Up- or down-regulated | Number of studies |
|---|---|---|---|---|
| GABRA5 | γ-aminobutyric acid-A receptor, alpha 5 | GABA-A receptor activity | Down | 4 |
| NRGN | Neurogranin | Calmodulin binding | Down | 4 |
| CCL2 | Chemokine (C-C motif) ligand 2 | CCR2 chemokine receptor binding | Up | 4 |
| GFAP | Glial fibrillary acidic protein | Protein binding | Up | 4 |
| CPLX2 | Complexin 2 | Syntaxin binding | Down | 3 |
| ENC1 | Ectodermal-neural cortex | Actin binding | Down | 3 |
| HPCAL4 | Hippocalcin like 4 | Calcium channel regulator activity | Down | 3 |
| INHBA | Inhibin, beta A | Cytokine activity | Down | 3 |
| PLCB1 | Phospholipase C, beta 1 | Calcium ion binding | Down | 3 |
| PSD | Pleckstrin and Sec7 domain containing | ARF guanyl-nucleotide exchange factor activity | Down | 3 |
| SNAP25 | Synaptosomal-associated protein, 25 kDa | SNARE binding | Down | 3 |
| STMN2 | Stathmin-like 2 | Protein binding | Down | 3 |
| CAPN3 | Calpain 3 (p94) | Calcium ion binding | Up | 3 |
| CD99 | CD99 molecule | Protein binding | Up | 3 |
| CDK2AP1 | CDK2-associated protein 1 | DNA binding | Up | 3 |
| DYNLT1 | Dynein, light chain, Tctex-type 1 | Motor activity | Up | 3 |
| OGG1 | 8-oxoguanine DNA glycosylase | Damaged DNA binding | Up | 3 |
| PABPC4 | Poly(A) binding protein, cytoplasmic 4 | RNA binding | Up | 3 |
| RDX | Radixin | Actin binding | Up | 3 |
| SPARC | Secreted protein, acidic, cysteine-rich | Calcium ion binding | Up | 3 |
| TF | Transferrin | Ferric iron binding | Up | 3 |
| ZFP36L1 | Zinc finger protein 36, C3H type-like 1 | AU-rich element binding | Up | 3 |
Gene ontology enrichment analysis
The top 25 significantly enriched gene ontology (GO) terms are listed in Table 4. The cellular component ontology-term enrichment suggests that the most important processes in the development of pharmacoresistance are occurring in neuronal projections, in the growth cone, at the pre- and post-synaptic terminal, in the cytoskeleton and in membrane-bound vesicles. The most important molecular functions are calcium transport and signalling, cytoskeletal function and transporter activity. The most significant biological processes are synaptic transmission and synaptic plasticity, regulation of the action potential, cellular cation homeostasis, axonal and dendritic morphogenesis and cytoskeletal organization.
Table 4.
GO-term enrichment
| Molecular function | Biological process | Cellular component |
|---|---|---|
| Protein binding | Transmission of nerve impulse | Neuron projection |
| Binding | Synaptic transmission | Synapse |
| Calmodulin binding | Cell projection organization | Vesicle |
| Cytoskeletal protein binding | Neuron development | Cytoplasmic vesicle |
| Kinase activity | Cell–cell signalling | Cell fraction |
| Protein kinase activity | Behaviour | Cytosol |
| Actin binding | Neuron differentiation | Cell projection |
| Nucleotide binding | Neuron projection development | Vesicle membrane |
| Protein serine/threonine kinase activity | Learning or memory | Membrane-bounded vesicle |
| Ribonucleotide binding | Cell projection morphogenesis | Plasma membrane part |
| Purine ribonucleotide binding | Response to organic substance | Axon |
| Transferase activity, transferring phosphorus-containing groups | Cell morphogenesis | Cytoplasmic membrane-bounded vesicle |
| Phosphotransferase activity, alcohol group as acceptor | Cellular component morphogenesis | Synapse part |
| Purine nucleotide binding | Cell part morphogenesis | Cell soma |
| Structural molecule activity | Cell motion | Insoluble fraction |
| Transporter activity | Neuron projection morphogenesis | Cytoplasmic vesicle part |
| Lipid binding | Regulation of synaptic plasticity | Cytoplasmic vesicle membrane |
| Adenyl ribonucleotide binding | Regulation of neuron projection development | Membrane fraction |
| Cation-transporting ATPase activity | Response to endogenous stimulus | Coated vesicle |
| Protein complex binding | Regulation of cell projection organization | Clathrin-coated vesicle |
| ATP binding | Intracellular signalling cascade | Plasma membrane |
| Protein tyrosine kinase activity | Glial cell development | Site of polarized growth |
| Di-, tri-valent inorganic cation transmembrane transporter activity | Cellular ion homeostasis | Synaptic vesicle |
| #24 not significant | Regulation of cell death | Clathrin-coated vesicle membrane |
| #25 not significant | Regulation of axonogenesis | Growth cone |
Pathway analysis
The top 25 significantly enriched canonical pathways and networks are shown in Table 5. Details of all enriched pathways and networks are provided in Supplementary Material, Table S3. An explanation of how some of the most significant pathways and networks might contribute to the development of pharmacoresistance is presented in the Discussion section.
Table 5.
Top 25 canonical pathways
| Canonical pathways |
|---|
| CCK/gastrin-mediated signalling |
| Synaptic long-term potentiation |
| Semaphorin signalling in neurons |
| Neuropathic pain signalling in dorsal horn neurons |
| Chemokine signalling |
| GNRH signalling |
| RhoA signalling |
| Renin-angiotensin signalling |
| NFAT in cardiac hypertrophy |
| Glutamate receptor signalling |
| Axonal guidance signalling |
| CDK5 signalling |
| Integrin signalling |
| CXCR4 signalling |
| Ephrin receptor signalling |
| Molecular mechanisms of cancer |
| Melatonin signalling |
| Thrombin signalling |
| CREB signalling in neurons |
| Protein kinase A signalling |
| IGF-1 signalling |
| Regulation of actin-based motility by rho |
| Calcium signalling |
| ERK/MAPK signalling |
| Corticotropin-releasing hormone signalling |
CREB, cAMP response element-binding; ERK, extracellular receptor kinase; GNRH, gonadotropin-releasing hormone; IGF-1, insulin-like growth factor 1; MAPK, mitogen-activated protein kinase; NFAT, nuclear factor of activated T-cells.
Comparison with an Alzheimer's disease brain microarray data set
To counter arguments that the genetic changes highlighted by this analysis are non-specific and could be found in any brain pathology, we compared our results with the results of a microarray study on Alzheimer's disease (AD)-affected hippocampi. From the AD study, we extracted 3738 unique genes which could be mapped to Entrez IDs. Ingenuity Systems Pathway Analysis was used to determine significantly enriched canonical pathways from this gene list. In Table 6, we compare the top 10 canonical pathways from the AD and the epilepsy data sets which convincingly demonstrated that there was no overlap between the identified top canonical pathways for the two data sets. In addition, the processes performed by the canonical pathways were also very different. For example, the top five canonical pathways in the pharmacoresistant epilepsy data set are related to restructuring of neuronal networks [cholecystokinin (CCK) signalling], modulation of synaptic transmission (synaptic long-term potentiation, neuropathic pain signalling) and neuroinflammation (semaphorin signalling, chemokine signalling), while the top five pathways in the AD data set are related to protein degradation (protein ubiquitination pathway), energy metabolism (mitochondrial dysfunction, oxidative phosphorylation) and translational regulation (regulation of eIF4 and p70S6K signalling, eIF2 signalling).
Table 6.
