Abstract
Recent genome-wide analyses have detected numerous cancer-specific alternative splicing (AS) events. Whether transcripts containing cancer-specific AS events are likely to be translated into functional proteins or simply reflect noisy splicing, thereby determining their clinical relevance, is not known. Here we show that consistent with a noisy-splicing model, cancer-specific AS events generally tend to be rare, containing more premature stop codons and have less identifiable functional domains in both the human and mouse. Interestingly, common cancer-derived AS transcripts from tumour suppressor and oncogenes show marked changes in premature stop-codon frequency; with tumour suppressor genes exhibiting increased levels of premature stop codons whereas oncogenes have the opposite pattern. We conclude that tumours tend to have faithful oncogene splicing and a higher incidence of premature stop codons among tumour suppressor and cancer-specific splice variants showing the importance of considering splicing noise when analysing cancer-specific splicing changes.
INTRODUCTION
Cancer cells are associated with profound changes at the transcriptome level with hundreds of genes being up- or down-regulated when compared with normal tissues (1). Transcription profiling of cancer samples has led to an increased understanding of cancer physiology and the identification of a number of transcriptional cancer markers. Alternative splicing (AS) is a post-transcriptional process in eukaryotic organisms by which multiple distinct functional transcripts are produced from a single gene. It is now known that most human genes undergo AS (2,3). Several studies have explored cancer-related changes in AS patterns (reviewed in 4–7) resulting in the identification of an increasing number of cancer-specific AS events in a variety of cancer tissues (8–12). Given the high number of AS events unique to cancer transcriptomes, cancer-specific transcripts have been proposed to play a key role in cancer physiology (6,12). Nevertheless, only a handful of cancer-specific AS events have been experimentally validated (8,10). Given that a significant proportion of alternatively spliced transcripts result from noisy splicing in normal human tissues (13–16), it is possible that most cancer-specific AS results from aberrant splicing in these abnormal cells and does not play any significant role in cancer onset or progression (6,9,11). Here, by examining human and mouse expressed sequenced tags (EST) libraries, we ask whether cancer transcriptomes show any differences in transcript quality compared with normal tissues.
RESULTS
Identification of cancer-specific AS events in human and mouse
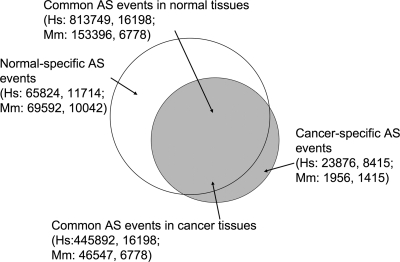
A total of 10 896 836 ESTs for the human and mouse were downloaded from UniGene (17). Of these, 3 384 826 ESTs had a clear disease state annotation and were split into 297 libraries representing normal 37 tissues and 362 cancer libraries for 32 tissues for the human and 164 normal libraries corresponding to 29 normal tissues and 42 cancer libraries from 14 tissues for the mouse (Table 1). To identify AS events, a complete exon template was constructed for each gene by mapping all partial and full transcripts available [using genomic mapping and alignment program (GMAP) software (18)]. Known nested genes as well as orphan exons, not present in any transcript extending beyond them, were removed from further analysis. Individual ESTs were then aligned to the resulting gene template to identify AS events. We identified a total of 1 349 341 and 271 491 AS transcripts containing AS events for the human and mouse, respectively. Of these, a total of 1 259 641 (93.3%) and 199 943 (73.6%) for the human and mouse, respectively, were found in both normal and cancer libraries, while 23 876 (1.8%) and 1956 (0.7%) were found only in cancer libraries. The remainder 65 824 (4.9%) and 69 592 (25.6%) transcripts were found to contain AS events exclusive to normal tissue-derived libraries (Fig. 1). The higher percentage of normal-specific AS events in mouse is explained by the limited cancer transcripts available for this species (Table 1).
Table 1.
Summary of transcripts from normal and cancer state
| Species name | Disease state | Tissue type | Development stage | Library count | EST count |
|---|---|---|---|---|---|
| Human | Normal | 37 | 7 | 297 | 1687 320 |
| Cancer | 34 | 5 | 362 | 920 844 | |
| Mouse | Normal | 29 | 15 | 164 | 628 506 |
| Cancer | 14 | 4 | 45 | 148 156 |
Figure 1.
