Abstract
Genome-wide association studies (GWAS) have revealed 19 common genetic variants that are associated with breast cancer risk. Testing of the index signals found through GWAS and fine-mapping of each locus in diverse populations will be necessary for characterizing the role of these risk regions in contributing to inherited susceptibility. In this large study of breast cancer in African-American women (3016 cases and 2745 controls), we tested the 19 known risk variants identified by GWAS and replicated associations (P < 0.05) with only 4 variants. Through fine-mapping, we identified markers in four regions that better capture the association with breast cancer risk in African Americans as defined by the index signal (2q35, 5q11, 10q26 and 19p13). We also identified statistically significant associations with markers in four separate regions (8q24, 10q22, 11q13 and 16q12) that are independent of the index signals and may represent putative novel risk variants. In aggregate, the more informative markers found in the study enhance the association of these risk regions with breast cancer in African Americans [per allele odds ratio (OR) = 1.18, P = 2.8 × 10−24 versus OR = 1.04, P = 6.1 × 10−5]. In this detailed analysis of the known breast cancer risk loci, we have validated and improved upon markers of risk that better characterize their association with breast cancer in women of African ancestry.
INTRODUCTION
Genome-wide association studies (GWAS) of breast cancer have identified at least 19 chromosomal regions that harbor common alleles that contribute to genetic susceptibility (1–10). These discoveries have allowed for improved understanding of genetic risk for this common cancer, although it is argued that many more markers will be needed to elucidate disease heritability, and in the clinical setting for disease prediction (11–13). Except for the breast cancer risk locus at 6q25 identified in a GWAS of Chinese women, the risk loci for breast cancer have been revealed in studies in women of European ancestry. We have recently shown in a multiethnic study that a summary score comprised of the index variants at many of these risk loci is statistically significantly associated with breast cancer risk in multiple populations [odds ratio (OR) per allele of >1.10], but not in African Americans (14). Similar studies in African-American women have also reported lack of replication with many of the reported index signals (15–17). Limited statistical power of these initial reports as well as variation in both allele frequency and patterns of linkage disequilibrium (LD) across populations may be contributing factors as to why the associations found in the GWAS populations may not be generalizable to African Americans. Association testing of the risk variants as well as fine-mapping in a sufficiently large sample of African Americans will be needed to identify and localize the subset of markers that best define risk of the functional allele(s) within known risk regions.
In the present study, we tested common genetic variation at the breast cancer risk loci identified in women of European and Asian descent in a large sample comprised of 3016 African-American breast cancer cases and 2745 controls to identify markers of risk that are relevant to this population. More specifically, we examined the index variants and conducted fine-mapping of the locus to both improve the current set of risk markers in African Americans as well as to identify new risk variants for breast cancer. We then applied this information to model breast cancer risk in African-American women in an attempt to characterize the spectrum of genetic risk in this population defined by common variants at the known risk loci.
RESULTS
The ages of cases and controls ranged from 22 to 87 years and 23 to 86 years, respectively, with cases and controls having similar mean ages (55 and 58 years, respectively; Supplementary Material, Table S1).
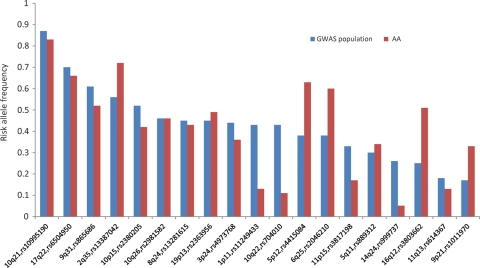
We tested 19 validated breast cancer risk variants (referred to as ‘index variants’ throughout the paper) at 1p11, 2q35, 3p24, 5p12, 5q11, 6q25, 8q24, 9p21, 9q31, 10p15, 10q21, 10q22, 10q26, 11p15, 11q13, 14q24, 16q12, 17q23 and 19p13 in models adjusted for age, study, global ancestry (the first 10 eigenvectors) and local ancestry (Table 1; Supplementary Material, Table S2) (1–10); 17 SNPs were directly genotyped, whereas 2 were imputed (r2> 0.98; see Materials and Methods). All 19 variants were common (≥0.05) in African Americans, with 11 variants being more common in Europeans than in African Americans (Table 1, Fig. 1). In previous GWAS, the index signals had modest ORs (1.05–1.29 per copy of the risk allele) and our sample size provided ≥70% statistical power to detect the reported effects for 12 of the 19 variants (at P < 0.05; Supplementary Material, Table S2).
Table 1.
Associations with common variants at known breast cancer risk regions in African Americans
| Chr., nearest genes | Index SNP from GWAS (3016 cases, 2745 controls) |
Best marker in African Americans (3016 cases, 2745 controls) |
|||
|---|---|---|---|---|---|
| Marker, position, alleles (risk/reference) | RAF in CEU/AAa, OR (95% CI), Ptrend | Marker, position, alleles (risk/reference) | RAF in CEU/AAa, OR (95% CI), Ptrend from stepwise analysis | r2 with index in CEU/YRIb | |
| 1p11 | rs11249433, 120982136, G/A | 0.43/0.13, 1.01 (0.90–1.14), 0.84 | |||
| 2q35 | rs13387042, 217614077, A/G | 0.56/0.72, 1.12 (1.03–1.21), 7.5 × 10−3 | rs13000023c, 217632639, G/A | 0.82/0.83, 1.20 (1.09–1.33), 5.8 × 10−4 | 0.35/0.53 |
| 3p24, NEK10 | rs4973768, 27391017, T/C | 0.44/0.36, 1.04 (0.96–1.13), 0.32 | |||
| 5p12, MRPS30 | rs4415084, 44698272, T/C | 0.38/0.63, 1.02 (0.95–1.11), 0.54 | |||
| 5q11, MAP3K1 | rs889312, 56067641, C/A | 0.30/0.34, 1.07 (0.99–1.18), 0.084 | rs16886165, 56058840, G/T | 0.16/0.31, 1.15 (1.06–1.25), 6.5 × 10−4 | 0.40/<0.01 |
| 6q25, C6orf97 | rs2046210c,d, 151990059, A/G | 0.38/0.60, 1.00 (0.93–1.09), 0.88 | |||
| 8q24 | rs13281615, 128424800, G/A | 0.45/0.43, 1.05 (0.97–1.13), 0.20 | |||
| 9p21, CDKN2B | rs1011970, 22052134, T/G | 0.17/0.33, 1.05 (0.97–1.14), 0.24 | |||
| 9q31 | rs865686, 109928199, T/G | 0.61/0.52, 1.08 (1.01–1.17), 0.034 | |||
| 10p15, ANKRD16 | rs2380205, 5926740, C/T | 0.52/0.42, 0.98 (0.91–1.06), 0.60 | |||
| 10q21, ZNF365 | rs10995190, 63948688, G/A | 0.87/0.83, 0.97 (0.88–1.08), 0.57 | |||
| 10q22, ZMIZ1 | rs704010, 80511154, T/C | 0.43/0.11, 0.99 (0.87–1.12), 0.83 | rs12355688, 80725632, T/C | 0.090/0.20, 1.24 (1.13–1.36), 6.8 × 10−6 | <0.01/<0.01 |
| 10q26, FGFR2 | rs2981582, 123342307, A/G | 0.46/0.46, 1.11 (1.03–1.19), 8.6 × 10−3 | rs2981578c, 123330301, C/T | 0.46/0.81, 1.24 (1.11–1.39), 1.7 × 10−4 | 0.66/0.059 |
| 11p15, LSP1 | rs3817198, 1865582, C/T | 0.33/0.17, 0.98 (0.88–1.08), 0.63 | |||
| 11q13 | rs614367, 69037945, T/C | 0.18/0.13, 0.96 (0.86–1.07), 0.45 | rs609275c, 69112096, C/T | 1.00/0.59, 1.20 (1.11–1.30), 1.0 × 10−5 | NA/<0.01 |
| 14q24, RAD51L1 | rs999737, 68104435, T/C | 0.26/0.051, 0.98 (0.82–1.17), 0.80 | |||
| 16q12, TNRC9 | rs3803662, 51143842, A/G | 0.25/0.51, 0.99 (0.92–1.08), 0.85 | rs3112572, 51157948, A/G | 0.020/0.20, 1.18 (1.08–1.30), 3.9 × 10−4 | 0.038/0.31 |
| 17q23, COX11 | rs6504950c, 50411470, G/A | 0.70/0.66, 1.05 (0.97–1.14), 0.19 | |||
| 19p13, ANKLE1 | rs2363956, 17255124, T/G | 0.45/0.49, 1.14 (1.05–1.22), 8.0 × 10−4 | rs3745185, 17245267, G/A | 0.52/0.75, 1.20 (1.10–1.32), 3.7 × 10−5 | 0.57/0.19 |
SNP positions are based on NCBI build 36.