Top 10 canonical pathways
| Pharmacoresistant epilepsy | Alzheimer's disease |
|---|---|
| CCK/gastrin-mediated signalling | Protein ubiquitination pathway |
| Synaptic long-term potentiation | Mitochondrial dysfunction |
| Semaphorin signalling in neurons | Regulation of eIF4 and p70S6K signalling |
| Neuropathic pain signalling in dorsal horn neurons | eIF2 signalling |
| Chemokine signalling | Oxidative phosphorylation |
| GNRH signalling | Purine metabolism |
| RhoA signalling | Breast cancer regulation by stathmin 1 |
| Renin-angiotensin signalling | NRF2-mediated oxidative stress response |
| Role of NFAT in cardiac hypertrophy | Huntington's disease signalling |
| Glutamate receptor signalling | Oestrogen receptor signalling |
DISCUSSION
Study heterogeneity and validation
While there is heterogeneity within the studies included in this review, there is also an underlying commonality in their design: comparing the gene expression between pharmacoresistant epileptic brain samples and non-epileptic or pharmacoresponsive brain samples. Although some of the differentially expressed genes identified in individual studies will be specific to the histological characteristics of the tissues being compared and to laboratory conditions, a significant subset of genes in each list represents the universal genetic changes linked with pharmacoresistance. As validation of this concept, our pairwise comparisons showed that there was a statistically significant overlap between gene lists from studies done on different types of brain tissue using different types of array technologies. For example, a study comparing various epileptic focus pathologies (focal cortical dysplasia, temporal lobe malacia, hippocampus sclerosis, neuron loss, neuron degeneration, gliosis, astrocytosis) with normal tissue from various brain regions of brain trauma patients using a Biostar H-40s microarray (16) had a statistically significant overlap (FDR = 1.2 × 10−2) with a study comparing sclerosed CA1 with histologically normal CA1 from MTLE patients using an Affymetrix U133A microarray (13).
In addition, a significant number of the 1174 genes which were shown to be differentially expressed in only one study (a ‘non-corroborated gene set’) are also likely to be relevant to epileptogenesis and pharmacoresistance. To demonstrate this, we compared the non-corroborated gene set with CarpeDB, a dynamic continuously updated epilepsy genetics database. Of the 278 genes extracted from CarpeDB, 54 genes overlap with our non-corroborated gene set. This overlap is highly significant (FDR = 1.2 × 10−24). This suggests that the non-corroborated gene list is also an important data source which should be appropriately studied and interpreted (see below).
It is of course possible that some of the gene expression changes identified in this analysis are the consequence, rather than the cause, of refractory seizures; this limitation is inherent in large-scale gene expression profiling studies and, hence, also in this analysis. The causative role of candidate genes identified as potentially playing a part in the development of pharmacoresistance will need to be established with further investigation, such as electrophysiological studies in hippocampal neuronal cultures or studies in rodent epilepsy models where the expression of the relevant gene has been appropriately altered. Another limitation which applies to all transcriptomic studies, and also to this analysis, is that mRNA concentration may not be a predictor of protein abundance or activity because of the many processes downstream of mRNA synthesis (e.g. alternative mRNA splicing and micro-RNA regulation) that may affect the total amount and activity of protein in tissue. It should also be noted that we have chosen not to include in the present work gene expression profiling studies on animal models of epilepsy as we are not aware of any microarray studies done on models of intractable epilepsy, such as the pharmacoresistant amygdale-kindled rat (21). Hence, the available gene expression profiling studies on animal models of epilepsy do not reflect the phenotype of interest.
Data integration
Microarray technology has become an important tool for biological research, but it can suffer from both low sensitivity and high false-positive rates (9). One way of overcoming these limitations is to use meta-analysis methods that integrate the results of separate microarray studies. Meta-analysis is a classical statistical methodology for combining results from different studies addressing the same scientific questions and has recently been applied to the analysis of microarray data, increasing both the sensitivity and the reliability of measurements of gene expression changes (9). Two basic meta-analysis methods have been applied to microarray studies: (1) combining P-values for each gene from the individual studies to estimate an overall P-value for each gene across all studies (9), and (2) integrating effect size estimates to obtain an overall estimate of the average effect size (22). However, the majority of studies included in this review did not provide the basic information that would allow either meta-analysis method to be applied. This being the case; we integrated the gene lists provided by each study to generate lists of genes consistently regulated in two, three or four studies. This integrative technique successfully improved the reliability of the regulated gene lists, without enhancement of the sensitivity. Hence, the 233 genes shown in this review to overlap between two or more studies are a ‘corroborated gene set’ unlikely to be specific to the histological characteristics of the tissues being compared or to laboratory conditions, and most likely to represent universal genetic changes linked with epileptogenesis and pharmacoresistance. For GO-term enrichment analysis, canonical pathway analysis and network analysis, we included both the corroborated and the non-corroborated gene sets. This is because a significant number of the non-corroborated genes are also likely to be relevant to pharmacoresistance, as demonstrated by the significant overlap with the CarpeDB database. Further, in terms of genes shown to be regulated in single studies, we took the view that different genes validate each other if they are significantly enriched together in a GO-term category, canonical pathway or an interaction network.
What does this analysis tell us about the mechanism of pharmacoresistance?
Current theories on pharmacoresistance in epilepsy include the MDT hypothesis, the drug target hypothesis and the inherent severity hypothesis (23). It has recently been suggested that these processes are likely to act not in a mutually exclusive manner, but rather an integrated manner, to cause pharmacoresistance (23). The current analysis supports this view, suggesting that all three hypotheses may be important. However, much of the integrated evidence generated by this analysis is weighted towards the intrinsic severity hypothesis.
MDT hypothesis
According to the MDT hypothesis, pharmacoresistance results from impaired drug penetration into the epileptic focus secondary to dysregulation of drug transporters (24). Studies on the role of MDTs in pharmacoresistant epilepsy have, hence far, been limited to a small number of selected adenosine triphosphate-binding cassette (ABC) transporters, with a predominant focus on ABCB1 (25). Five ABC transporters were found to be up-regulated in the studies included in this analysis: ABCA6, ABCA8, ABCC4, ABCA2 and ABCB6. Of these five transporters, only ABCC4 has previously been studied in pharmacoresistant epilepsy (26,27). The identification in this study of hitherto-overlooked MDTs suggests that the search for drug transporters relevant to epilepsy pharmacoresistance needs to be expanded beyond the selected few candidates which have been studied so far. It is interesting to note that ABCB1, which has been the focus of the majority of research on the role of MDTs in epilepsy pharmacoresistance (28), was not shown to be regulated in any of the included studies, even though a probe for ABCB1 was present on the arrays of at least four of the nine studies. The role non-ABC drug transporters may play in epilepsy pharmacoresistance is also much neglected. For example, the solute carrier (SLC) group of membrane transporters has been shown to play a role in cancer pharmacoresistance (29), but their potential role in epilepsy pharmacoresistance has not been studied. Twenty-seven different SLC transporters were shown to be dysregulated in the studies included in this analysis.
Drug target hypothesis
According to the drug target hypothesis, pharmacoresistance is caused by the modification of one or more drug target molecules.
An important AED target is the voltage-gated sodium channel. These channels consist of α- and β-subunits. To date, nine α-subunits and four β-subunits have been identified. It has been suggested that in pharmacoresistant epilepsy subunit composition of these channels is altered, such that the expression of AED-insensitive subunit combinations is promoted. The SCN1A subunit gene has been the focus of most research so far (30). This analysis showed that the expression of SCN2A, SCN3A, SCN3B and SCN8A was also altered in pharmacoresistant epilepsy. SCN2A was shown to be down-regulated in two different microarray studies included in this analysis. In support of this finding, SCN2A was also found to be down-regulated in epileptic foci in a radioactive in situ hybridisation study (31). In addition, two different SCN2A polymorphisms have been reported to be associated with epilepsy pharmacoresistance (32,33). The role of SCN2A, and the other subunits identified in this analysis, should be confirmed in future studies.