Schematic representation of the proportion of transcripts containing AS events common in both normal and cancer libraries, or cancer/normal specific. First number in each label represents the total number of distinct AS events detected and the second the number of genes represented for human (Hs) and mouse (Mm).
Cancer transcripts show signatures consistent with splicing noise
We then assessed whether cancer libraries and in particular cancer-specific transcripts show signatures consistent with increased rates of splicing noise. If so, we expect cancer transcripts to: (A) have a higher incidence of nonsense or frameshift mutations which introduce a premature translation termination codons to mRNAs resulting in truncated proteins or more often rendering them vulnerable to nonsense-mediated decay (13,14). In the case of cancer-specific transcripts, we can expect them to: (B) have reduced identifiable functional components consistent with higher rates of aberrant incorporation of non-coding regions into the transcript (see Materials and Methods); (C) be found mostly as single copy and (D) be present in only one library thus not being part of the core cancer transcription profile as these are more likely to result from splicing errors (15).
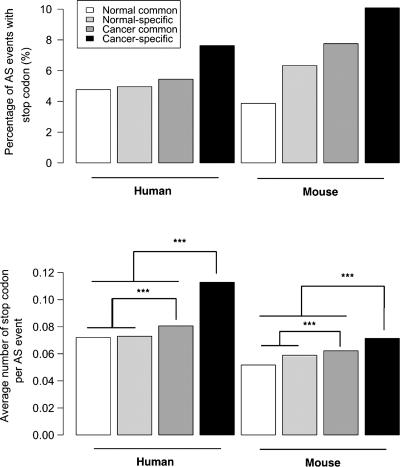
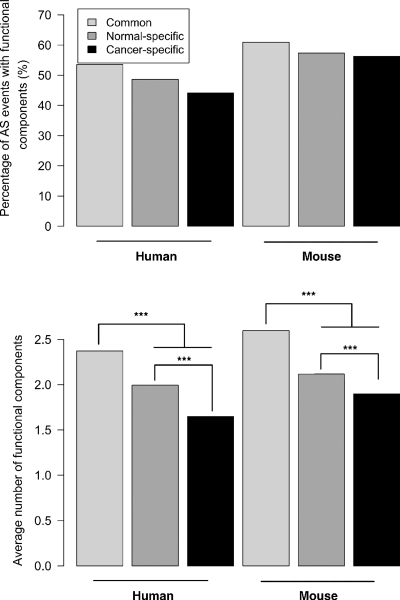
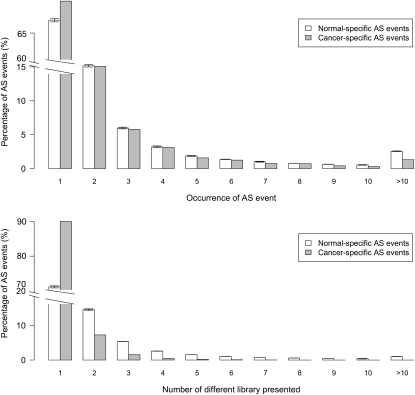
Transcripts were classified according to whether they contained AS events found in both normal and cancer tissues or unique to either resulting in four groups: (1) normal common, with transcripts containing AS events also found in at least one cancer library; (2) normal-specific, whose AS events are only found in normal tissue samples; (3) cancer common, containing transcripts from cancer libraries with AS events also found in at least one normal tissue library; and (4) cancer-specific with transcripts with AS events unique to cancer libraries. Our results show, compared with normal tissue-derived transcripts, an increased incidence of premature stop codons among cancer-derived transcripts which are higher for cancer-specific transcripts (Fig. 2, P < 0.0001) in both the human and mouse. In both species, cancer-specific events were also found to have a significantly lower number of identifiable functional components (P < 0.0001; Fig. 3). In addition, we found that the vast majority (79.0%) have been sequenced only once with 90.5% identified in a single EST library in the human (Fig. 4). In contrast, normal-specific transcripts show less-pronounced differences in premature stop codons and functional components compared with transcripts with normal-common AS events (Figs 2 and 3). We also found that transcripts containing AS events particular to normal tissues are significantly less likely to be found as a single copy or confined to a single library (P ≤ 0.0001; Fig. 4).
Figure 2.