ORs are per allele odds ratios adjusted for age, study, the first 10 eigenvectors and local ancestry at each risk locus.
Ptrend values are based on test of trend (1 d.f.).
aRAF, risk allele frequencies in the original GWAS population (HapMap CEU, or CHB for rs2046210) and AA (African American) controls in this study. Risk allele is the allele associated with increased risk in previous GWAS.
bPairwise correlations (r2) between the index signal and the best marker are from the CEU (CHB for rs2046210) and YRI populations in the 1000 Genomes Project (March 2010 release).
cImputed SNPs.
dIndex signal reported in Han Chinese. RAFs based on HapMap CHB and r2 based on CHB in the 1000 Genomes Project (March 2010 release).
Figure 1.
RAFs in Europeans and African Americans. The distribution of RAFs for the 19 index SNPs (from Table 1) in HapMap CEU (CHB for rs2046210) and African Americans (AA). The variants are sorted based on the RAF in the GWAS population.
We observed positive associations with 11 of the 19 variants (OR > 1); however, only 4 were statistically significant (P < 0.05 at 2q35, 9q31, 10q26 and 19p13; Table 1). Of the 15 variants that were not replicated at P < 0.05, statistical power was <70% for only 7 of the variants. Although power was more limited, we also evaluated associations by estrogen receptor (ER) status as some risk variants have been found to be more strongly associated with ER-positive (ER+) or ER-negative (ER−) breast cancer (2,18). We observed positive associations with 12 variants (2 at P < 0.05) for ER+ disease (n = 1520) and with 9 variants for ER− (3 at P < 0.05; n = 988) (Supplementary Material, Table S3). For only one variant did we observe statistically significant risk heterogeneity by ER status (rs13387042 at 2q35, P = 0.013) (Supplementary Material, Table S3).
Local ancestry was included in all models, as it was found to be associated with breast cancer risk in many regions (Supplementary Material, Table S4). We observed nominally significant associations between local ancestry and overall breast cancer, ER+ or ER− disease risk at 5 loci (5p12, 6q25, 8q24, 10p15, 10q26). The most statistically significant association was between European ancestry and ER+ breast cancer risk at 6q25 (OR per European allele chromosome = 1.19, P = 6.2 × 10−3). The inverse association observed between European ancestry and ER+ disease risk at 10q26 (OR per European chromosome = 0.85, P = 0.011) is consistent with previous reports of over-representation of African ancestry at this locus in many of these same cases (19,20).
Aside from statistical power, the lack of a statistically significant association with an index variant (OR > 1 and P < 0.05) suggests that the particular variant revealed in the GWAS populations may not be adequately correlated with the biologically relevant allele in African Americans. In an attempt to identify a better genetic marker of risk in African Americans, we conducted fine-mapping across all risk regions, using genotyped SNPs on the Illumina 1M array and imputed SNPs to Phase 2 HapMap populations (see Materials and Methods). If a marker associated with risk in African Americans represents the same signal as that reported in the initial GWAS, then it should be correlated to some degree with the index signal in the GWAS population. Using HapMap data for the populations in which the risk variant was identified [Utah residents with ancestry from northern and western Europe (CEU), or Han Chinese in Beijing, China (CHB)], we catalogued and tested all SNPs that were correlated (r2≥ 0.2) with the index signal (within 250 kb), applying an αa of 3.2 × 10−3 which was estimated to be 0.05 divided by the average number of tags needed to capture (r2≥ 0.8) the common risk alleles correlated with the index allele in each region in the Yoruba HapMap population [in Ibadan, Nigeria (YRI); Supplementary Material, Table S5]. We also tested for novel independent associations, focusing on SNPs that were uncorrelated with the index signal in the initial GWAS populations. Here, we applied a Bonferroni correction for defining novel associations as statistically significant in each region, with αb estimated to be 0.05 divided by the total number of tags needed to capture (r2≥ 0.8) all common risk alleles in the 19 regions in the YRI population (αb= 1.0 × 10−5; similar to the genome-wide-type correction of 5 × 10−8, which accounts for the number of tags needed to capture all common alleles in the genome; Supplementary Material, Table S5). For each region, stepwise logistic regression was used with SNPs kept in the final model based on αa or αb (results for each model are provided in Supplementary Material, Tables S6 and S7). These procedures were applied to all cases and controls as well as in hypothesis-generating analyses stratified by ER status.
At nine loci, we detected variants that were statistically significantly associated with breast cancer risk in African Americans. These regions include 9q31, where the sole marker of risk was the index signal (rs865686: OR = 1.08, P = 0.034; Table 1). In five of these nine regions, the index marker itself was not statistically significantly associated with disease risk. Through fine-mapping, we revealed markers in four regions that were more significantly associated with risk than the index signal (>1 order of magnitude change in the P-value) and are likely to capture the same signal (2q35, 5q11, 10q26 and 19p13). We also identified markers in four regions that are not correlated with the index signal in the GWAS populations (8q24, 10q22, 11q13 and 16q12) and may represent putative novel risk variants, with one being specific for ER+ disease (8q24) (Table 1, Fig. 2 and Supplementary Material, Table S8). These regions are discussed in what follows.
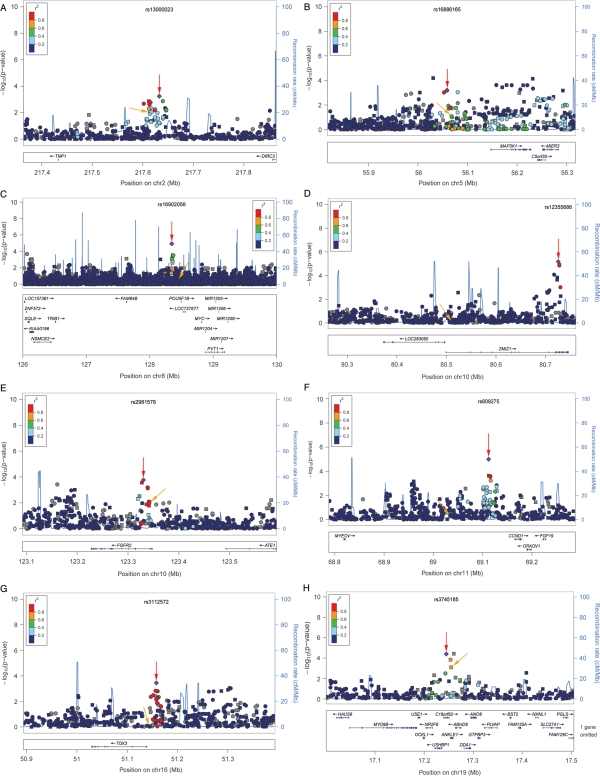
Figure 2.