Another important AED target is the γ-aminobutyric acid-A (GABAA) receptor (34). The pharmacological properties of GABAA receptors depend on the combination of subunits of which there are more than 20 to choose from (35). Three GABAA receptor subunit genes were shown to be down-regulated in the studies included in this analysis: GABRA5, GABRB3 and GABRG2. GABRA5 was down-regulated in four of the included studies. Using immunohistochemical labelling, Bethmann et al. (36) showed that GABRA5 subunit protein expression in the hippocampi of AED resistant rats was significantly lower than in responsive rats. Bonin et al. (37) have shown that the depolarizing current required to generate an action potential was 2-fold greater in neurons from wild-type than from GABRA5 knockout mice. Phenytoin (38) and carbamazepine (39) have been shown to up-regulate GABRA5, and this has been suggested to be involved in their antiepileptic mechanism. GABRA5-selective inverse agonists have been shown to have convulsant or proconvulsant effect in mice in a dose-dependent manner and proportional to the extent of GABRA5 efficacy (40). The current analysis reveals strong evidence of down-regulation of GABRA5 in human epileptic tissue. Further studies are needed to look for associations between polymorphisms in the GABRA5 gene and pharmacoresistance and for ways to reverse the down-regulation of GABRA5.
Intrinsic severity hypothesis
The observation that a high frequency of seizures prior to onset of treatment is a prognostic signal of increased severity and future drug failure suggests that common neurobiological factors may underlie both disease severity and pharmacoresistance (23). A number of processes are thought to contribute to the development of pharmacoresistant epilepsy through promoting severity: neuroinflammation, enduring increases in excitatory synaptic transmission, changes in GABAergic inhibition, neuronal cell death and the development of aberrant innervation patterns in part arising from reactive axonal growth (41). Although this review lends supports to all these processes playing a part, three basic themes were most prominently overrepresented in the gene list assembled:
neuroinflammation,
modulation of synaptic transmission and
restructuring of neuronal networks.
It is worth noting that although the important processes highlighted by this analysis may appear somewhat non-specific, the pattern of genetic changes demonstrated is distinctly different from those seen in other common brain pathologies, as shown by our comparison with an AD data set.
What follows is a brief summary of the aforementioned three themes, as represented by our analysis. From the themes, we highlight novel and important genes and pathways. This is not a comprehensive précis, and we invite readers to peruse the gene lists and enriched GO terms, pathways and networks to find other genes, pathways and networks of interest.
Neuroinflammation
It is being increasingly recognized that inflammation plays an important role in the pathogenesis of epilepsy (42). The role of chemokine- and semaphorin-induced inflammation in the development of refractory epilepsy is little studied (43). Our analysis suggests that chemokine and semaphorin signalling plays a significant role in epilepsy pharmacoresistance. We have also identified particular chemokines and semaphorins that are potentially important therapeutic targets.
Chemokines
The chemokine signalling canonical pathway was shown to be dysregulated in our analysis, and the chemokine CCL2 and chemokine target inhibin β-A were prominent in our gene list. Chemokines can induce neuronal hypersynchronization and neuronal epileptiform activity (44–46), leading to seizure generation in animal models of epilepsy (47). CCL2 up-regulation has been linked to increased susceptibility to seizures in a ‘two-hit’ seizure model in rats (48), and valproic acid has been shown to down-regulate CCL2 mRNA in rat brain (49). Inhibin β-A is a member of the transforming growth factor-β superfamily (50) and may mediate neuroprotective actions of basic fibroblast growth factor (51). Zhang et al. (52) have identified inhibin β-A as one of a set of neuroprotective genes, termed activity-regulated inhibitor of death (AID) genes, which promote the survival of hippocampal neurons after growth factor withdrawal or staurosporine treatment in vitro and after kainic acid-induced status epilepticus in vivo.
In addition, the canonical pathway of chemokine receptor CXCR4 was dysregulated. Increased expression of CXCR4 allows for increased CXCL12 binding. CXCL12 induces a slow inward current followed by a spontaneous synaptic activity via ionotropic glutamatergic receptors (53) and induces microglia to release tumour necrosis factor-α, which potentiates prostaglandin-dependent Ca2+ activation and glutamate release (13). Other dysregulated chemokines in our analysis were CCL3, CCL4, CCL28, CCL3L1, CX3CL1 and CXCL14.
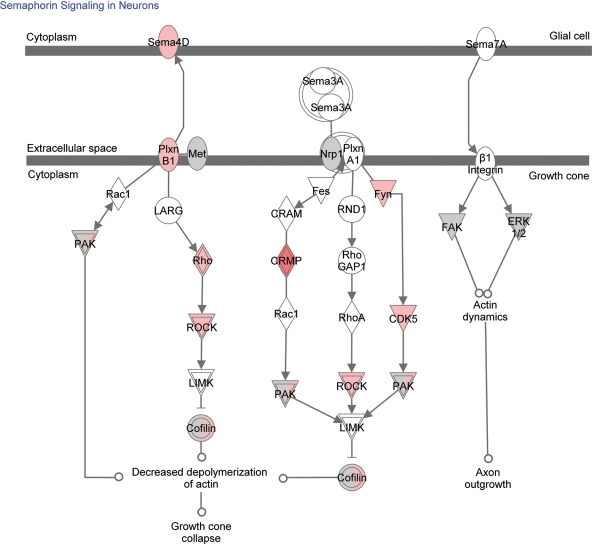
Semaphorin signalling in neurons
The current review reveals the up-regulation of the semaphorin pathway (Fig. 1), most importantly Sema4D and Plexin B1. Semaphorins are a large family of secreted and transmembrane molecules that function as repulsive axon guidance factors (54) and modulate dendritic and axonal arborizations of developing neurons. In addition, semaphorins play a significant role in microglia activation and in neuroinflammation. A recent study by Okuno et al. (55) has shown that Sema4D promoted inducible nitric oxide synthase expression by primary mouse microglia. This expression was Plexin B1-dependent as it was abolished in Plexin B1-deficient cells. Moreover, during the development of myelin oligodendrocyte glycoprotein peptide-induced experimental autoimmune encephalomyelitis (EAE), the expression of Sema4D and Plexin B1 was induced in infiltrating mononuclear cells and microglia, respectively (55). Wild-type myelin oligodendrocyte glycoprotein-specific T cells adoptively transferred into Plexin B1-deficient mice were not able to induce the disease. Similarly, bone marrow chimeric mice with Plexin B1-deficient central nervous system resident cells were not able to develop EAE. Furthermore, blocking antibodies against Sema4D significantly inhibited neuroinflammation during EAE development. The role semaphorins play in the inflammatory processes associated with epilepsy is not yet known, but deserves further study.
Figure 1.
Up-regulated genes in the semaphorin pathway. The intensity of the colour indicates the number of studies in which the gene is up-regulated.
Modulation of synaptic transmission
The cellular component ontology-term enrichment shows that some of the most important processes in the development of pharmacoresistance are occurring in membrane-bound vesicles and at the pre- and post-synaptic terminal. The most significant biological processes are cellular cation homeostasis, synaptic transmission and synaptic plasticity. The enriched GO terms suggest that the most important molecular functions in the development of pharmacoresistance are calcium transport and signalling and transporter activity. Therefore, changes in calcium signalling and in synaptic structure and function were shown to be important in our analysis.