Premature stop codons in normal and cancer AS events. Top panel shows the percentage of premature stop codon containing AS events for normal and cancer tissues subdivided into those containing AS events unique to normal/cancer libraries or found in both. Bottom panel shows average number of premature stop codons with events divided in the same way as top panel. Stars represent significant differences among groups from top panels (Chi-square tests; P > 0.05 for all comparisons) and bottom panels (Wilcoxon test; all significant comparisons with P < 0.001 denoted by ***).
Figure 3.
Identifiable functional components in AS events in cancer and normal tissues. Top panel shows the percentage of AS events with at least one identifiable functional component (see Materials and Methods). Bottom panel shows average number of identifiable functional components per AS area. In both panels, transcripts were divided as in Figure 2. Stars represent significant differences among groups from top panels (Chi-square tests; P > 0.05 for all comparisons) and bottom panels (Wilcoxon tests; P < 0.001 for all significant comparisons denoted by ***).
Figure 4.
Normal and cancer-specific AS events frequency distributions. Top panel shows the number of times each AS event is found and bottom panel shows the number of libraries where an AS event is found. Error bars in distributions from normal-specific transcripts represent 100 randomly selected samples from normal-specific transcripts of equal transcript and library number to the number of cancer-specific transcripts and libraries available.
Tumour suppressor and oncogenes reveal contrasting transcript quality reductions in cancer libraries
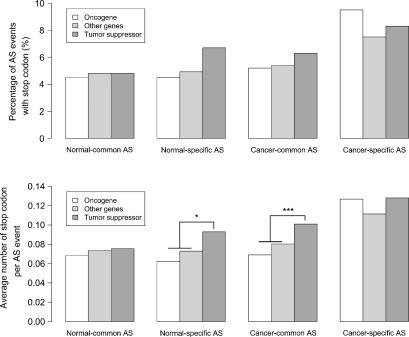
Because tumour suppressor and oncogenes play a key role in tumour progression, we tested whether these gene categories presented any differences in the frequencies of disabled transcripts. Inactivation of tumour suppressor genes NFI, FHIT and TSG101 and strengthening oncogenes CD44 and RON by AS have been reported (reviewed in 4,6). To test whether splicing noise signatures affect tumour suppressor and oncogenes differently, we divided all genes into oncogenes (648), tumour suppressor (850) and other genes according to the CancerGenes database (19). We found that even if as a whole cancer-derived transcripts are more likely to contain premature stop codons consistent with missplicing (Fig. 2), this increase is not equally distributed between gene categories (Fig. 5). Common cancer-derived oncogene transcripts show only marginal changes in the rate of premature stop codons compared with transcripts derived from normal tissues (Fig. 5). In contrast, tumour suppressor genes show a marked increase in the incidence of premature stop codons in cancer libraries (Fig. 5, P < 0.001). These differences in transcript quality among gene categories are not observed in normal libraries.
Figure 5.
Premature stop-codon frequency in oncogenes, tumour suppressor and other genes. Top panel shows the percentage of premature stop-codon containing AS events. Bottom panel shows the average number of stop codons per AS events. AS events were classified depending on whether they were derived from oncogenes tumour suppressor and other genes. Broader groupings from Figure 2 and Figure 3 are also labelled. Stars represent significant differences among groups from top panels (Chi-square tests; no significant differences found among groups with P < 0.05) and bottom panels (Wilcoxon test; significant comparisons at 0.05 are denoted by * and those with P < 0.001 are denoted by ***).