–Log P plots for common alleles at eight breast cancer risk loci in African Americans. –Log P-values for risk-associated alleles in African Americans from logistic regression models adjusted for age, study, global ancestry (the first 10 eigenvectors) and local ancestry. P-values are for overall breast cancer risk except for 8q24, which is for ER+ breast cancer. Pairwise correlations (r2) in the HapMap CEU population are shown in relation to markers identified through fine-mapping in African Americans (diamond), except for 11q13, where r2 is shown in HapMap YRI as the marker is monomorphic in CEU. Squares denote genotyped SNPs; circles, imputed SNPs. Gray squares and circles denote that r2 cannot be estimated (not in HapMap or monomorphic in CEU). Red arrows denote markers identified in African Americans; yellow arrows, GWAS index variants. Each panel shows a –log P plot for common alleles for regions: (A) 2q35; (B) 5q11; (C) 8q24; (D) 10q22; (E)10q26; (F) 11q13; (G) 16q12; (H) 19p13. The plots were generated using LocusZoom (55).
Risk variants that better define the index signal in African Americans
2q35
The index signal at 2q35 was statistically significantly associated with risk of overall breast cancer (rs13387042: OR = 1.12, P = 7.5 × 10−3; Table 1) and ER+ disease (OR = 1.22, P = 2.6 × 10−4; Supplementary Material, Table S3). However, we found stronger associations with two markers that are each modestly correlated with the index signal in CEU and YRI: rs13000023 with overall breast cancer (OR = 1.20, P = 5.8 × 10−4) and rs12998806 with ER+ disease (OR = 1.39, P = 3.3 × 10−6) (Table 1 and Supplementary Material, Table S8). As shown in Supplementary Material, Figure S1, the signal in this region appeared limited to ER+ breast cancer, which is consistent with the initial report of this risk locus (2) but not with subsequent large-scale replication efforts in European populations (21).
5q11
We found a positive non-significant association with the index signal at 5q11, which is located 79 kb centromeric of the MAP3K1 gene (rs889312: OR = 1.07, P = 0.084; Table 1). Fine-mapping revealed statistically significant associations with markers, rs16886165 for overall breast cancer (OR = 1.15, P = 6.5 × 10−4) and rs832529 for ER− disease (OR = 1.22, P = 1.3 × 10−3; Table 1 and Supplementary Material, Table S8). These SNPs show greater correlation with the index signal in Europeans (CEU, r2= 0.40 and 0.46) than in Africans (YRI, r2< 0.01 and r2= 0.09), which suggests that they may be better markers of the biologically functional variant in African Americans (Table 1, Fig. 2).
10q26
Both the index signal, rs2981582 (OR = 1.11, P = 8.6 × 10−3; Table 1) and rs2981578, which was identified previously through fine-mapping in African Americans (which some of these studies contributed to) (22), were statistically significantly associated with risk (OR = 1.24, P = 1.7 × 10−4, Table 1). Variant rs2981578 was the most strongly associated marker in the region for overall breast cancer and for ER+ disease, which is consistent with previous reports of variation in this region being more strongly associated with ER+ breast cancer (Supplementary Material, Table S8) (18). In fine-mapping the locus, we observed a suggestive association with a correlated marker and ER− disease (rs2912774: OR = 1.19, P = 2.1 × 10−3; Supplementary Material, Table S8); however, the association was also noted with ER+ disease (OR = 1.10, P = 0.041; Supplementary Material, Table S9) and is likely to capture the same signal as rs2981578.
19p13
19p13 was the first risk locus reported to harbor a variant that may be specific for ER− disease (9). In African Americans, the index variant was statistically significantly associated with risk of overall breast cancer (rs2363956: OR = 1.14, P = 8.0 × 10−4), as well as ER+ (OR = 1.12, P = 0.016) and ER− disease (OR = 1.14, P = 0.018; Table 1 and Supplementary Material, Table S3). The most significant association in the region for overall breast cancer and ER+ disease was with rs3745185 (P = 3.7 × 10−5 and P = 8.2 × 10−4, respectively), which is likely to capture the same functional variant (r2= 0.57 in CEU and 0.19 in YRI; Table 1 and Supplementary Material, Table S8). The most significant marker for ER− breast cancer was correlated with both rs2363956 and rs3745185 (rs11668840: OR = 1.25, P = 5.1 × 10−5; Supplementary Material, Tables S8 and S10).
Novel risk-associated markers at breast cancer susceptibility loci
8q24
Given the importance of the 8q24 locus in cancer, we conducted association testing across the entire cancer risk region (126.0–130.0 Mb) (23–25). The index signal (rs13281615) was not statistically significantly associated with risk in African Americans (Table 1 and Supplementary Material, Table S3), nor did we identify significant associations with correlated SNPs. However, we did detect a significant association with rs16902056 and ER+ breast cancer [risk allele frequency (RAF) 0.95; P = 6.7 × 10−6; ER−: P = 0.66; Supplementary Material, Table S8]. This SNP is located 78 kb centromeric of the index variant and is not correlated with the index variant (r2< 0.01 in CEU and r2= 0.027 in YRI). No statistically significant associations were observed with variants found previously in association with cancers of the bladder and ovary, or leukemia (rs9642880: OR = 1.03, P = 0.58; rs10088218: OR = 1.02, P = 0.62; rs2456449: OR = 1.07, P = 0.14) (26–28). Of the known risk variants for prostate cancer (29–35), we found a single nominally significant (P < 0.05) association with the same risk allele of rs1016343 (P = 0.015) which is located >260 kb centromeric of the breast cancer risk region and is not correlated with rs13281615 or rs16902056.
10q22
We observed no association with the index signal at 10q22 (rs704010) which is located in intron 1 of the gene ZMIZ1, or with any correlated markers. However, we did detect strong evidence of a second signal located 215 kb telomeric in intron 12 of the gene ZMIZ1 (rs12355688: OR = 1.24, P = 6.8 × 10−6). As is shown in Table 1 and Figure 2, this putative novel risk variant is not correlated with the index variant in the CEU or YRI populations (r2< 0.01).
11q13
No positive association was noted with the index variant at 11q13. However, we did detect evidence of a second independent signal (rs609275: OR = 1.20, P = 1.0 × 10−5), located 74 kb telomeric, and 53 kb centromeric of CCND1. The variant is monomorphic and uncorrelated with the index signal in the CEU population; and r2 with the index signal in the YRI population is <0.01 (Table 1).
16q12
As in previous studies of African Americans, we were not able to replicate the association signal defined by the index variant rs3803662 (Table 1) (15,16). A recent study of African Americans reported a suggestive association with SNP rs3104746, which is located 15 kb telomeric of rs3803662 (16). This SNP has a minor allele frequency (MAF) of 0.04 in the HapMap CEU population, 0.19 in our African-American controls, and is modestly correlated with rs3803662 in Africans (r2= 0.31 in YRI), but not in Europeans (r2= 0.038; Supplementary Material, Table S10). Fine-mapping around this putative signal revealed a perfect proxy (r2= 1) for rs3104746, rs3112572, which is significantly associated with breast cancer risk in African Americans (OR = 1.18, P = 3.9 × 10−4), with the association noted to be stronger for ER+ breast cancer (OR = 1.27, P = 3.1 × 10−5; Table 1 and Supplementary Material, Table S8).