Calcium signalling
Network 3 shows that changes in calcium-mediated cell signalling leading to alterations in neuronal excitability are likely to be important in the development of pharmacoresistance in epilepsy. The network reveals dysregulation of proteins regulating calcium channels (calcium-binding protein 1 and calpastatin), of proteins involved in calcium homeostasis (inositol 1,4,5-triphosphate receptor, type 1, ATPase Ca2+ transporting cardiac muscle slow twitch 2, ATPase Ca2+ transporting plasma membrane 1, SLC family 8 sodium/calcium exchanger member 1, calcium channel voltage-dependent alpha 2/delta subunit 1, calcium channel voltage-dependent gamma subunit 3, calcium channel voltage-dependent alpha 2/delta subunit 1, adenosylhomocysteinase-like 1) and of calcium-dependent signalling proteins (calpain 3 and calcium-binding protein 1). Calcium signalling is among the top 25 enriched canonical pathways in our gene list. Calcium signalling, therefore, plays a significant part in pharmacoresistance and its manipulation may be an important therapeutic strategy—two potential target genes which overlap in three or more studies are neurogranin and SNAP-25.
Neurogranin has been implicated in the modulation of post-synaptic signal transduction pathways, in synaptic plasticity (56) and in the enhancement of long-term potentiation by promoting calcium-mediated signalling (57), and it is hypothesized that it participates in the action of valproate (58). SNAP-25 modulates neurotransmitter exocytosis (59–62) and negatively regulates voltage-gated calcium channels (VGCCs) in glutamatergic neurons (63). Silencing of endogenous SNAP-25 in glutamatergic neurons leads to an augmentation of VGCC activity, which would increase network excitability. Reductions in SNAP-25 expression have been correlated with neurological conditions characterized by increased network excitability. For example, VGCC currents are up-regulated resulting in absence-like epilepsy and hyperactivity (64) in the Coloboma mouse mutant characterized by the heterozygous deletion of the SNAP-25 gene. Also, alterations in SNAP-25 expression have been described in human patients with attention deficit hyperactivity disorder and schizophrenia (62).
Alterations in synaptic structure and function
As stated above, GO-term enrichment analysis suggests that some of the most important processes in the development of pharmacoresistance are occurring in membrane-bound vesicles and at the pre- and post-synaptic terminal and some of the most significant biological processes are synaptic transmission and synaptic plasticity.
Network 1 reveals that there is dysregulation of proteins involved in maintaining synaptic structure and function. There is dysregulation of pre-synaptic cytomatrix proteins (bassoon, ELKS/RAB6-interacting/CAST family member 2 and piccolo), an active zone protein (PTPRF interacting protein, binding protein 1), a protein which controls presynaptic residual Ca2+ concentrations (ATPase Ca2+ transporting plasma membrane 2), proteins in post-synaptic sites which form a multimeric scaffold for the clustering of receptors, ion channels and associated signalling proteins (calcium/calmodulin-dependent serine protein kinase and DLG2), and proteins which may play a role in clustering of NMDA receptors at excitatory synapses (DLG3). Complexin, a part of the SNARE complex (65) which is the core of the synaptic release machinery (66), was down-regulated in three studies and is particularly deserving of further study. Perturbations of the complexin proteins cause an increase in spontaneous and asynchronous release of neurotransmitter in some types of synapses, indicating an inhibitory role of the proteins (65). Hence, the reduction in complexin 2 expression in the epileptic focus, as demonstrated in this review, could lead to increases in excitatory synaptic transmission and deserves further study.
Restructuring of neuronal networks
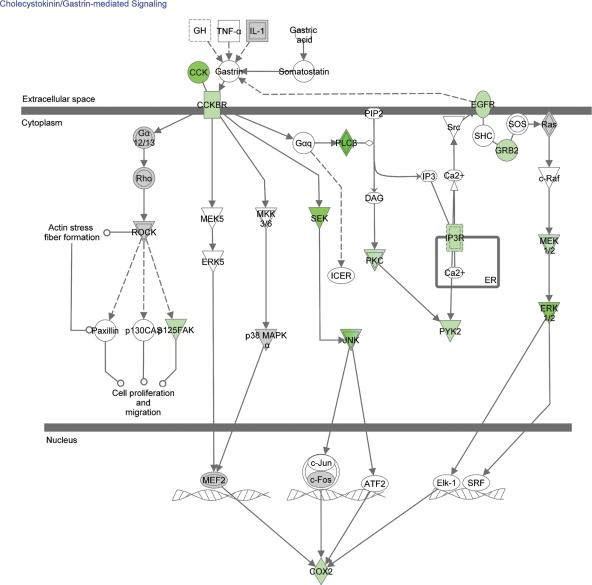
The GO enrichment analysis suggests that some of the most important processes in the development of pharmacoresistance are occurring in the cytoskeleton, the dendrite, the axon and the growth cone. Some of the most significant biological processes are apoptosis, cytoskeletal organization, neuronal development and differentiation, axonal and dendritic morphogenesis, and one of the most important molecular functions in the development of pharmacoresistance is cytoskeletal function. In support of this, Network 2 reveals dysregulation of proteins involved in apoptotic mechanisms (translocase of inner mitochondrial membrane 23, histone cluster 1 H1c, peptidylprolyl isomerase F, dedicator of cytokinesis 1 and Jumonji domain containing 6), in cell growth (protein tyrosine kinase 2, protein tyrosine phosphatase non-receptor type 12 and neural precursor cell expressed developmentally down-regulated 9) and in cell shape maintenance (tensin 1). Network 3 reveals dysregulation of proteins involved in neurogenesis, neurite outgrowth and axon guidance (neurogenic differentiation 2, neurofascin and dihydropyrimidinase-like 3). Network 1 reveals that there is also dysregulation of proteins involved in axon guidance (protein tyrosine phosphatase receptor type f and protein tyrosine phosphatase receptor type D). Similarly, the enriched canonical pathways include axonal guidance signalling, ras homolog gene family, member A (RhoA) signalling (which plays a significant role in actin stress fibres formation), CDK5 signalling (which regulates neurite outgrowth), and ephrin receptor signalling (which is involved in nervous system development). The CCK signalling pathway was the most significantly enriched canonical pathway in our analysis (Fig. 2) and has an important, yet understudied, role in the development of aberrant neuronal networks.
Figure 2.
Down-regulated genes in the CCK pathway. The intensity of the colour indicates the number of studies in which the gene is down-regulated.
CCK signalling
CCK controls the perisomatically targeting inhibitory interneurons of the hippocampus (67). In kindling rodent models of epilepsy, there is a reduction in GABA-evoked inhibitory post-synaptic currents in the hippocampus, and this loss corresponds with a reduction in CCK-labelled interneurons (68). In humans, the CCK content of cortical tissue from which active epileptic spiking was recorded at the time of surgery was significantly decreased in comparison to tissue samples from patients in whom the lateral temporal cortex was electrographically free of epileptiform spikes (69). CCK may have anticonvulsant and neuroprotective properties. Pretreatment of hippocampal slices with sulphated CCK blocked the effect of kainic acid on synaptic transmission (70). Seizures in a breed of rat with congenital audiogenic seizure were suppressed by CCK octapeptide-injected intraperitoneally (71) and by intracerebral injection of a CCK gene vector (72). Seizures induced by picrotoxin and electroshock are inhibited by intracerebroventricular administration of CCK in rats (73). There is also a positive influence of CCK on the anticonvulsant efficacy of vigabatrin (74). CCK, therefore, is another potential therapeutic agent that should be further studied.
CONCLUSIONS
Large-scale gene expression profiling studies on brain tissue from epilepsy surgery have been performed largely with the aim of generating hypotheses about the causes of epileptogenesis and pharmacoresistance. Although there have been at least 12 such studies in the last 10 years, they have failed to make a significant impact on our understanding of pharmacoresistance in epilepsy for the reasons which have been outlined in this paper. Our analysis shows that there is a statistically significant overlap between the gene lists of studies performed on different kind of epileptic foci using different types of microarrays. We have used an inter-study cross-validation technique to simultaneously verify the expression changes of large numbers of genes. We have performed an integrative analysis of the gene lists from different studies to identify the cellular components, biological processes, molecular functions, pathways, networks and individual genes most important in the development of refractory epilepsy. We invite investigators to use the data presented with this analysis to support or counter prevalent hypotheses and to develop new ones.
