Abstract
Germline mutations in SDHD, a mitochondrial complex II (succinate dehydrogenase) subunit gene at chromosome band 11q23, cause highly penetrant paraganglioma (PGL) tumors when transmitted through fathers. In contrast, maternal transmission rarely, if ever, leads to tumor development. The mechanism underlying this unusual monogenic tumor predisposition pattern is poorly understood. Here, we describe identification of imprinted methylation within an alternative promoter for a large intergenic non-coding RNA located at a distant gene desert boundary flanking SDHD. Methylation at this site primarily occurs within two consecutive HpaII restriction enzyme sites in a tissue-specific manner, most commonly in the adrenal gland. Informative fetal tissues and PGL tumors demonstrate maternal allelic hypermethylation. While a strong binding site for the enhancer-blocking protein CTCF within the alternative promoter shows no evidence of methylation, hyper-methylated adrenal tissues show increased binding of the chromatin-looping factor cohesin relative to the hypo-methylated tissues. These results suggest that the differential allelic methylation we observe at this locus is associated with altered chromatin architectures. These results provide molecular evidence for imprinting at a boundary element flanking the SDHD locus and suggest that epigenetic suppression of the maternal allele is the underlying mechanism of the imprinted penetrance of SDHD mutations.
INTRODUCTION
Hereditary paraganglioma (PGL) is characterized by the development of highly vascularized neuroendocrine tumors, including pheochromocytomas, derived from paraganglia of neural crest origin. Paraganglia, particularly the carotid body, are highly sensitive to oxygen-deprivation and hence play an important role in physiological adaptation to hypoxia. PGL is caused by inherited mitochondrial defects in succinate dehydrogenase structural or regulatory subunits. The mutations often involve SDHB on chromosome band 1p35 (PGL4), SDHC on chromosome band 1q13 (PGL3), SDHD on chromosome band 11q23 (PGL1) or SDHAF2 (SDH5) on chromosome band 11q13 (PGL2) (1,2). PGL mimics the paraganglia's neoplastic response to chronic hypoxia and PGL tumors develop following somatic loss of the wild-type SDH allele, consistent with a classical tumor suppressor role for the SDH genes (3).
PGL is unusual among monogenic cancer predisposition syndromes in that the tumor risk strictly depends on a paternal transmission of mutations in SDHD and SDHAF2 (2,4–6). The evidence of parent-of-origin-dependent penetrance is especially strong for SDHD mutations. Although diverse mutations were described in many large pedigrees, the development of histopathologically proven PGLs has yet to be demonstrated upon maternal transmission of an SDHD mutation (7). This transmission pattern suggests maternal genomic imprinting (suppression) of SDHD. However, molecular evidence of imprinting at the long arm of human chromosome 11 is lacking. Although sex-specific imprinted expression of Sdhd has been implicated in the mouse brain (8), the penetrance of human SDHD mutations is not sex-specific and the SDHD heterozygous knockout mice show no phenotypic evidence of PGL or imprinting (9,10). It has been shown that as little as a 20% change in the expression of a tumor suppressor gene can alter tumor predisposition (11). Thus, we hypothesized that tissue-specific quantitative imprinting of SDHD conferring subtle allele-specific expression differences could explain the parent-of-origin-dependent penetrance of its mutations. Therefore, in this study, we sought epigenetic evidence of imprinting that was identified in other well-studied imprinted loci, including differentially methylated CpG island (CGI)-like regions (DMRs), highly expressed large non-coding RNAs and strong CTCF-cohesin binding sites (12).