For index SNPs found to be nominally associated with breast cancer risk, as well as risk-associated markers identified through fine-mapping, we also tested for associations by genotype. Results from the genotype-specific model were consistent with log-additive associations (Supplementary Material, Tables S9 and S11). Risk variants at 2q35 and 8q24 were also found to have significantly stronger associations with ER+ breast cancer than ER− disease (Supplementary Material, Table S7), which is consistent with previous studies (2,18).
We observed no statistically significant associations with common variation at 10 risk loci on 1p11, 3p24, 5p12, 6q25, 9p21, 10p15, 10q21, 11p15, 14q24 and 17q23 (Supplementary Material, Fig. S2). We also could not replicate the association with the recently identified SNP rs9397435 at 6q25 that was found through fine-mapping in European, African and Asian population samples (17) (P = 0.26 for overall breast cancer, P = 0.71 for ER+ and P = 0.36 for ER− tumor subtypes). Neither could we replicate the association with SNP rs4784227 at 16q12, which was identified by a recent multi-stage GWAS in women of Asian ancestry (36) in our African-American sample (P = 0.51 overall, P = 0.35 and P = 0.65 for ER+ and ER− subtypes, respectively).
Risk modeling
We next estimated the cumulative effect of all breast cancer risk variants, and compared a summary risk score comprised of unweighted counts of all GWAS-reported risk variants with a risk score that included variants we identified as being associated with risk in African Americans (Table 2). Using the 19 index signals from GWAS (see Materials and Methods), the risk per allele was 1.04 [95% confidence interval (CI) 1.02–1.06; P = 6.1 × 10−5], and individuals in the top quintile of the risk allele distribution were at 1.4-fold greater risk (P = 7.4 × 10−5) of breast cancer compared with those in the lowest quintile (Table 2). As expected, the risk score was improved when utilizing the markers that we identified at the known risk loci as being more relevant to African Americans (eight markers for overall breast cancer: 2q35, 5q11, 9q31, 10q22, 10q26, 11q13, 16q12 and 19p13; OR = 1.18; 95% CI 1.14–1.22; P = 2.8 × 10−24), with risk for those in the top quartile being 2.2 times that observed in the lowest quintile (P = 3.6 × 10−17). This score was significantly associated with risk of both ER+ (OR = 1.20, P = 1.7 × 10−19) and ER− (OR = 1.15, P = 2.8 × 10−9) disease (Phet= 0.12) (Supplementary Material, Table S12).
Table 2.
The association of the total risk score with breast cancer risk in African Americans
| Index markers from GWAS (19 markers) | Risk-associated best markers in African Americansa (8 markers) |
|||
|---|---|---|---|---|
| Mean number of risk alleles in controls (range) | 15.7 (6–25) | 8.4 (3–14) | ||
| Per allele OR (95% CI) | 1.04 (1.02–1.06) | 1.18 (1.14–1.22) | ||
| Ptrend | 6.1 × 10−5 | 2.8 × 10−24 | ||
| First-degree family history negativeb | First-degree family history positiveb | |||
| Subjects, n cases/n controls | 3016/2745 | 3016/2745 | 2387/2349 | 554/303 |
| Risk quintilesc | ||||
| Q1 | ||||
| n cases/n controls | 536/549 | 352/462 | 281/387 | 62/57 |
| OR (95%CI) | 1.00 (ref.) | 1.00 (ref.) | 1.00 (ref.) | 1.58 (1.06–2.37) |
| P-value | — | — | — | 0.025 |
| Q2 | ||||
| n cases/n controls | 722/742 | 430/505 | 344/437 | 77/47 |
| OR (95% CI) | 0.99 (0.84–1.16) | 1.17 (0.96–1.42) | 1.15 (0.93–1.43) | 2.18 (1.46–3.26) |
| P-value | 0.88 | 0.11 | 0.18 | 1.5 × 10−4 |
| Q3 | ||||
| n cases/n controls | 435/382 | 632/625 | 503/549 | 115/53 |
| OR (95%CI) | 1.15 (0.96–1.39) | 1.37 (1.14–1.64) | 1.31 (1.07–1.60) | 3.14 (2.17–4.53) |
| P-value | 0.14 | 7.2 × 10−4 | 8.0 × 10−3 | 1.2 × 10−9 |
| Q4 | ||||
| n cases/n controls | 753/669 | 665/566 | 517/476 | 132/75 |
| OR (95%CI) | 1.16 (0.98–1.36) | 1.56 (1.30–1.87) | 1.51 (1.24–1.86) | 2.52 (1.81–3.52) |
| P-value | 0.080 | 2.3 × 10−6 | 6.2 × 10−5 | 4.0 × 10−8 |
| Q5 | ||||
| n cases/n controls | 570/403 | 937/587 | 742/500 | 168/71 |
| OR (95%CI) | 1.44 (1.20–1.72) | 2.16 (1.80–2.58) | 2.11 (1.73–2.56) | 3.44 (2.47–4.77) |
| P-value | 7.4 × 10−5 | 3.6 × 10−17 | 1.3 × 10−13 | 9.9 × 10−14 |
ORs are adjusted for age, study and the first 10 eigenvectors.
Ptrend values are based on test of trend (1 d.f.).
aThe most significant markers from the stepwise analysis for overall breast cancer in each region from Table 1.
bInformation about first-degree family history of breast cancer is available on 97.5% of cases and 96.6% of controls.
cBased on distribution in controls (cut points for index markers aggregate: 13.3, 15, 16, 18; cut points for best markers aggregate: 7, 8, 9, 10).
Stratifying by first-degree family history of breast cancer differentiated risk further with those with a family history and in the top quintile of the risk score distribution (4% of the population) having a 3.4-fold greater risk (P = 9.9 × 10−14) compared with those without a family history and in the lowest quintile of the risk score (Table 2).
In hypothesis-generating analyses, we also developed risk scores for ER+ and ER− breast tumor subtypes, utilizing the most informative markers revealed through fine-mapping of each phenotype. These phenotype-specific scores were highly significant (ER+: OR = 1.30, P = 6.0 × 10−18; ER−: OR = 1.20, P = 2.3 × 10−10) with statistically significant heterogeneity noted when the scores were applied to the other subtype (Phet= 1.7 × 10−5 and 5.0 × 10−3 for ER+ and ER− scores, respectively) (Supplementary Material, Table S12).
DISCUSSION
In this large study of breast cancer in African-American women, we were able to replicate associations with 4 of the 19 index variants (at P < 0.05). Through fine-mapping, we observed that overall breast cancer risk was statistically significantly associated with markers in four regions which are likely to capture the GWAS-reported signal and to serve as better markers of the functional allele and risk in African Americans. We also detected putative novel associations that are independent of the index signals in three regions for overall breast cancer (10q22, 11q13 and 16q12) and in one region for ER+ disease (8q24). In 10 of the risk regions, however, we were not able to replicate the GWAS index signals, nor did we detect statistically significant associations of common SNPs with breast cancer risk at the levels of statistical significance we set for fine-mapping. The inability to replicate associations with the index signals despite adequate statistical power (>70% power for 12 of 19 variants) suggests that they are unlikely to be functional variants or capture the functional variants as efficiently in this population. Our ability to find associated markers in five regions where index signals were not significantly associated with risk also demonstrates the value of testing common variation at GWAS-identified risk loci in additional populations (14,16,17,22,37,38).
In four regions, we observed risk markers that are correlated with, and in the same LD block as the index markers in CEU (rs13000023 at 2q35, rs16886165 at 5q11, rs2981578 at 10q26 and rs3745185 at 19p13). It is likely that these risk markers capture the same signal as defined by the index markers based on the r2 values between these markers and the index markers (≥0.35). We cannot rule out the possibility, though, that some of them may represent a second, independent signal in the same region.