METHODS
Inclusion criteria
We aimed to include all large-scale gene expression profiling studies fulfilling the following criteria:
recruited patients had clearly defined refractory epilepsy;
the epileptic focus was ascertained on the basis of clinical features, electroencephalography, brain imaging and post-operative tissue histology as necessary;
‘pharmacoresistant’ tissue was compared with suitable control (non-epileptic or ‘pharmacoresponsive’) tissue;
epileptic tissue analysed was not cancerous or neoplastic;
at least 500 genes from across the genome were assayed and
MIAME-compliant data were provided or, at the very least, a list of significantly regulated genes and the direction of change (up- or down-regulated) was provided.
Search strategy
We searched records in Medline and Embase, without language restriction, between January 1987 and January 2011. Large-scale microarray technology was first described in 1987 (75); hence 1987 formed the starting point for our search. We used the following search terms: (1) ‘gene-expression profiling’, (2) ‘microarray analysis’, (3) ‘transcription profiling’, (4) ‘cluster analysis’, (5) ‘Affymetrix’, (6) ‘GeneChip’, (7) ‘serial analysis of gene expression’ or (8) ‘SAGE’ and (1) ‘epilepsy’, (2) ‘TLE’ or (3) ‘MTLE’. We also hand-searched the reference lists of every primary study, previously published systematic reviews and other review articles.
We also searched public repositories of microarray data sets: ArrayExpress (www.ebi.ac.uk/arrayexpress), GEO (www.ncbi.nlm.nih.gov/geo), Stanford Microarray Database (smd.stanford.edu), University of Pennsylvania RAD (www.cbil.upenn.edu/RAD/php/index.php), UNC Microarray Database (genome.unc.edu), MUSC Microarray Database (proteogenomics.musc.edu/musc_madb.html), Genevestigator (www.genevestigator.com), Princeton University MicroArray database (puma.princeton.edu), University of Tennessee Microarray Database (genome.ws.utk.edu, last accessed date May 1, 2011).
Data gathering: raw data
We found 12 published genome-wide gene expression profiling studies on non-neoplastic tissue from epilepsy surgery. Raw data for the study by Ozbas-Gerceker et al. (15) were available from the GEO database. MIAME-compliant data for the study by van Gassen et al. (11) were available on the ArrayExpress database. MIAME-compliant data were not publically available for any other study. The corresponding author from each of the remaining 10 studies was contacted to request MIAME-compliant microarray results data, but MIAME-compliant data were not provided by any author. As raw data were only available for two studies performed on two entirely different platforms (Serial Analysis of Gene Expression and two-channel oligonucleotide microarray analysis), we judged that a useful integrative analysis of the raw data could not be performed, and raw data were not used in the present study. Instead, we obtained differentially regulated gene lists from individual studies, as detailed below.
Data gathering: gene lists
Six studies published complete lists of all genes which were differentially regulated in epileptic tissue according to each study's individually defined criteria (11,13,15,17–20). Xi et al. (14) published a partial list of gene regulated in their study. Xiao et al. (16) directly provided a list of all genes that were differentially regulated in epileptic tissue according to their individually defined criteria. Three studies did not publish a list of regulated genes and their authors did not provide this on request (76–78), and hence were excluded from further analysis in this systematic review.
Data integration
To allow inter-study comparison, gene identifiers from each study were converted to Entrez gene numbers, unless these were provided in the publication. Affymetrix probe numbers were converted to Entrez gene numbers using NetAffx Analysis Center (www.affymetrix.com/analysis). All other gene identifiers were converted to Entrez gene numbers using MatchMiner (build 137, discover.nci.nih.gov/matchminer).
For each gene list, changes in gene expression were changed to a binary score: any gene up-regulated was given a score of +1, while any gene down-regulated was given a score of −1. The nine lists of regulated genes from the nine included studies were integrated in a MS Excel spreadsheet. Forty-two genes were excluded because of conflicting scores (i.e. they were down-regulated in one study but up-regulated in another), leaving 1407 regulated genes in the integrated list incorporating all included studies. The total score was calculated for each gene.
Data validation
The overlapping probabilities of differentially regulated gene lists were calculated using ConceptGen (conceptgen.ncibi.org/core/conceptGen), a gene set enrichment testing tool. ConceptGen uses the Expression Analysis Systematic Explorer (EASE) score (a modified Fisher's exact test widely employed in published literature) to test pairs of gene lists to determine if there exists a larger number of overlapping genes than is expected by chance (79). P-values are then adjusted for multiple testing by calculating the FDR (80), values <0.05 being deemed significant. All of the gene lists from the included studies were tested in pairs in this manner. In addition, to test the validity of the list of genes differentially regulated in only one study, we compared this with a list of all genes from CarpeDB (www.carpedb.ua.edu), a dynamic continuously updated epilepsy genetics database. We extracted the human genes in the CarpeDB database. In addition, we extracted the rat and mouse genes and mapped them to human homologues using ConceptGen. Two hundred and seventy-eight CarpeDB human, mouse and rat genes in total mapped to ConceptGen.
GO enrichment analysis
GO terms enrichment analysis was performed in the Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.7 (david.abcc.ncifcrf.gov). DAVID performs an enrichment test based on the EASE score, and an FDR <0.05 is deemed significant. For biological process and cellular component terms, there were a large number of enriched broad GO terms, which made practical interpretation difficult. Hence, for biological process and cellular component domains, ‘Go Fat’ analysis was performed. Go Fat filters the broadest terms so that they do not overshadow the more specific terms.
Pathway analysis
Ingenuity Systems (www.ingenuity.com) Core Analysis was used to determine significantly enriched canonical pathways and to build significantly enriched networks from the full list of differentially regulated genes. The significance of the association between our data set genes and the canonical pathways was measured in two ways: (1) a ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway is displayed, and (2) a Fisher's exact test was used to calculate a P-value, determining the probability that the association between the genes in the data set and the canonical pathway was explained by chance alone. Networks were ranked by a score: the higher the score, the lower the probability of finding the observed data set genes in a given network by chance. The score takes into account the number of data set genes and the size of the network and is the negative log of the P-value.
Comparison with an AD brain microarray data set
To counter arguments that the genetic changes highlighted by this analysis are non-specific and could be found in any brain pathology, we compared our results with the results of a large-scale gene expression profiling study on brain samples from 34 individuals with AD (81). From this study, we extracted the list of genes exhibiting differential expression between AD-affected and normal hippocampi—an AD hippocampus data set was used as most of the included epilepsy studies utilized hippocampal tissue. Three thousand seven hundred and thirty eight unique genes could be mapped to Entrez gene IDs. Ingenuity Systems Pathway Analysis was used to determine significantly enriched canonical pathways from this gene list. The top 10 canonical pathways from the epilepsy and AD data sets were then compared.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
N.M. is a MRC Research Training Fellow. M.P. is a NIHR Senior Investigator and acknowledges the support of the UK Department of Health (NHS Chair of Pharmacogenetics), Wolfson Foundation, MRC and Wellcome Trust.