RESULTS
Analysis of domain imprinting at the SDHD locus
SDHD is located in a 1038 kb genomic region that contains at least 30 genes (UCSC hg19 version). At its telomeric end, this gene-rich region is flanked by a 669 kb gene desert that lacks coding genes, CGIs and appreciable transcriptional activity. Gene-poor regions >500 kb compose ∼25% of the human genome and are thought to contain important regulatory elements (13). Because imprinted genes tend to cluster in domains, we first tested for domain imprinting in the gene-rich region containing SDHD. We screened several CGIs including promoters of DIXDC1, SIK2 and SDHD in fetal tissues by methylation-sensitive Southern blot analyses and found no evidence of methylation (Supplementary Material, Table S1 and Fig. S1). Bisulfite sequencing of the SDHD promoter showed only focal and low-level methylation (Supplementary Material, Fig. S1). Similarly, expressed polymorphism analyses of several regional genes including POU2AF1, SIK2 and IL18 showed biallelic expression (Supplementary Material, Table S1). These results did not support domain-wide imprinting effects at the SDHD locus and are consistent with the lack of known imprinted phenotypes mapped to this domain in the human or mouse.
Identification of a large non-coding RNA at telomeric gene-desert boundary
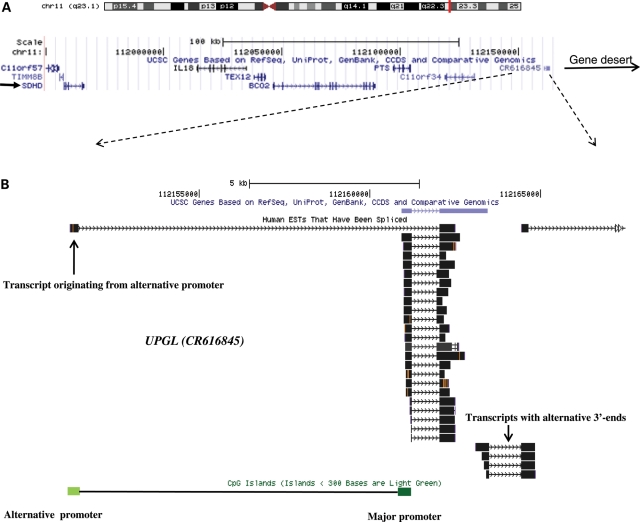
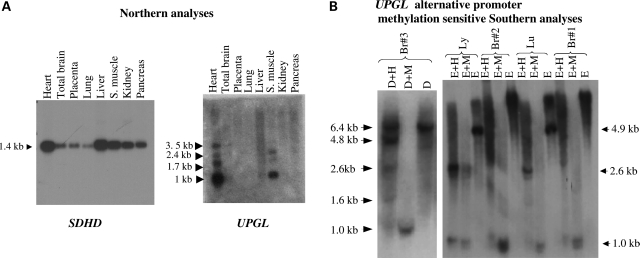
Large regulatory non-coding RNAs transcribed downstream of imprinted coding gene promoters are a characteristic of well-studied Igf2/H19 and Igf2r/Airn loci (12). Thus, we searched for a non-coding RNA in the 3′-region of the SDHD promoter. We identified a large intergenic non-coding RNA (lincRNA) which is located 206 kb telomeric to SDHD and borders the 669 kb distal gene desert (Fig. 1A). The lincRNA (CR616845) is transcribed in the same direction as SDHD (i.e. centromeric to telomeric), lacks long open reading frames, is primarily comprised of two exons, and is alternatively spliced and polyadenylated (Fig. 1B). We termed this lincRNA as UPGL (Untranslated in ParaGanglioma Locus). Northern analysis showed robust expression of UPGL with a major transcript being 1 kb in length (Fig. 2A). We tested the major promoter CGI of the UPGL gene and found no evidence of significant methylation by methylation-sensitive Southern analysis or by bisulfite sequencing in five fetal adrenal glands (Supplementary Material, Fig. S2A).
Figure 1.
(A) Gene-rich SDHD domain is separated from a flanking gene desert by UPGL, a large non-coding RNA. The non-coding UPGL gene (CR616845) is located at the telomeric boundary of a gene-rich region containing SDHD on chromosome band 11q23. The locations of SDHD and the gene desert are indicated by arrows. (B) UPGL primary transcript is composed of two alternatively spliced exons. The first exon originates from the CGI major promoter, although rare transcripts originate from an alternative CGI promoter. Many truncated transcripts downstream of UPGL suggest alternative transcript processing at 3′-end of UPGL.