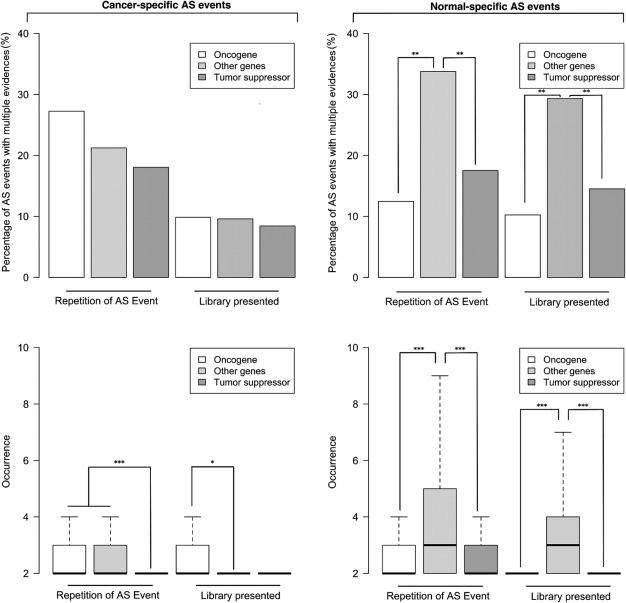
Analyses of transcripts specific to cancer or normal tissues showed that cancer-specific AS events have an elevated rate of premature stop codons in all three categories, further suggesting that a significant proportion of cancer-specific AS events containing transcripts are likely to result from splicing errors. We also found an elevated frequency in premature stop codons among tumour suppressor-derived normal-specific AS transcripts (Fig. 5; P = 0.016 and P = 0.014) which is not explained by the fact that these genes have a slightly longer average coding region (Supplementary Material, Figs S1 and S2). When comparing transcript abundance in cancer-specific AS events (Fig. 6), we found that oncogenes are more likely to produce cancer-specific AS events with more than one copy and to be found in more than one library than other genes (P = 0.037; Fig. 6). This pattern is not found for normal-specific AS transcripts where the group of other genes were far more likely to be present in multiple copies and multiple libraries than both tumour suppressor and oncogenes (P < 0.0001; Fig. 6).
Figure 6.
AS event frequency for normal and cancer transcripts divided into oncogene tumour suppressor and other genes. Left and right panels represent cancer-specific and normal-specific AS events, respectively. Distributions for normal-specific AS events are the average results from 100 randomly selected samples of equal size to the number of cancer-specific AS events. Top panels present the percentage of AS events which are present in more than one copy and or more than one library. Bottom panels are a box plot of the average number of copies per AS event or the number of libraries where each AS event is present. Stars represent significant differences among groups from top panels (Chi-square test) and bottom panels (Wilcoxon test) with P < 0.05 (*), 0.001 < P < 0.01 (**) and P < 0.001 (***).
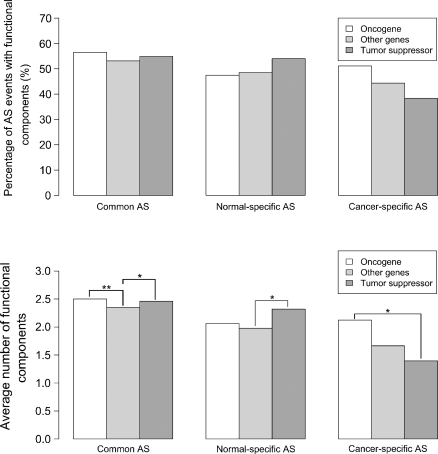
In order to assess functional content, we examined the distribution of functional components for oncogenes, tumour suppressor and other genes in both cancer and normal AS transcripts. For AS events found in both cancer and normal libraries, oncogenes and tumour suppressor-derived transcripts had higher frequencies of functional components compared with other genes (Fig. 7, P = 0.008 and P = 0.04), suggesting that AS areas contribute significantly to the functional properties of these genes protein products. While among normal-specific AS areas, there is a reduction in the functional content from oncogenes, in cancer-specific AS areas it is tumour suppressor genes which show a marked reduction in functional content. No such reduction is observed among AS areas of oncogenes (Fig. 7, P = 0.019).
Figure 7.
Identifiable functional components in AS events in cancer and normal transcripts divided into oncogene, tumour suppressor and other gene-derived. Top panel shows the percentage of AS events with at least one identifiable functional component (see Materials and Methods). Bottom panel shows average number of identifiable functional components per AS area. In both panels, AS events were divided into groups as in Figure 3 and further subdivided into oncogene, tumour suppressor and other genes. Stars represent significant differences among groups from top panels (Chi-square tests; no significant comparisons found) and bottom panels (Wilcoxon tests; with P < 0.01 denoted by ** and P < 0.05 denoted by *).
DISCUSSION
We have shown that transcripts derived from cancer libraries have an elevated rate of stop codons consistent with increased rates of missplicing in cancer transcriptomes. Transcripts with alternatively splicing events unique to cancer libraries showed an even greater enrichment in premature stop codons (Fig. 2) as well as containing fewer identifiable functional domains (Fig. 3). Importantly, all cancer-specific transcripts were found in fewer than 10 cancer libraries (out of a total of 362) with almost 80% of them found as a single copy (Fig. 4). These features suggest that a significant proportion of these transcripts are unlikely to produce a functional protein product and given that no cancer-specific transcripts were found to be ubiquitous to all cancer libraries or even a cancer type, we believe that the majority of cancer-specific transcripts, although probably functional, are unlikely to form part of a core cancer transcriptome. Thus, we estimate that the clinical and diagnostic relevance of particular cancer-specific transcripts may prove rather limited.