Supplementary Material
REFERENCES
- 1.Sander J.W. The natural history of epilepsy in the era of new antiepileptic drugs and surgical treatment. Epilepsia. 2003;44(Suppl. 1)):17–20. doi: 10.1046/j.1528-1157.44.s.1.1.x. [DOI] [PubMed] [Google Scholar]
- 2.Shorvon S.D. The epidemiology and treatment of chronic and refractory epilepsy. Epilepsia. 1996;37(Suppl. 2):S1–S3. doi: 10.1111/j.1528-1157.1996.tb06027.x. [DOI] [PubMed] [Google Scholar]
- 3.Engel J., Jr A greater role for surgical treatment of epilepsy: why and when? Epilepsy Curr. 2003;3:37–40. doi: 10.1046/j.1535-7597.2003.03201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogawski M.A., Johnson M.R. Intrinsic severity as a determinant of antiepileptic drug refractoriness. Epilepsy Curr. 2008;8:127–130. doi: 10.1111/j.1535-7511.2008.00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engel T., Murphy B.M., Schindler C.K., Henshall D.C. Elevated p53 and lower MDM2 expression in hippocampus from patients with intractable temporal lobe epilepsy. Epilepsy Res. 2007;77:151–156. doi: 10.1016/j.eplepsyres.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong M. Stabilizing dendritic structure as a novel therapeutic approach for epilepsy. Expert Rev. Neurother. 2008;8:907–915. doi: 10.1586/14737175.8.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siebzehnrubl F.A., Blumcke I. Neurogenesis in the human hippocampus and its relevance to temporal lobe epilepsies. Epilepsia. 2008;49(Suppl. 5):55–65. doi: 10.1111/j.1528-1167.2008.01638.x. [DOI] [PubMed] [Google Scholar]
- 8.King H.C., Sinha A.A. Gene expression profile analysis by DNA microarrays: promise and pitfalls. JAMA. 2001;286:2280–2288. doi: 10.1001/jama.286.18.2280. [DOI] [PubMed] [Google Scholar]
- 9.Rhodes D.R., Barrette T.R., Rubin M.A., Ghosh D., Chinnaiyan A.M. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 10.Lukasiuk K., Pitkanen A. Large-scale analysis of gene expression in epilepsy research: is synthesis already possible? Neurochem. Res. 2004;29:1169–1178. doi: 10.1023/b:nere.0000023604.91584.6c. [DOI] [PubMed] [Google Scholar]
- 11.van Gassen K.L., de Wit M., Koerkamp M.J., Rensen M.G., van Rijen P.C., Holstege F.C., Lindhout D., de Graan P.N. Possible role of the innate immunity in temporal lobe epilepsy. Epilepsia. 2008;49:1055–1065. doi: 10.1111/j.1528-1167.2007.01470.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y.Y., Smith P., Murphy M., Cook M. Global expression profiling in epileptogenesis: does it add to the confusion? Brain Pathol. 2010;20:1–16. doi: 10.1111/j.1750-3639.2008.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee T.S., Mane S., Eid T., Zhao H., Lin A., Guan Z., Kim J.H., Schweitzer J., King-Stevens D., Weber P., et al. Gene expression in temporal lobe epilepsy is consistent with increased release of glutamate by astrocytes. Mol. Med. 2007;13:1–13. doi: 10.2119/2006-00079.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xi Z.Q., Xiao F., Yuan J., Wang X.F., Wang L., Quan F.Y., Liu G.W. Gene expression analysis on anterior temporal neocortex of patients with intractable epilepsy. Synapse. 2009;63:1017–1028. doi: 10.1002/syn.20681. [DOI] [PubMed] [Google Scholar]
- 15.Ozbas-Gerceker F., Redeker S., Boer K., Ozguc M., Saygi S., Dalkara T., Soylemezoglu F., Akalan N., Baayen J.C., Gorter J.A., et al. Serial analysis of gene expression in the hippocampus of patients with mesial temporal lobe epilepsy. Neuroscience. 2006;138:457–474. doi: 10.1016/j.neuroscience.2005.11.043. [DOI] [PubMed] [Google Scholar]
- 16.Xiao F., Wang X.F., Li J.M., Xi Z.Q., Lu Y., Wang L., Zeng Y., Guan L.F., Yuan J. Overexpression of N-WASP in the brain of human epilepsy. Brain Res. 2008;1233:168–175. doi: 10.1016/j.brainres.2008.07.101. [DOI] [PubMed] [Google Scholar]
- 17.Arion D., Sabatini M., Unger T., Pastor J., Alonso-Nanclares L., Ballesteros-Yanez I., Garcia Sola R., Munoz A., Mirnics K., DeFelipe J. Correlation of transcriptome profile with electrical activity in temporal lobe epilepsy. Neurobiol. Dis. 2006;22:374–387. doi: 10.1016/j.nbd.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Becker A.J., Chen J., Zien A., Sochivko D., Normann S., Schramm J., Elger C.E., Wiestler O.D., Blumcke I. Correlated stage- and subfield-associated hippocampal gene expression patterns in experimental and human temporal lobe epilepsy. Eur. J. Neurosci. 2003;18:2792–2802. doi: 10.1111/j.1460-9568.2003.02993.x. [DOI] [PubMed] [Google Scholar]
- 19.Becker A.J., Chen J., Paus S., Normann S., Beck H., Elger C.E., Wiestler O.D., Blumcke I. Transcriptional profiling in human epilepsy: expression array and single cell real-time qRT-PCR analysis reveal distinct cellular gene regulation. Neuroreport. 2002;13:1327–1333. doi: 10.1097/00001756-200207190-00023. [DOI] [PubMed] [Google Scholar]
- 20.Jamali S., Bartolomei F., Robaglia-Schlupp A., Massacrier A., Peragut J.C., Regis J., Dufour H., Ravid R., Roll P., Pereira S., et al. Large-scale expression study of human mesial temporal lobe epilepsy: evidence for dysregulation of the neurotransmission and complement systems in the entorhinal cortex. Brain. 2006;129:625–641. doi: 10.1093/brain/awl001. [DOI] [PubMed] [Google Scholar]
- 21.Loscher W. Animal models of drug-resistant epilepsy. Novartis Found. Symp. 2002;243:149–159. discussion 159–166, 180–145. [PubMed] [Google Scholar]
- 22.Choi J.K., Yu U., Kim S., Yoo O.J. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics. 2003;19(Suppl. 1):i84–i90. doi: 10.1093/bioinformatics/btg1010. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt D., Loscher W. New developments in antiepileptic drug resistance: an integrative view. Epilepsy Curr. 2009;9:47–52. doi: 10.1111/j.1535-7511.2008.01289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chayasirisobhon S. The mechanisms of medically refractory temporal lobe epilepsy. Acta Neurol. Taiwan. 2009;18:155–160. [PubMed] [Google Scholar]
- 25.Lazarowski A., Czornyj L., Lubienieki F., Girardi E., Vazquez S., D'Giano C. ABC transporters during epilepsy and mechanisms underlying multidrug resistance in refractory epilepsy. Epilepsia. 2007;48(Suppl. 5):140–149. doi: 10.1111/j.1528-1167.2007.01302.x. [DOI] [PubMed] [Google Scholar]
- 26.Kubota H., Ishihara H., Langmann T., Schmitz G., Stieger B., Wieser H.G., Yonekawa Y., Frei K. Distribution and functional activity of P-glycoprotein and multidrug resistance-associated proteins in human brain microvascular endothelial cells in hippocampal sclerosis. Epilepsy Res. 2006;68:213–228. doi: 10.1016/j.eplepsyres.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Kennerly E.M., Idaghdour Y., Olby N.J., Munana K.R., Gibson G. Pharmacogenetic association study of 30 genes with phenobarbital drug response in epileptic dogs. Pharmacogenet. Genomics. 2009;19:911–922. doi: 10.1097/FPC.0b013e3283307cba. [DOI] [PubMed] [Google Scholar]