Figure 2.
Expression and alternative-promoter methylation of UPGL (A). Northern blot analyses of the SDHD and UPGL genes. SDHD is expressed ubiquitously at a single major transcript size, whereas UPGL is expressed predominantly in the heart, skeletal muscle and brain and at multiple transcript sizes. Database analysis of expressed sequence tags (ESTs) suggests that the most common UPGL transcript (∼1 kb) is composed of two exons and originates from the major promoter. The ESTs show variation in the 3′ termini, including a third alternatively spliced 3′-exon. These transcript variations might account for the longer bands of weaker intensity observed in the UPGL northern blot. (B) Methylation-sensitive southern blot analysis (right panel) of genomic DNA shows partial methylation of EcoRI (E) and HpaII (H) sites in the upstream CGI (the UPGL alternative promoter) in fetal brain (Br) but not in lung (lu) and lymphoblastoid DNAs (ly). Partial methylation of HpaII sites in the fetal brain in UPGL alternative promoter is confirmed (left panel) by first digesting with DraI, a methylation-insensitive enzyme, followed by HpaII and MspI (M). (See Supplementary Material, Fig. S1 for a more detailed interpretation of the Southern blots).
Because the SDHD/UPGL arrangement resembles that of the imprinted Igf2/H19 locus in that the upstream gene is protein-encoding and paternally (over)expressed, whereas the downstream gene is a highly expressed and spliced lincRNA, we hypothesized that a DMR exists upstream of the UPGL promoter similar to that found at the H19 locus. We identified a CGI-like sequence ∼9.5 kb upstream of the UPGL promoter. Database and reverse transcriptase–PCR (RT–PCR) analyses indicate that this CGI functions as an alternative UPGL promoter where transcripts with an alternate exon 1 originate (Fig. 1B).
Evidence of imprinting at a boundary element
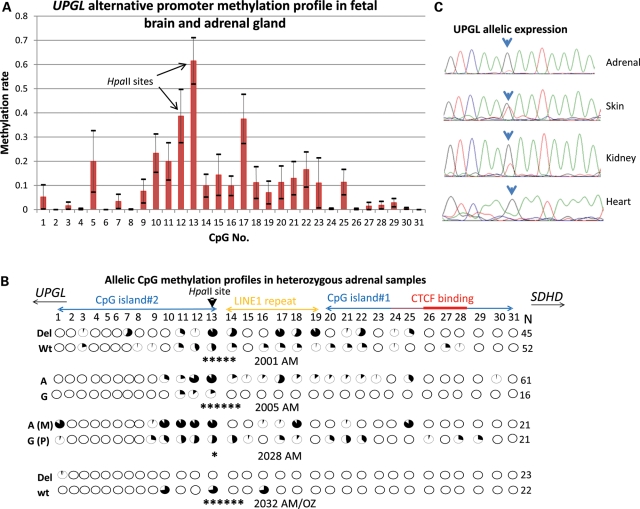
We evaluated 31 CpG dinucleotides within this CGI-like sequence that spans two canonical CGIs measuring 292 bp (CGI#1) and 294 bp (CGI#2), respectively, and an intervening truncated long interspersed nuclear elements (LINE) repeat measuring 251 bp. Methylation-sensitive Southern blot analysis revealed partial methylation at two HpaII restriction sites in CGI#2 (CpG#12 and CpG#13) in fetal brain, but not in lung or lymphoblastoid cell lines (Fig. 2B and Supplementary Material, Fig. S2B). High-resolution methylation analysis by bisulfite PCR, cloning and sequencing confirmed significant methylation (>30% of the clones) of one or both HpaII restriction sites in four of four fetal adrenal glands and in four of five fetal brains, but not in fetal lung (n= 1), heart (n= 1) and adult brain (n= 1) (Fig. 3B and Supplementary Material, Fig. S3). Joint analysis of the brain and adrenal glands showed that the CpG methylation primarily targeted the two tandem HpaII sites at CpG#12 and CpG#13 (Fig. 3A), which approximately correspond to the alternative exon 1 transcript start sites. We also confirmed partial methylation of these two CpGs in purified adult adrenal medulla tissues by bisulfite PCR and direct sequencing (Supplementary Material, Fig. S4). Notably, the fetal adrenal gland, brain and adult adrenal medulla are related to paraganglionic tissues by function, embryology and histology.