In contrast, analyses of transcripts only found in normal tissue samples did not reveal a similar increase in noise signatures (Figs 2 and 3) and a significantly greater proportion were found in multiple libraries (Fig. 4). Mutations leading to the absence of these transcripts in cancer libraries may have a role in cancer establishment and its progression and may therefore warrant further studies examining their clinical potential.
Interestingly, when dividing genes into oncogenes, tumour suppressors and other genes, we found marginal increases in stop codons in oncogene-derived transcripts in cancer libraries, while tumour suppressor genes showed a strong increase in premature stop codons. We found a higher incidence of premature stop codons among of tumour suppressor genes in both normal-specific and cancer-common AS (Fig. 5). This is not explained by differences in coding region length (Supplementary Material, Figs S1 and S2). The fact that cancer-specific oncogene transcripts have a higher functional content compared to those normal specific suggests that, in some instances, oncogene-derived cancer-specific transcripts may confer novel functional properties to protein products potentially having a role in cancer cells. Given that this set of transcripts are mostly found in single libraries, it is likely that their functional contribution is likely to be specific to cancers of individual patients.
We conclude that cancer states are associated with an elevated rate of aberrant transcripts particularly pronounced in tumour suppressor genes but from which oncogenes are spared. We therefore suggest that splicing noise should be considered when evaluating cancer-specific splicing events as they have a significant higher incidence of premature stop codons. Given that nonsense mutations affect only a minority of transcripts, it is feasible to assume that most cancer- and normal-specific transcripts may be transcribed into functional proteins and may contribute significantly to the cancerous phenotype. Nevertheless, the fact that most cancer-specific splice variants we identified are found as single copies in one EST library may somewhat limit their value as wide spectrum diagnostic probes and/or treatment targets. Assessment of global AS signatures by gene category may be more promising. Finally, we propose that the roles of normal specific and mutation in common AS variants should be examined in addition to cancer-specific transcripts; analyses of these absent AS transcripts may further aid in the understanding of the cancer physiology.
MATERIALS AND METHODS
Data sources
Sequence and genome annotations were obtained from Ensembl. EST sequences and library information were downloaded from UniGene (17).
Identification of AS events
To estimate AS events in different organisms, a novel procedure was applied as follows: (i) mapping predicted genes and ESTs to genome and grouping ESTs for each gene. Overlapping and nested genes were identified and removed from further analyses. GMAP (18) was used to align full transcripts and high-quality ESTs to their corresponding predicted genes. Genes with no matching transcripts were removed from further analyses. (ii) Template building. To obtain a gene template as complete as possible, full transcripts and ESTs were overlaid onto the genomic sequence. This was done as follows: first the longest partial or full transcript available forms the base of the template. All other mRNAs and ESTs are then aligned to the genomic sequence and boundaries with the previously included transcripts are revised to extend exons or include new ones. If a transcript only encompasses a single exon then it will be discarded. This allows identifying any single exon which has not been previously annotated and discarding any non-supported exons annotated in ‘predicted gene’. (iii) Detecting AS events. We developed an algorithm for AS event detection to compare the exon boundaries of any transcript to its corresponding template. Discrepancies of <15 bp in the length were discarded. To identify AS isoforms, transcripts were first sorted according to the number of AS events they contain. Then transcripts containing identical or similar AS events were classed as redundant. Each AS event was classified depending on whether it derives from cancer or normal libraries. Those AS events not found in either normal or cancer libraries were deemed cancer or normal specific, respectively, while AS events shared in both normal and cancer libraries were defined as normal common and cancer common, respectively.