In the four regions where we observed independent signals, the risk alleles (rs16902056 at 8q24, rs12355688 at 10q22, rs609275 at 11q13 and rs3112572 at 16q12) were uncorrelated with, and not in, the same LD block as the index variant in Europeans (CEU, r2< 0.04)) (distances from the index signal ranged from 14 kb at 16q12 to 215 kb at 10q22) (Supplementary Material, Fig. S3). Therefore, these variants are likely to pick up a novel signal independent of the index signal. However, because of different LD patterns in European and African ancestry populations, they may each mark the same functional variant, and if the functional variant is less common it may not be well captured by either common marker alone. At 10q22, both the index SNP and the novel variant are located within introns of the ZMIZ1 gene. ZMIZ1 encodes zinc finger MIZ-type containing 1, which regulates the activity of various transcription factors (39–41). At 11q13, rs609275 lies 74kb telomeric of the index signal and in closer proximity to a number of candidate genes, including CCND1 (encoding cyclin D1, a protein crucial for cell-cycle control), ORAOV1 (encoding oral cancer overexpressed 1) and FGF19 (encoding fibroblast growth factor 19). The association at 16q12 confirms the findings of a previous, smaller study of African Americans (16), and is consistent with a previous fine-mapping study suggesting that African Americans may harbor a separate causal variant in this region (42). Whether this variant is influencing the same genes/pathways as the index variant rs3803662 is not known; however, the stronger associations noted for both variants with ER+ disease (2,18) suggest that they may affect the same biological process.
Notably, at region 19p13, which was originally reported in association with ER− breast cancer (9), the index signal was statistically significantly associated with both ER+ and ER− subtypes in African Americans. In addition, we found a stronger marker in this region (rs3745185) for ER+ as well as overall breast cancer risk (Table 1 and Supplementary Material, Table S8). We also found stronger associations with ER+ than ER− disease for variants in many regions, including 2q35, 8q24, 10q26 and 16q12, which is consistent with previous reports (2,18). In the study, we also found strong signals for ER− disease in regions 5q11, 10q26 and 19p13. It is possible that these signals may explain some of the excess risk for ER− disease in African Americans, since these risk alleles have higher frequencies in this population than they do in European-ancestry populations. However, our understanding of their contribution to racial and ethnic differences in disease incidence will only be determined once the functional variants have been identified and tested across populations. Unfortunately, we were not able to assess associations with triple-negative (ER/PR/HER2-negative; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2) breast cancer, since HER2 status was available for only a limited number of cases. However, in a large study of women of European ancestry which tested many of these same index variants, further stratification on tumor subtype using HER2 status was not additionally informative for ER/PR-negative breast cancer (43).
The observation of secondary signals at many loci, and associations of variants with different tumor subtypes that have not yet been reported in European-ancestry populations could indicate a different genetic architecture of breast cancer across populations. For example, the index signal at TNRC9 does not replicate in African Americans, but there appears to be a second risk variant that is unique to this population. At FGFR2, which was originally reported to be associated with ER+ disease in women of European ancestry, we found a signal for ER− disease with a marker correlated with the index variant. Similarly, for chromosome 19p13, which was reported as an ER− locus, we observed an association with ER+ breast cancer. However, these findings and their implications require further validation.
We investigated local ancestry as a potential confounding factor in the analysis of each risk locus. At five loci, we observed nominally significant evidence of association between local ancestry and breast cancer risk, with the most statistically significant association observed at 6q25 between European ancestry and ER+ breast cancer risk. Although the association of local ancestry and breast cancer risk needs to be validated in additional large studies, the inability to identify a risk variant that is differentiated in frequency between populations of European and African ancestry implies that either the association with local ancestry at many regions is a false-positive signal and/or we have not tested an adequate surrogate of the functional alleles.
The majority of the variants identified by GWAS for common cancers are of low risk (relative risks <1.30) and in aggregate are not yet informative for risk prediction (11–13). Until the functional alleles at each susceptibility locus are identified and their effects are accurately estimated, modeling of the genetic risk will rely on markers that best capture risk for a given population. Many of the markers we identified at these risk loci appear to have stronger associations with breast cancer risk compared with the GWAS-identified variants in African-American women. The risk score for overall breast cancer was also equally efficient for ER+ and ER− tumors. However, our hypothesis-generating model suggests that identification of tumor subtype-specific variants will improve the fit of these models.
While this is the largest study of African Americans to date to investigate genetic risk at known breast cancer susceptibility loci, statistical power was still limited. We had only 35% power to detect an OR of 1.10 for a risk allele of 0.10 frequency which may account for our inability to replicate GWAS signals or risk-associated markers in 10 of the regions. While attempting to apply a strict threshold for declaring significance through fine-mapping, we did not take into account testing for multiple phenotypes (overall breast as well as ER+ and ER− disease). As a result, the α-levels used as selection criteria may be too liberal. However, our risk modeling focused on the variants revealed for overall breast cancer, whereas we consider the associations observed for markers identified for ER+ or ER− disease and used in the subtype-specific risk modeling as hypothesis-generating. Since all of the cases and controls used for fine-mapping/discovery were also included in the risk modeling, the risk model is likely to over-estimate the level of association due to winner's curse. Instead of partitioning the sample into test and validation sets, we felt it was necessary to use all of the subjects in the association testing of known variants and in fine-mapping to increase the statistical power to detect associations in each region. Therefore, other studies with reasonable power in African Americans must be performed in the future to test the model presented.
In summary, through fine-mapping of the breast cancer susceptibility regions in a large sample of African-American women, we identified markers with enhanced association with breast cancer in this population. Validation and augmentation of this model are needed before risk modeling based on genetic variants of low risk can be implemented in the clinical setting.
MATERIALS AND METHODS
Ethics statement
The Institutional Review Board at the University of Southern California approved the study protocol.
Study populations
This study included 9 epidemiological studies of breast cancer among African-American women, which comprise a total of 3153 cases and 2831 controls. Sample size and selected characteristics for these studies are summarized in Supplementary Material, Table S1. What follows is a brief description of these studies.
The Multiethnic Cohort Study (MEC)
The MEC is a prospective cohort study of 215 000 men and women in Hawaii and Los Angeles (44) between the ages of 45 and 75 years at baseline (1993–1996). Through 31 December 2007, a nested breast cancer case–control study in the MEC included 556 African-American cases (544 invasive and 12 in situ) and 1003 African-American controls. An additional 178 African-American breast cancer cases (ages: 50–84) diagnosed between 1 June 2006 and 31 December 2007 in Los Angeles County (but outside of the MEC) were included in the study.
The Los Angeles component of The Women's Contraceptive and Reproductive Experiences (CARE) Study
The CARE Study is a large multi-center, population-based case–control study that was designed to examine the effects of oral contraceptive use on invasive breast cancer risk among African-American women and white women aged 35–64 years in five US locations (45). Cases in Los Angeles County were diagnosed from 1 July 1994 through 30 April 1998, and controls were sampled by random-digit dialing (RDD) from the same population and time period; 380 African-American cases and 224 African-American controls were included in the study.
The Women's Circle of Health Study (WCHS)
The WCHS is an ongoing case–control study of breast cancer among European women and African-American women in the New York City boroughs and in seven counties in New Jersey (46). Eligible cases included women with invasive breast cancer between 20 and 74 years of age; controls were identified through RDD. The WCHS contributed 272 invasive African-American cases and 240 African-American controls.
The San Francisco Bay Area Breast Cancer Study (SFBCS)
The SFBCS is a population-based case–control study of invasive breast cancer in Hispanic, African-American and non-Hispanic white women conducted between 1995 and 2003 in the San Francisco Bay Area (47). African-American cases, aged 35–79 years, were diagnosed between 1 April 1995 and 30 April 1999, with controls identified through RDD. Included from this study were 172 invasive African-American cases and 231 African-American controls.