Figure 3.
Evidence of imprinting at the UPGL alternative promoter. (A) Average CpG methylation rates in the UPGL alternative promoter in the fetal adrenal gland (n= 4) and fetal brain (n= 5) reveals highest methylation in an HpaII restriction enzyme site (CpG#13). CpG graph displays mean ± standard error of the mean (horizontal bars) for average methylation rates for each of the 31 CpGs. (B) Allelic methylation profiles in four heterozygous adrenal glands indicate statistically significant allelic methylation differences at CpG#13 in four informative cases (Fisher's exact test: *P< 0.05, *****P< 10−5, ******P< 10−6). Allelic sequence variations are shown on the left. Each row demonstrates methylation rates in the CpGs in the allele defined by the sequence variation on left (Del/Wt or A/G). The del(GAA) variant removes one of the GAA triplets following CpG#13 without eliminating the CpG or the HpaII restriction enzyme site. In sample 2028 AM, maternal and paternal alleles could be assigned by the availability of an informative (homozygous AA) maternal decidual tissue. In fetus 2032, the methylation profile of each allele was derived from a different paraganglionic tissue: AM, adrenal medulla/gland, OZ, organ of Zuckerkandl (peri-aortic paraganglia). N indicates the number of PCR clones sequenced for each allele. Core CTCF-binding site is shown by a thick red bar, the extended binding site is shown by a thin red bar. (C) An expressed SNP (G/T) in UPGL exon 2 in one fetus reveals marked imbalance in allelic expression levels, revealing monoallelic expression of the G allele in the fetal adrenal gland and heart, imbalanced expression in the kidney and balanced expression in the skin. Because all tissues are confirmed to be heterozygous at the genomic level by sequencing, the allelic imbalances likely stem from allelic epigenetic differences.
We further evaluated the most methylated CpG, CpG#13, for imprinting. Bioinformatic tools predict that CpG#13 is located within a consensus sequence (5′-CCGGAAG-3′) for ‘E-twenty six’ family of transcription factors including GA binding protein/nuclear respiratory factor 2 (GABPα/NRF2) (14), which regulates many nuclear-encoded mitochondrial genes (15) and whose loss leads to depression of mitochondrial activity (16). Although the SDHD promoter strongly binds GABPα/NRF2, a recent chromatin immunoprecipitation assay coupled with massively parallel sequencing (ChIP-Seq) shows no evidence of binding to the UPGL alternative promoter (17). Notably, CpG#13 is juxtaposed to a LINE repeat, which are often enriched in imprinted loci (18). We found statistically significant differences in allelic methylation rates in all four polymorphic fetal adrenal samples (Fig. 3B). Maternal decidual tissue informative for one of the heterozygous samples established that the hypermethylated allele was maternal (Supplementary Material, Table S2).
Previously, SDHD was shown to be biallellically expressed in the fetal brain, kidney and lung (4). We also observed biallelic SDHD expression in adrenal gland (n= 4) and in placenta (n= 3). To test allelic expression of the UPGL gene, we performed RT–PCR and sequencing analyses using expressed polymorphisms identified in seven fetuses. We observed biallelic expression in the adrenal gland, lung, kidney, skin and brain tissues from six fetuses. However, significant allelic imbalance and mono-allelic expression were observed in the fetal kidney, adrenal gland and heart, but not in skin in one fetus (Fig. 3C). Maternal decidual tissue was not available to determine the parental origin of the expressed UPGL allele.