Identification of premature stop codons, functional and structural protein components per AS event
As transcripts supporting the same AS event may contain premature stop-codon causing mutations, stop-codon presence was characterized and counted on a per transcript basis. Other features such as functional components were jointly analysed for each splicing event. To calculate the proportion of AS transcripts with stop codons, BLASTX (20) was run to search for open reading frames according to protein sequences. From the BLASTX alignment files, amino acid sequences of AS area were extracted and stop codons were identified and counted. To functionally characterize AS events, we used InterProScan which contains 14 applications for the prediction of protein domains (21), including Pfam for the prediction of protein domains (22), SignalP 3.0 for signal peptide predictions (23) and TMHMM (24) for the predictions of transmembrane domains. PSORT II (25) was used to identify the likely sub-cellular localization of protein products. Secondary protein structures were predicted by CLC Main Workbench 5.7, which is based on extracted protein sequences from the protein databank (http://www.rcsb.org/pdb/).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was funded by UK-China scholarship for excellence and University of Bath research studentship to L.C., a CONACyT scholarship to J.M.T.-C. and a La Ligue Contre le Cancer postdoctoral bursary, a L'Oreal UK Women in Science Award, a Royal Society Dorothy Hodgkin Research Fellowship, Royal Society research grant and a Royal Society research grant for fellows to A.U.O. Funding to pay the Open Access publication charges for this article was provided by the Royal Society.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Laurence Hurst for comments on an earlier version of this manuscript.
REFERENCES
- 1.Martinez O., Reyes-Valdes M.H., Herrera-Estrella L. Cancer reduces transcriptome specialization. PLoS ONE. 2010;5:e10398. doi: 10.1371/journal.pone.0010398. doi:10.1371/journal.pone.0010398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Q., Shai O., Lee L.J., Frey J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. doi:10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 3.Wang E.T., Sandberg R., Luo S.J., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. doi:10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalnina Z., Zayakin P., Silina K., Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. doi:10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 5.Venables J.P. Unbalanced alternative splicing and its significance in cancer. BioEssays. 2006;28:378–386. doi: 10.1002/bies.20390. doi:10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 6.Skotheim R.I., Nees M. Alternative splicing in cancer: noise, functional, or systematic? Int. J. Biochem. Cell Biol. 2007;39:1432–1449. doi: 10.1016/j.biocel.2007.02.016. doi:10.1016/j.biocel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Wang G.S., Cooper T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. doi:10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z., Lo H.S., Yang H., Gere S., Hu Y., Buetow K.H., Lee M.P. Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. [PubMed] [Google Scholar]
- 9.Xu Q. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. doi:10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hui L., Zhang X., Wu X., Lin Z., Wang Q., Li Y., Hu G. Identification of alternatively spliced mRNA variants related to cancers by genome-wide ESTs alignment. Oncogene. 2004;23:3013–3023. doi: 10.1038/sj.onc.1207362. doi:10.1038/sj.onc.1207362. [DOI] [PubMed] [Google Scholar]
- 11.Kim E., Goren A., Ast G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2008;24:7–10. doi: 10.1016/j.tig.2007.10.001. doi:10.1016/j.tig.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 12.He C., Zhou F., Zuo Z., Cheng H., Zhou R. A global view of cancer-specific transcript variants by subtractive transcriptome-wide analysis. PloS ONE. 2009;4:e4732. doi: 10.1371/journal.pone.0004732. doi:10.1371/journal.pone.0004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green R.E., Lewis B.P., Hillman R.T., Blanchette M., Lareau L.F., Garnett A.T., Rio D.C., Brenner S.E. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19:i118–i121. doi: 10.1093/bioinformatics/btg1015. doi:10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- 14.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. doi:10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z.G., Xin D.D., Wang P., Zhou L., Hu L.D., Kong X.Y., Hurst L.D. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:13. doi: 10.1186/1741-7007-7-23. doi:10.1186/1741-7007-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickrell J.K., Pai A.A., Gilad Y., Pritchard J.K. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet. 2010;6:e1001236. doi: 10.1371/journal.pgen.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayers E.W., Barrett T., Benson D.A., Bolton E., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;38:D5–D16. doi: 10.1093/nar/gkp967. doi:10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu T.D., Watanabe C.K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. doi:10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 19.Higgins M.E., Claremont M., Major J.E., Sander C., Lash A.E. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35:D721–D726. doi: 10.1093/nar/gkl811. doi:10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J.H., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zdobnov E.M., Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. doi:10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 22.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. doi:10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. doi:10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 24.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. doi:10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 25.Nakai K., Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. doi:10.1016/S0968-0004(98)01336-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.