The Northern California Breast Cancer Family Registry (NC-BCFR)
The NC-BCFR is a population-based family study conducted in the Greater San Francisco Bay Area, and one of six sites of the Breast Cancer Family Registry (BCFR) (48). African-American breast cancer cases in NC-BCFR were diagnosed after 1 January 1995 and between the ages of 18 and 64 years; population controls were identified through RDD. Genotyping was conducted for 440 invasive African-American cases and 53 African-American controls.
The Carolina Breast Cancer Study (CBCS)
The CBCS is a population-based case–control study conducted between 1993 and 2001 in 24 counties of central and eastern North Carolina (49). Cases were identified by rapid case ascertainment system in cooperation with the North Carolina Central Cancer Registry, and controls were selected from the North Carolina Division of Motor Vehicle and United States Health Care Financing Administration beneficiary lists. Participants' ages ranged from 20 to 74 years. DNA samples were provided from 656 African-American cases with invasive breast cancer and 608 African-American controls.
The Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) Cohort
PLCO, coordinated by the US National Cancer Institute (NCI) in 10 US centers, enrolled approximately 155 000 men and women aged 55–74 years during 1993–2001 in a randomized, two-arm trial to evaluate the efficacy of screening for these four cancers (50). A total of 64 African-American invasive breast cancer cases and 133 African-American controls contributed to this study.
The Nashville Breast Health Study (NBHS)
The NBHS is a population-based case–control study of incident breast cancer conducted in Tennessee (15). The study was initiated in 2001 to recruit patients with invasive breast cancer or ductal carcinoma in situ, and controls, recruited through RDD between the ages of 25 and 75 years. NBHS contributed 310 African-American cases (57 in situ) and 186 African-American controls.
Wake Forest University Breast Cancer Study (WFBC)
African-American breast cancer cases and controls in WFBC were recruited at Wake Forest University Health Sciences from November 1998 through December 2008 (51). Controls were recruited from the patient population receiving routine mammography at the Breast Screening and Diagnostic Center. Age range of participants was 30–86 years. WFBC contributed 125 cases (116 invasive and 9 in situ) and 153 controls to the analysis.
Genotyping and quality control
Genotyping in stage 1 was conducted using the Illumina Human1M-Duo BeadChip. Of the 5984 samples from these studies (3153 cases and 2831 controls), we attempted genotyping of 5932, removing samples (n = 52) with DNA concentrations <20 ng/μl. Following genotyping, we removed samples based on the following exclusion criteria: (i) unknown replicates (≥98.9% genetically identical) that we were able to confirm (only one of each duplicate was removed, n = 15); (ii) unknown replicates that we were not able to confirm through discussions with study investigators (pair or triplicate removed, n = 14); (iii) samples with call rates <95% after a second attempt (n = 100); (iv) samples with ≤5% African ancestry (n = 36) (discussed in what follows); and (v) samples with <15% mean heterozygosity of SNPs on the X chromosome and/or similar mean allele intensities of SNPs on the X and Y chromosomes (n = 6) (these are likely to be males).
In the analysis, we removed SNPs with <95% call rates (n = 21 732) or MAFs <1% (n = 80 193). To assess genotyping reproducibility, we included 138 replicate samples; the average concordance rate was 99.95% (>99.93% for all pairs). We also eliminated SNPs with genotyping concordance rates <98% based on the replicates (n = 11 701). The final analysis data set included 1 043 036 SNPs genotyped on 3016 cases (1520 ER+, 988 ER− and the remaining 508 cases with unknown ER status) and 2745 controls, with an average SNP call rate of 99.7% and average sample call rate of 99.8%.
Statistical analysis
Ancestry estimation
We used principal components analysis (52) to estimate global ancestry among the 5761 individuals, using 2546 ancestry informative markers. Eigenvector 1 was highly correlated (ρ = 0.997, P < 1 × 10−16) with percentage of European ancestry, estimated in HAPMIX (53), and accounted for 10.1% of the variation between subjects; subsequent eigenvectors accounted for no more than 0.5%. At each locus and for each participant, we also estimated local ancestry [i.e. the number of European chromosomes (continuous between 0 and 2) carried by the participant], using the HAPMIX program (53). To summarize local ancestry at each region, for each individual we averaged across all local ancestry estimates that were within the start and end points of the region (Supplementary Material, Table S5). To address the potential for confounding by genetic ancestry, we adjusted for both global and local ancestry in all analyses.
SNP imputation
In order to generate a data set suitable for fine-mapping, we carried out genome-wide imputation using the software MACH (54). Phased haplotype data from the founders of the CEU and YRI HapMap Phase 2 samples were used to infer LD patterns in order to impute ungenotyped markers. The r2 metric, defined as the observed variance divided by the expected variance, provides a measure of the quality of the imputation at any SNP, and was used as a threshold in determining which SNPs to filter from analysis (r2< 0.3). Of the 1 539 328 common SNPs (MAF ≥ 0.05) in the YRI population in HapMap Phase 2, we could impute 1 392 294 (90%) with r2≥ 0.8. For all the imputed SNPs presented in Results and the tables reported herein, the average r2 was 0.92 (estimated in MACH).
Association testing
For each typed and imputed SNP, ORs and 95% CIs were estimated using unconditional logistic regression adjusting for age at diagnosis (or age at the reference date for controls), study, the first 10 eigenvalues and local ancestry. For each SNP, we tested for allele dosage effects through a 1 d.f. Wald χ2 trend test.
We fine-mapped each risk locus using the combined genotyped and imputed SNPs in search of (i) an SNP that is more associated with risk in African Americans than the index signal; and (ii) a novel signal that is independent of the index signal. As some risk loci have been found to be more strongly associated with breast cancer subtypes, we investigated three outcomes: (i) overall breast cancer, (ii) ER+ breast cancer, and (iii) ER− breast cancer, with the latter two being hypothesis-generating. These analyses included SNPs (genotyped and imputed) spanning 250 kb upstream and 250 kb downstream of each index signal. If the index signal was contained within an LD block (based on the D′ statistic) of >250 kb, then the region was extended to include the entire region of LD.
Stepwise regression was performed by region to select the most informative risk variants as discussed in what follows, in models adjusted for age, study, global ancestry (the first 10 eigenvectors) and local ancestry. In the stepwise regression, we preserved the original sample size by using the mean genotype of typed subjects in place of ‘no-calls’ for SNPs with <100% genotyping completion rate.
Within each known risk locus, it is expected that markers that are associated with risk in African Americans will be correlated with the index signal reported in Europeans. Thus, we identified and tested SNPs that are correlated (r2> 0.2) with the index signals in the GWAS populations (HapMap CEU or CHB for 6q25). For each region, we determined the number of tags needed to capture all the SNPs correlated with the index signal in the YRI population (Phase 2 HapMap). The average number of tags in each region was then used as the correction factor for Bonferroni correction. An α-level of 0.05 divided by average number of tags needed in each region was applied in the stepwise regression process. For all of the remaining markers that were not correlated with the index signal (in Europeans), we applied a more stringent α-level for defining statistical significance. In each risk region, we determined the number of tag SNPs needed to capture all common alleles (MAF > 0.05, with r2> 0.8) in the YRI HapMap population. The total number of tags across the 19 regions was then used as a correction factor, as they define the number of independent tests in each region. An α of 0.05 divided by the number of tags was applied to assess statistical significance for any putative novel, independent signal in each region. For correlated SNPs that were selected to be better markers, we also assessed phase to ensure that the new risk allele is on the same haplotype as the GWAS-reported risk allele in the HapMap CEU population.