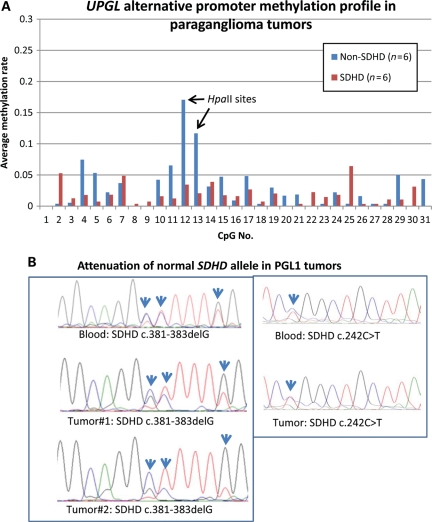
SDHD-related PGL tumors provide an alternative tool to evaluate parental origin of the hypermethylated allele in the UPGL alternative promoter because these tumors develop after loss of the normal maternal SDHD allele, often by deletion of the whole maternal chromosome 11 (3). We compared the UPGL alternative-promoter methylation patterns in SDHD and non-SDHD-related PGL tumors. Direct sequencing and loss of heterozygosity (LOH) analyses confirmed loss of normal (maternal) alleles on chromosome 11q23 in SDHD-mutation positive tumors consistent with earlier data (Fig. 4B). Bisulfite PCR, cloning and sequencing analyses of six SDHD-related, four SDHB-related and two apparently sporadic SDHD/SDHB-negative PGL tumors showed the UPGL alternative-promoter CpG#13 methylation in one SDHB-related (35% methylation) and one sporadic tumor (24% methylation). All SDHD tumors showed low levels (<6%) of methylation (Fig. 4A). These results suggest that the UPGL alternative-promoter CpG#13 retains methylation in a subset of non-SDHD-related tumors but not in SDHD-related tumors consistent with hypermethylation of the maternal allele which is lost during SDHD-related tumor development.
Figure 4.
Methylation profile of the UPGL alternative promoter in PGL tumors (A). Comparison of average CpG methylation rates in non-SDHD (n= 6) and SDHD PGLs (n= 6) shows statistically significantly more clones methylated at the UPGL alternative promoter (CpG#13) (47/378 versus 5/268 methylated clones; Fisher's exact test: P< 10−8) among non-SDHD tumors. (B) Comparison of sequence fluorograms between blood and tumor shows attenuation of the normal unmutated alleles (arrows) in the tumors consistent with LOH.
Differential methylation in the UPGL alternative promoter is associated with different chromatin architecture
CTCF-binding regions coinciding with differentially methylated CGIs, especially those at antisense or alternative promoters, are significantly enriched in imprinted genes (19). For example, the DMR upstream of the H19 locus is a methylation-sensitive CTCF-binding site and determines outcome of an enhancer competition between the Igf2 and H19 genes (20,21). A previous genome-wide chromatin immunoprecipitation and genomic tiling arrays (ChIP-chip) analysis uncovered a strong CTCF-binding site over a very high scoring CTCF recognition sequence within the UPGL CGI#1, 350 bp upstream (centromeric) to the CpG#13 (Fig. 3B) (22). Here, the specific chromatin immunoprecipitation signal was as large as the imprint control region near the H19 promoter. We performed ChIP analyses and confirmed strong CTCF binding in CGI#1 (Fig. 5A) in both adrenal and lung tissues. However, in contrast to the expectations from the Igf2/H19 locus, we found no evidence of significant CpG methylation in the CTCF-binding site in any normal tissue or in PGL tumors (Figs 3A, B and 4A).
Figure 5.