- 28.Hughes J.R. One of the hottest topics in epileptology: ABC proteins. Their inhibition may be the future for patients with intractable seizures. Neurol. Res. 2008;30:920–925. doi: 10.1179/174313208X319116. [DOI] [PubMed] [Google Scholar]
- 29.Gupta S., Burckhardt G., Hagos Y. SLC22 transporter family proteins as targets for cytostatic uptake into tumor cells. Biol. Chem. 2011;392:117–124. doi: 10.1515/BC.2011.014. [DOI] [PubMed] [Google Scholar]
- 30.Loscher W., Klotz U., Zimprich F., Schmidt D. The clinical impact of pharmacogenetics on the treatment of epilepsy. Epilepsia. 2009;50:1–23. doi: 10.1111/j.1528-1167.2008.01716.x. [DOI] [PubMed] [Google Scholar]
- 31.Whitaker W.R., Faull R.L., Dragunow M., Mee E.W., Emson P.C., Clare J.J. Changes in the mRNAs encoding voltage-gated sodium channel types II and III in human epileptic hippocampus. Neuroscience. 2001;106:275–285. doi: 10.1016/s0306-4522(01)00212-3. [DOI] [PubMed] [Google Scholar]
- 32.Lakhan R., Kumari R., Misra U.K., Kalita J., Pradhan S., Mittal B. Differential role of sodium channels SCN1A and SCN2A gene polymorphisms with epilepsy and multiple drug resistance in the north Indian population. Br. J. Clin. Pharmacol. 2009;68:214–220. doi: 10.1111/j.1365-2125.2009.03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwan P., Poon W.S., Ng H.K., Kang D.E., Wong V., Ng P.W., Lui C.H., Sin N.C., Wong K.S., Baum L. Multidrug resistance in epilepsy and polymorphisms in the voltage-gated sodium channel genes SCN1A, SCN2A, and SCN3A: correlation among phenotype, genotype, and mRNA expression. Pharmacogenet. Genomics. 2008;18:989–998. doi: 10.1097/FPC.0b013e3283117d67. [DOI] [PubMed] [Google Scholar]
- 34.Rogawski M.A., Loscher W. The neurobiology of antiepileptic drugs. Nat. Rev. Neurosci. 2004;5:553–564. doi: 10.1038/nrn1430. [DOI] [PubMed] [Google Scholar]
- 35.Brooks-Kayal A.R., Shumate M.D., Jin H., Rikhter T.Y., Coulter D.A. Selective changes in single cell GABA(A) receptor subunit expression and function in temporal lobe epilepsy. Nat. Med. 1998;4:1166–1172. doi: 10.1038/2661. [DOI] [PubMed] [Google Scholar]
- 36.Bethmann K., Fritschy J.M., Brandt C., Loscher W. Antiepileptic drug resistant rats differ from drug responsive rats in GABA A receptor subunit expression in a model of temporal lobe epilepsy. Neurobiol. Dis. 2008;31:169–187. doi: 10.1016/j.nbd.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Bonin R.P., Martin L.J., MacDonald J.F., Orser B.A. Alpha5GABAA receptors regulate the intrinsic excitability of mouse hippocampal pyramidal neurons. J. Neurophysiol. 2007;98:2244–2254. doi: 10.1152/jn.00482.2007. [DOI] [PubMed] [Google Scholar]
- 38.Mariotti V., Melissari E., Amar S., Conte A., Belmaker R.H., Agam G., Pellegrini S. Effect of prolonged phenytoin administration on rat brain gene expression assessed by DNA microarrays. Exp. Biol. Med. 2010;235:300–310. doi: 10.1258/ebm.2009.009225. [DOI] [PubMed] [Google Scholar]
- 39.Almgren M., Nyengaard J.R., Persson B., Lavebratt C. Carbamazepine protects against neuronal hyperplasia and abnormal gene expression in the megencephaly mouse. Neurobiol. Dis. 2008;32:364–376. doi: 10.1016/j.nbd.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 40.Atack J.R., Bayley P.J., Fletcher S.R., McKernan R.M., Wafford K.A., Dawson G.R. The proconvulsant effects of the GABAA α5 subtype-selective compound RY-080 may not be α5-mediated. Eur. J. Pharmacol. 2006;548:77–82. doi: 10.1016/j.ejphar.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 41.Gall C.M., Lynch G. Integrins, synaptic plasticity and epileptogenesis. Adv. Exp. Med. Biol. 2004;548:12–33. doi: 10.1007/978-1-4757-6376-8_2. [DOI] [PubMed] [Google Scholar]
- 42.Vezzani A., Granata T. Brain inflammation in epilepsy: experimental and clinical evidence. Epilepsia. 2005;46:1724–1743. doi: 10.1111/j.1528-1167.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 43.Fabene P.F., Bramanti P., Constantin G. The emerging role for chemokines in epilepsy. J. Neuroimmunol. 2010;224:22–27. doi: 10.1016/j.jneuroim.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 44.Seiffert E., Dreier J.P., Ivens S., Bechmann I., Tomkins O., Heinemann U., Friedman A. Lasting blood-brain barrier disruption induces epileptic focus in the rat somatosensory cortex. J. Neurosci. 2004;24:7829–7836. doi: 10.1523/JNEUROSCI.1751-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivens S., Kaufer D., Flores L.P., Bechmann I., Zumsteg D., Tomkins O., Seiffert E., Heinemann U., Friedman A. TGF-beta receptor-mediated albumin uptake into astrocytes is involved in neocortical epileptogenesis. Brain. 2007;130:535–547. doi: 10.1093/brain/awl317. [DOI] [PubMed] [Google Scholar]
- 46.Marchi N., Angelov L., Masaryk T., Fazio V., Granata T., Hernandez N., Hallene K., Diglaw T., Franic L., Najm I., et al. Seizure-promoting effect of blood-brain barrier disruption. Epilepsia. 2007;48:732–742. doi: 10.1111/j.1528-1167.2007.00988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabene P.F., Navarro Mora G., Martinello M., Rossi B., Merigo F., Ottoboni L., Bach S., Angiari S., Benati D., Chakir A., et al. A role for leukocyte-endothelial adhesion mechanisms in epilepsy. Nat. Med. 2008;14:1377–1383. doi: 10.1038/nm.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somera-Molina K.C., Nair S., Van Eldik L.J., Watterson D.M., Wainwright M.S. Enhanced microglial activation and proinflammatory cytokine upregulation are linked to increased susceptibility to seizures and neurologic injury in a ‘two-hit’ seizure model. Brain Res. 2009;1282:162–172. doi: 10.1016/j.brainres.2009.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinn D.I., Kim S.J., Chu K., Jung K.H., Lee S.T., Song E.C., Kim J.M., Park D.K., Kun Lee S., Kim M., et al. Valproic acid-mediated neuroprotection in intracerebral hemorrhage via histone deacetylase inhibition and transcriptional activation. Neurobiol. Dis. 2007;26:464–472. doi: 10.1016/j.nbd.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 50.Unsicker K., Krieglstein K. TGF-betas and their roles in the regulation of neuron survival. Adv. Exp. Med. Biol. 2002;513:353–374. doi: 10.1007/978-1-4615-0123-7_13. [DOI] [PubMed] [Google Scholar]
- 51.Alzheimer C., Werner S. Fibroblast growth factors and neuroprotection. Adv. Exp. Med. Biol. 2002;513:335–351. doi: 10.1007/978-1-4615-0123-7_12. [DOI] [PubMed] [Google Scholar]
- 52.Zhang S.J., Zou M., Lu L., Lau D., Ditzel D.A., Delucinge-Vivier C., Aso Y., Descombes P., Bading H. Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 2009;5:e1000604. doi: 10.1371/journal.pgen.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ragozzino D., Renzi M., Giovannelli A., Eusebi F. Stimulation of chemokine CXC receptor 4 induces synaptic depression of evoked parallel fibers inputs onto Purkinje neurons in mouse cerebellum. J. Neuroimmunol. 2002;127:30–36. doi: 10.1016/s0165-5728(02)00093-0. [DOI] [PubMed] [Google Scholar]