Risk modeling
We modeled the cumulative genetic risk of breast cancer using the risk variants reported in previous GWAS (total = 19). We compared the results with a model of the SNPs found to be significantly associated with risk in African Americans, which included SNPs identified from the stepwise procedures at all loci for overall breast cancer risk (presented in Table 1). More specifically, in each case we summed the number of risk alleles for each individual and estimated the OR per allele for this aggregate-unweighted allele count variable as an approximate risk score appropriate for unlinked variants with independent effects of approximately the same magnitude for each allele. We then applied this risk score to overall breast cancer as well as ER+/ER− breast cancer subtypes. We also constructed risk scores based on risk alleles for ER+ and ER− tumor subtypes separately, and, as hypothesis-generating, applied both risk scores to overall and ER+/ER− breast cancer subtypes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by a Department of Defense Breast Cancer Research Program Era of Hope Scholar Award to C.A.H. (W81XWH-08-1-0383), a National Institute of Health grant to C.A.H. (R01-CA132839), the Norris Foundation, and a grant from the California Breast Cancer Research Program to D.O.S. (15UB-8402). Each of the participating studies was supported by the following grants: MEC: by National Institutes of Health (R01-CA63464 and R37-CA54281); CARE: by National Institute for Child Health and Development (NO1-HD-3-3175), WCHS: by US Army Medical Research and Material Command (USAMRMC) (DAMD-17-01-0-0334); the National Institutes of Health (R01-CA100598); and the Breast Cancer Research Foundation; SFBCS: by National Institutes of Health (R01-CA77305) and United States Army Medical Research Program (DAMD17-96-6071); NC-BCFR: by National Institutes of Health (U01-CA69417); CBCS: by National Institutes of Health Specialized Program of Research Excellence in Breast Cancer (P50-CA58223) and Center for Environmental Health and Susceptibility, National Institute of Environmental Health Sciences, National Institutes of Health (P30-ES10126); PLCO: by Intramural Research Program, National Cancer Institute, National Institutes of Health; NBHS: by National Institutes of Health (R01-CA100374); WFBC: by National Institutes of Health (R01-CA73629). The Breast Cancer Family Registry (BCFR) was supported by the National Cancer Institute, National Institutes of Health under (RFA CA-95-011) and through cooperative agreements with members of the Breast Cancer Family Registry and Principal Investigators.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the women who volunteered to participate in each study. We also thank Madhavi Eranti, Andrea Holbrook, Paul Poznaik, Loreall Pooler, Xin Sheng and David Wong from the University of Southern California for their technical support. We would also like to acknowledge co-investigators from the WCHS study: Dana H. Bovbjerg (University of Pittsburgh), Lina Jandorf (Mount Sinai School of Medicine) and Gregory Ciupak, Warren Davis, Gary Zirpoli, Song Yao and Michelle Roberts from Roswell Park Cancer Institute.
REFERENCES
- 1.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. doi:10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A., et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. doi:10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 3.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Huntchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey S.N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S.A., Jonsson G.F., Jakobsdottir M., Bergthorsson J.T., Gudmundsson J., Aben K.K., et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. doi:10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 5.Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L., et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. doi:10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas G., Jacobs K.B., Kraft P., Yeager M., Wacholder S., Cox D.G., Hankinson S.E., Hutchinson A., Wang Z., Yu K., et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. doi:10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed S., Thomas G., Ghoussaini M., Healey C.S., Humphreys M.K., Platte R., Morrison J., Maranian M., Pooley K.A., Luben R., et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. doi:10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbull C., Shahana A., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S., et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. doi:10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoniou A.C., Wang X., Fredericksen Z.S., McGuffog L., Tarrell R., Sinilnikova O.M., Healey S., Morrison J., Kartsonaki C., Lesnick T., et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet. 2010;42:885–892. doi: 10.1038/ng.669. doi:10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher O., Johnson N., Orr N., Hosking F.J., Gibson L.J., Walker K., Zelenika D., Gut I., Heath S., Palles C., et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J. Natl Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. doi:10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 11.Pepe M.S., Janes H.E. Gauging the performance of SNPs, biomarkers, and clinical factors for predicting risk of breast cancer. J. Natl Cancer Inst. 2008;100:978–979. doi: 10.1093/jnci/djn215. doi:10.1093/jnci/djn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gail M.H. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J. Natl Cancer Inst. 2008;100:1037–1041. doi: 10.1093/jnci/djn180. doi:10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pharoah P.D., Antoniou A., Bobrow M., Zimmern R.L., Easton D.F., Ponder B.A. Polygenic susceptibility to breast cancer and implications for prevention. Nat. Genet. 2002;31:33–36. doi: 10.1038/ng853. doi:10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 14.Chen F., Stram D.O., Le Marchand L., Monroe K.R., Kolonel L.N., Henderson B.E., Haiman C.A. Caution in generalizing known genetic risk markers for breast cancer across all ethnic/racial populations. Eur. J. Hum. Genet. 2010;19:243–245. doi: 10.1038/ejhg.2010.185. doi:10.1038/ejhg.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng W., Cai Q., Signorello L.B., Long J., Hargreaves M.K., Deming S.L., Li G., Li C., Cui Y., Blot W.J. Evaluation of 11 breast cancer susceptibility loci in African-American women. Cancer Epidemiol. Biomarkers Prev. 2009;18:2761–2764. doi: 10.1158/1055-9965.EPI-09-0624. doi:10.1158/1055-9965.EPI-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz-Narvaez E.A., Rosenberg L., Cozier Y.C., Cupples L.A., Adams-Campbell L.L., Palmer J.R. Polymorphisms in the TOX3/LOC643714 locus and risk of breast cancer in African-American women. Cancer Epidemiol. Biomarkers Prev. 2010;19:1320–1327. doi: 10.1158/1055-9965.EPI-09-1250. doi:10.1158/1055-9965.EPI-09-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stacey S.N., Sulem P., Zanon C., Gudjonsson S.A., Thorleifsson G., Helgason A., Jonasdottir A., Besenbacher S., Kostic J.P., Fackenthal J.D., et al. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6:e1001029. doi: 10.1371/journal.pgen.1001029. doi:10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Closas M., Hall P., Nevanlinna H., Pooley K., Morrison J., Richesson D.A., Bojesen S.E., Nordestgaard B.G., Axelsson C.K., Arias J.I., et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. doi:10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasaniuc B., Zaitlen N., Lettre G., Chen G., Tandon A., Kao L., Ruczinski I., Fornage M., Siscovick D., Zhu X., et al. Enhanced statistical tests for GWAS in admixed populations: assessment using African Americans from CARe and a breast cancer consortium. PLoS Genet. 2011;7:e1001371. doi: 10.1371/journal.pgen.1001371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fejerman L., Haiman C.A., Reich D., Tandon A., Deo R.C., John E.M., Ingles S.A., Ambrosone C.B., Bovbjerg D.H., Jandorf L.H., et al. An admixture scan in 1484 African American women with breast cancer. Cancer Epidemiol. Biomarkers Prev. 2009;18:3110–3117. doi: 10.1158/1055-9965.EPI-09-0464. doi:10.1158/1055-9965.EPI-09-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milne R.L., Benitez J., Nevanlinna H., Heikkinen T., Aittomaki K., Blomqvist C., Arias J.I., Zamora M.P., Burwinkel B., Bartram C.R., et al. Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J. Natl Cancer Inst. 2009;101:1012–1018. doi: 10.1093/jnci/djp167. doi:10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Udler M.S., Meyer K.B., Pooley K.A., Karlins E., Struewing J.P., Zhang J., Doody D.R., MacArthur S., Tyrer J., Pharoah P.D., et al. FGFR2 variants and breast cancer risk: fine-scale mapping using African American studies and analysis of chromatin conformation. Hum. Mol. Genet. 2009;18:1692–1703. doi: 10.1093/hmg/ddp078. doi:10.1093/hmg/ddp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia L., Landan G., Pomerantz M., Jaschek R., Herman P., Reich D., Yan C., Khalid O., Kantoff P., Oh W., et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. doi:10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freedman M.L. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. doi:10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghoussaini M., Song H., Koessler T., Al Olama A.A., Kote-Jarai Z., Driver K.E., Pooley K.A., Ramus S.J., Kjaer S.K., Hogdall E., et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. doi:10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiemeney L.A., Thorlacius S., Sulem P., Geller F., Aben K.K., Stacey S.N., Gudmundsson J., Jakobsdottir M., Bergthorsson J.T., Sigurdsson A., et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. doi:10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goode E.L., Chenevix-Trench G., Song H., Ramus S.J., Notaridou M., Lawrenson K., Widschwendter M., Vierkant R.A., Larson M.C., Kjaer S.K., et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet. 2010;42:874–879. doi: 10.1038/ng.668. doi:10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crowther-Swanepoel D., Broderick P., Di Bernardo M.C., Dobbins S.E., Torres M., Mansouri M., Ruiz-Ponte C., Enjuanes A., Rosenquist R., Carracedo A., et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat. Genet. 2010;42:132–136. doi: 10.1038/ng.510. doi:10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salinas C.A., Kwon E., Carlson C.S., Koopmeiners J.S., Feng Z., Karyadi D.M., Ostrander E.A., Stanford J.L. Multiple independent genetic variants in the 8q24 region are associated with prostate cancer risk. Cancer Epidemiol. Biomarkers Prev. 2008;17:1203–1213. doi: 10.1158/1055-9965.EPI-07-2811. doi:10.1158/1055-9965.EPI-07-2811. [DOI] [PubMed] [Google Scholar]
- 30.Gudmundsson J., Sulem P., Gudbjartsson D.F., Blondal T., Gylfason A., Agnarsson B.A., Benediktsdottir K.R., Magnusdottir D.N., Orlygsdottir G., Jakobsdottir M., et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. doi:10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gudmundsson J., Sulem P., Manolescu A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. doi:10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 32.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. doi:10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 33.Yeager M., Chatterjee N., Ciampa J., Jacobs K.B., Gonzalez-Bosquet J., Hayes R.B., Kraft P., Wacholder S., Orr N., Berndt S., et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. doi:10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al Olama A.A., Kote-Jarai Z., Giles G.G., Guy M., Morrison J., Severi G., Leongamornlert D.A., Tymrakiewicz M., Jhavar S., Saunders E., et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. doi:10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 35.Haiman C.A., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. doi:10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long J., Cai Q., Shu X.O., Qu S., Li C., Zheng Y., Gu K., Wang W., Xiang Y.B., Cheng J., et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet. 2010;6:e1001002. doi: 10.1371/journal.pgen.1001002. doi:10.1371/journal.pgen.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters K.M., Stram D.O., Hassanein M.T., Le Marchand L., Wilkens L.R., Maskarinec G., Monroe K.R., Kolonel L.N., Altshuler D., Henderson B.E., et al. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6:e1001078. doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waters K.M., Le Marchand L., Kolonel L.N., Monroe K.R., Stram D.O., Henderson B.E., Haiman C.A. Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol. Biomarkers Prev. 2009;18:1285–1289. doi: 10.1158/1055-9965.EPI-08-1142. doi:10.1158/1055-9965.EPI-08-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma M., Li X., Wang Y., Zarnegar M., Huang C.-Y., Palvimo J.J., Lim B., Sun Z. hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 2003;22:6101–6114. doi: 10.1093/emboj/cdg585. doi:10.1093/emboj/cdg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., Thyssen G., Beliakoff J., Sun Z. The novel PIAS-like protein hZimp10 enhances Smad transcriptional activity. J. Biol. Chem. 2006;281:23748–23756. doi: 10.1074/jbc.M508365200. doi:10.1074/jbc.M508365200. [DOI] [PubMed] [Google Scholar]
- 41.Lee J., Beliakoff J., Sun Z. The novel PIAS-like protein hZimp10 is a transcriptional co-activator of the p53 tumor suppressor. Nucleic Acids Res. 2007;35:4523–4534. doi: 10.1093/nar/gkm476. doi:10.1093/nar/gkm476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Udler M.S., Ahmed S., Healey C.S., Meyer K., Struewing J., Maranian M., Kwon E.M., Zhang J., Tyrer J., Karlins E., et al. Fine scale mapping of the breast cancer 16q12 locus. Hum. Mol. Genet. 2010;19:2507–2515. doi: 10.1093/hmg/ddq122. doi:10.1093/hmg/ddq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broeks A., Schmidt M.K., Sherman M.E., Couch F.J., Hopper J.L., Dite G.S., Apicella C., Smith L.D., Hammet F., Southey M.C., et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum. Mol. Genet. 2011;20:3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolonel L.N., Henderson B.E., Hankin J.H., Nomura A.M., Wilkens L.R., Pike M.C., Stram D.O., Monroe K.R., Earle M.E., Nagamine F.S. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchbanks P.A., McDonald J.A., Wilson H.G., Burnett N.M., Daling J.R., Bernstein L., Malone K.E., Strom B.L., Norman S.A., Weiss L.K., et al. The NICHD Women's Contraceptive and Reproductive Experiences Study: methods and operational results. Ann. Epidemiol. 2002;12:213–221. doi: 10.1016/s1047-2797(01)00274-5. doi:10.1016/S1047-2797(01)00274-5. [DOI] [PubMed] [Google Scholar]
- 46.Ambrosone C.B., Ciupak G.L., Bandera E.V., Jandorf L., Bovbjerg D.H., Zirpoli G., Pawlish K., Godbold J., Furberg H., Fatone A., et al. Conducting molecular epidemiological research in the age of HIPAA: a multi-institutional case-control study of breast cancer in African-American and European-American women. J. Oncol. 2009;2009:871250. doi: 10.1155/2009/871250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.John E.M., Schwartz G.G., Koo J., Wang W., Ingles S.A. Sun exposure, vitamin D receptor gene polymorphisms, and breast cancer risk in a multiethnic population. Am. J. Epidemiol. 2007;166:1409–1419. doi: 10.1093/aje/kwm259. doi:10.1093/aje/kwm259. [DOI] [PubMed] [Google Scholar]
- 48.John E.M., Hopper J.L., Beck J.C., Knight J.A., Neuhausen S.L., Senie R.T., Ziogas A., Andrulis I.L., Anton-Culver H., Boyd N., et al. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. doi:10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman B., Moorman P.G., Millikan R., Qaqish B.F., Geradts J., Aldrich T.E., Liu E.T. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res. Treat. 1995;35:51–60. doi: 10.1007/BF00694745. doi:10.1007/BF00694745. [DOI] [PubMed] [Google Scholar]
- 50.Prorok P.C., Andriole G.L., Bresalier R.S., Buys S.S., Chia D., Crawford E.D., Fogel R., Gelmann E.P., Gilbert F., Hasson M.A., et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin. Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. doi:10.1016/S0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 51.Smith T.R., Levine E.A., Freimanis R.I., Akman S.A., Allen G.O., Hoang K.N., Liu-Mares W., Hu J.J. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132–2138. doi: 10.1093/carcin/bgn193. doi:10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 53.Price A.L., Tandon A., Patterson N., Barnes K.C., Rafaels N., Ruczinski I., Beaty T.H., Mathias R., Reich D., Myers S. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. doi:10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. doi:10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. doi:10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.