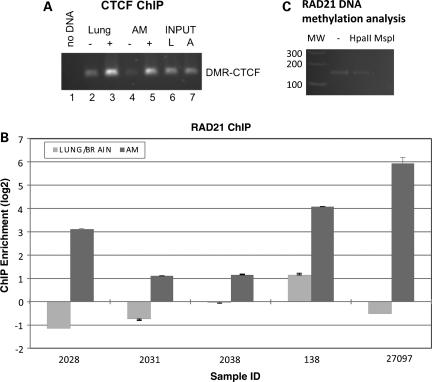
CTCF and cohesin binding in the UPGL alternative promoter. (A) ChIP-PCR analysis of CTCF in adrenal gland (noted AM or A) or lung (noted LUNG or L) tissues from the 2028 sample. − and + indicate mock- or CTCF-specific immunoprecipitation. Input indicates total chromatin prior to immunoprecipitation. The ChIP-PCR is analyzed by agarose gel electrophoresis. (B) The results from ChIP-qPCR analysis of cohesin binding at the DMR using an antibody against RAD21, a component of cohesin. Three paired fetal lung and adrenal medulla tissues from three fetuses (numbers 2028, 2031, 2038) and two paired fetal brain and adrenal medulla tissues from two fetuses (numbers 138 and 27097) were analyzed by cohesin ChIP. The fold enrichment values are determined by determining the threshold cycle numbers for the ChIP DNA and input control DNA. In all experiments, the adrenal tissue shows increased RAD21 binding. Increased RAD21 binding is also noted in one of two fetal brains (number 138) consistent with frequent methylation at HpaII restriction sites observed in fetal brains. (C) Restriction enzyme digestion of the RAD21 ChIP DNA with HpaII or MspI (which both cut DNA at CCGG sites, but HpaII is blocked by CG methylation) and PCR analysis shows that Rad21 ChIP DNA is resistant to HpaII digestion, while PCR failed to amplify the specific product from MspI-digested ChIP DNA, suggesting that significant fraction of RAD21 bound DNA is methylated. (MW, molecular weight marker in base pairs. Lane ‘–’ was not treated by restriction enzyme.)
Cohesin is a chromatin-associated protein complex that co-localizes with CTCF and enables isolation of enhancer and promoters in the Igf2/H19 region (23). We found that RAD21, a cohesin subunit, shows increased binding to CGI#1 in the hypermethylated fetal adrenal tissue relative to the fetal lung and brain (Fig. 5B). PCR amplification of RAD21 immuno-precipitated DNA from one fetal adrenal gland (2028) following HpaII or MspI restriction enzyme digestion showed amplification only after HpaII digestion, suggesting that cohesin binds to methylated DNA (Fig. 5C). Because the lung and brain show lower rates of methylation in the UPGL alternative promoter than the adrenal gland (Fig. 3B and Supplementary Material, Fig. S3), these results suggest that the unmethylated HpaII sites may have role in preventing cohesin loading at this CTCF-binding site.
The boundary elements are conserved among primates but not in the mouse
The imprinting status of most genes is conserved between the mouse and human (24). However, we found no evidence of the UPGL alternative promoter or UPGL orthologues in the mouse syntenic region on central chromosome 9 (25), although the gene desert and the flanking protein-coding genes, Plet1 and Ncam, were conserved between the mouse and human. In contrast, UPGL, the UPGL alternative promoter and the CTCF-binding sites are present in primates (Supplementary Material, Fig. S5). These results indicate that the UPGL alternative promoter and UPGL are non-conserved genomic elements which could play a role in the imprinted penetrance of SDHD mutations observed in humans but not in mice.
DISCUSSION
Genomic imprinting confers different functional properties to maternal and paternal alleles, resulting in asymmetric parental contributions to progeny. The influence of genomic imprinting in disease susceptibility is well recognized in developmental disorders, such as Prader-Willi/Angelman syndrome (26). The tumor susceptibility associated with the Beckwith–Wiedemann syndrome is also influenced by parent-of-origin effects (27). The imprinting defects associated with these disorders are often caused by gross genetic changes such as large deletions, chromosomal translocations and/or epigenetic changes which alter the imprinting status of multiple genes in large imprinted domains. The mechanisms underlying the imprinted penetrance of SDHD and SDHAF2 mutations, both on the long arm of chromosome 11, are poorly understood, because these PGL genes do not reside in known imprinted domains and molecular evidence of imprinting has been lacking.