- 54.Kolodkin A.L., Matthes D.J., Goodman C.S. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75:1389–1399. doi: 10.1016/0092-8674(93)90625-z. [DOI] [PubMed] [Google Scholar]
- 55.Okuno T., Nakatsuji Y., Moriya M., Takamatsu H., Nojima S., Takegahara N., Toyofuku T., Nakagawa Y., Kang S., Friedel R.H., et al. Roles of Sema4D–plexin-B1 interactions in the central nervous system for pathogenesis of experimental autoimmune encephalomyelitis. J. Immunol. 2010;184:1499–1506. doi: 10.4049/jimmunol.0903302. [DOI] [PubMed] [Google Scholar]
- 56.Kubota Y., Putkey J.A., Shouval H.Z., Waxham M.N. IQ-motif proteins influence intracellular free Ca2+ in hippocampal neurons through their interactions with calmodulin. J. Neurophysiol. 2008;99:264–276. doi: 10.1152/jn.00876.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang K.P., Huang F.L., Jager T., Li J., Reymann K.G., Balschun D. Neurogranin/RC3 enhances long-term potentiation and learning by promoting calcium-mediated signaling. J. Neurosci. 2004;24:10660–10669. doi: 10.1523/JNEUROSCI.2213-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu L., Peng J., Wei C., Liu G., Wang G., Li K., Yin F. Characterization, using comparative proteomics, of differentially expressed proteins in the hippocampus of the mesial temporal lobe of epileptic rats following treatment with valproate. Amino Acids. 2011;40:221–238. doi: 10.1007/s00726-010-0638-8. [DOI] [PubMed] [Google Scholar]
- 59.Schiavo G., Stenbeck G., Rothman J.E., Sollner T.H. Binding of the synaptic vesicle v-SNARE, synaptotagmin, to the plasma membrane t-SNARE, SNAP-25, can explain docked vesicles at neurotoxin-treated synapses. Proc. Natl Acad. Sci. USA. 1997;94:997–1001. doi: 10.1073/pnas.94.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Augustine G.J. How does calcium trigger neurotransmitter release? Curr. Opin. Neurobiol. 2001;11:320–326. doi: 10.1016/s0959-4388(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 61.Chapman E.R. Synaptotagmin: a Ca(2+) sensor that triggers exocytosis? Nat. Rev. Mol. Cell Biol. 2002;3:498–508. doi: 10.1038/nrm855. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X., Kim-Miller M.J., Fukuda M., Kowalchyk J.A., Martin T.F. Ca2+-dependent synaptotagmin binding to SNAP-25 is essential for Ca2+-triggered exocytosis. Neuron. 2002;34:599–611. doi: 10.1016/s0896-6273(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 63.Condliffe S.B., Corradini I., Pozzi D., Verderio C., Matteoli M. Endogenous SNAP-25 regulates native voltage-gated calcium channels in glutamatergic neurons. J. Biol. Chem. 2010;285:24968–24976. doi: 10.1074/jbc.M110.145813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Risinger C., Bennett M.K. Differential phosphorylation of syntaxin and synaptosome-associated protein of 25 kDa (SNAP-25) isoforms. J. Neurochem. 1999;72:614–624. doi: 10.1046/j.1471-4159.1999.0720614.x. [DOI] [PubMed] [Google Scholar]
- 65.Neher E. Complexin: does it deserve its name? Neuron. 2010;68:803–806. doi: 10.1016/j.neuron.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 66.Sudhof T.C., Rothman J.E. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee S.Y., Soltesz I. Cholecystokinin: a multi-functional molecular switch of neuronal circuits. Dev. Neurobiol. 2011;71:83–91. doi: 10.1002/dneu.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sayin U., Osting S., Hagen J., Rutecki P., Sutula T. Spontaneous seizures and loss of axo-axonic and axo-somatic inhibition induced by repeated brief seizures in kindled rats. J. Neurosci. 2003;23:2759–2768. doi: 10.1523/JNEUROSCI.23-07-02759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iadarola M.J., Sherwin A.L. Alterations in cholecystokinin peptide and mRNA in actively epileptic human temporal cortical foci. Epilepsy Res. 1991;8:58–63. doi: 10.1016/0920-1211(91)90036-f. [DOI] [PubMed] [Google Scholar]
- 70.Aitken P.G., Jaffe D.B., Nadler J.V. Cholecystokinin blocks some effects of kainic acid in CA3 region of hippocampal slices. Peptides. 1991;12:127–129. doi: 10.1016/0196-9781(91)90178-r. [DOI] [PubMed] [Google Scholar]
- 71.Zhang L.X., Zhou Y., Du Y., Han J.S. Effect of CCK-8 on audiogenic epileptic seizure in P77PMC rats. Neuropeptides. 1993;25:73–76. doi: 10.1016/0143-4179(93)90072-i. [DOI] [PubMed] [Google Scholar]
- 72.Zhang L.X., Wu M., Han J.S. Suppression of audiogenic epileptic seizures by intracerebral injection of a CCK gene vector. Neuroreport. 1992;3:700–702. [PubMed] [Google Scholar]
- 73.Kadar T., Pesti A., Penke B., Telegdy G. Inhibition of seizures induced by picrotoxin and electroshock by cholecystokinin octapeptides and their fragments in rats after intracerebroventricular administration. Neuropharmacology. 1984;23:955–961. doi: 10.1016/0028-3908(84)90010-8. [DOI] [PubMed] [Google Scholar]
- 74.Ferraro G., Sardo P. Cholecystokinin-8 sulfate modulates the anticonvulsant efficacy of vigabatrin in an experimental model of partial complex epilepsy in the rat. Epilepsia. 2009;50:721–730. doi: 10.1111/j.1528-1167.2008.01956.x. [DOI] [PubMed] [Google Scholar]
- 75.Kulesh D.A., Clive D.R., Zarlenga D.S., Greene J.J. Identification of interferon-modulated proliferation-related cDNA sequences. Proc. Natl. Acad. Sci. USA. 1987;84:8453–8457. doi: 10.1073/pnas.84.23.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee T.S., Eid T., Mane S., Kim J.H., Spencer D.D., Ottersen O.P., de Lanerolle N.C. Aquaporin-4 is increased in the sclerotic hippocampus in human temporal lobe epilepsy. Acta Neuropathol. 2004;108:493–502. doi: 10.1007/s00401-004-0910-7. [DOI] [PubMed] [Google Scholar]
- 77.Liu F.Y., Wang X.F., Li M.W., Li J.M., Xi Z.Q., Luan G.M., Zhang J.G., Wang Y.P., Sun J.J., Li Y.L. Upregulated expression of postsynaptic density-93 and N-methyl-D-aspartate receptors subunits 2B mRNA in temporal lobe tissue of epilepsy. Biochem. Biophys. Res. Commun. 2007;358:825–830. doi: 10.1016/j.bbrc.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 78.Li J.M., Wang X.F., Xi Z.Q., Gong Y., Liu F.Y., Sun J.J., Wu Y., Luan G.M., Wang Y.P., Li Y.L., et al. Decreased expression of thyroid receptor-associated protein 220 in temporal lobe tissue of patients with refractory epilepsy. Biochem. Biophys. Res. Commun. 2006;348:1389–1397. doi: 10.1016/j.bbrc.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 79.Hosack D.A., Dennis G., Jr, Sherman B.T., Lane H.C., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. 1995;57:289–300. [Google Scholar]
- 81.Liang W.S., Dunckley T., Beach T.G., Grover A., Mastroeni D., Ramsey K., Caselli R.J., Kukull W.A., McKeel D., Morris J.C., et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set. Physiol. Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.