In this study, we have identified several factors which collectively support imprinting within an alternative promoter of a lincRNA (UPGL) located at the boundary between the SDHD locus and a flanking gene desert. These factors include differential methylation and an outstanding CTCF-binding site within a CGI as well as increased cohesin binding in the methylated adrenal tissue, suggesting that differential methylation at the UPGL alternative promoter is associated with different chromatin architectures at this locus. In a recent study, the co-occurrence of differential methylation, CTCF binding and distinct chromatin signatures in a locus resulted in marked enrichment for human imprinted genes (19). In addition, we obtained direct evidence of imprinting by demonstrating asymmetric allelic methylation in four heterozygous specimens, one of which allowed us to establish maternal hypermethylation at the UPGL alternative promoter. Parental allelic asymmetry was also supported by analyses of SDHD tumors which showed no significant methylation at the UPGL alternative promoter, consistent with somatic maternal chromosomal losses that often occur as a second-hit in SDHD tumors (3). In contrast, two of the six non-SDHD PGL tumors showed partial methylation at the UPGL alternative-promoter HpaII sites. Finally, in one of the seven heterozygous UPGL samples, mono-allelic expression is observed.
The epigenetic hallmarks of imprinting identified in this study draw similarities between SDHD/UPGL and the reciprocally imprinted Igf2/H19 genes, where enhancer competition between the upstream coding and the downstream non-coding genes is regulated by methylation at CTCF sites upstream of the non-coding H19 locus (20). Unexpectedly, we found that the tissue-specific methylation at the UPGL alternative promoter does not involve a nearby strong CTCF site. Nevertheless, the preferential binding of cohesin to the methylated DNA in the adrenal tissue suggests that the methylation might regulate composition of proteins bound to this CGI promoter resulting in different allelic chromatin architecture. The methylation might either prevent binding of a transcription factor or attract a methyl-binding protein, resulting in a conformational change that allows cohesin binding. Alternatively, cohesin binding in the adrenal gland might reflect tissue-specific differences in composition of transcription factors bound to the alternative CGI. In support of this model, it has been recently shown that pluripotency factor OCT4 antagonizes cohesin loading at the CTCF-binding site in the HOXA locus and that loss of OCT4 in the differentiated neuronal progenitor cells results in loading of cohesin and chromosome looping (28). Regardless of the exact mechanism, our results suggest an association between the UPGL alternative-promoter HpaII site methylation and cohesin binding.
Though more mechanistic work is needed, we speculate that CTCF-cohesin binding on the hypermethylated maternal allele creates an insulator that interferes with functional engagement of the SDHD promoter and a cis-acting element (enhancer) located within the gene desert. When the UPGL alternative promoter is hypomethylated on the paternal allele, lack of cohesin permits long-range interaction between an enhancer and the SDHD promoter. This differential allelic interaction may result in maternal downregulation of SDHD and the parent-of-origin-dependent tumor susceptibility. Partial inactivation of SDHD by imprinting might provide an advantage for early detection of hypoxia, because PGL tumors, where SDH is completely inactive, show constitutive activation of hypoxia-inducible pathways (1,29,30). Further research is required to determine chromatin interactions involving SDHD and whether alterations of the epigenetic elements described here play a role in mitochondrial alterations and hypoxic adaptation in tumors.
MATERIALS AND METHODS
Samples and subjects
Normal fetal and adult tissues and PGL tumors were collected at University of Pittsburgh School of Medicine, Yale School of Medicine and National Institute of Health following protocol approvals by Institutional Review Board and Human Investigative Committees. Additional fetal adrenal glands were obtained from Maryland Tissue Bank. Adult adrenal medullary tissues were obtained from kidney resection specimens after gross dissection of the adrenal medullary tissues. A total of 75 fetal adrenal glands, 10 fetal brains, 30 adult adrenal medullas and 1 adult brain were available for polymorphism search and imprinting analysis in the UPGL alternative promoter, SDHD promoter and SDHD and UPGL transcripts. A subset of the fetal samples also included the fetal kidney, lung, heart, skin and muscle tissues. Because collecting normal paraganglionic tissues distributed in small clusters along major arteries and nerves is technically and practically difficult, we used primarily fetal brain and adrenal gland, as these tissues are embryologically, anatomically and functionally related to normal paraganglia. Notably, fetal adrenal gland is thought to function as a direct oxygen-sensor in utero (31–33). Adrenal glands were sampled from internal part of the organ to enrich for adrenal medulla.
Maternal decidual tissues were also available for a subset of samples and were grossly obtained from the maternal side of fresh placentas. Maternal tissue enrichment in placental sections and the maternity is confirmed by genotyping with five highly polymorphic microsatellite repeats using a commercial kit from Corriell Institute for Medical Research (New Jersey, USA).
Mutation, polymorphism and methylation analyses
DNA was extracted from tissues using a commercial kit (DNeasy, Qiagen) and quantified using a Nanodrop (Thermofisher Scientific). Polymorphisms and mutations were identified by PCR amplification and sequencing of the UPGL alternative promoter, SDHD promoters and UPGL, SDHD exons from genomic DNA using standard protocols. The UPGL alternative promoter was amplified using primers F5.U and R5.U (Supplementary Material, Table S3). The sequence files were analyzed using Sequencher software (Gene Codes Corporation, Ann Arbor, MI, USA).
Methylation analyses were initially performed by methylation-sensitive Southern blot analysis. Briefly, 10–20 μg of genomic DNA were first completely digested by a 6bp cutter restriction enzyme (e.g. EcoRI, DraI) using 10–20 U enzyme per microgram. Subsequent to phenol/chloroform extraction, the genomic DNA was re-digested either by CpG methylation-sensitive enzyme HpaII, or its methylation-insensitive isoschizomer MspI. Northern analyses were performed in commercial blots that contain ∼2 μg of RNA for each tissue (Clontech Laboratories). Radioactive hybridization probes were generated by PCR amplification of tested CGI and labeled by random priming.
High-resolution methylation analyses of CGIs were performed by first treating the genomic DNA with bisulfite and then subsequently amplifying one of the converted DNA strands by specific post-conversion primers. Bisulfite conversion was carried out with the EZ DNA Methylation Kit (Zymo Research) according to the manufacturer's instructions. Briefly, 1 μg genomic DNA was treated with the bisulfite reagent in a MJ Research Thermal Cycler using the following protocol: 10 min at 98°C (denaturation), 30 min at 58°C, eight cycles of 6 min at 53°C and 30 min at 37°C. The converted DNA was de-sulfonated, column purified and quantified for use as a PCR template. When necessary, the amplified DNA strand was chosen so that the alleles of heterozygous samples could still be distinguished after the bisulfite treatment. Bisulfite-treated DNA was amplified by two rounds of PCR using nested or semi-nested primer pairs. Primers were designed using sequences that spanned putative CGIs but contained 0–1 CpG dinucleotides within the primer region. No CpG dinucleotides were allowed at the 3′ end of any primer. All cytosine residues in the primer sequence were coded as thymidine. The UPGL alternative promoter, where methylation was identified, was amplified in a nested PCR amplification using primers CGIALLMethy-F5/CGIALLMethy-R5 in the first PCR and CGIALLMethy-F6/CGIALLMethy-R6 in the second nested PCR (Supplementary Material, Table S3). The products were cloned into the pGEM-T Easy vector (Promega) and plated on LB-Amp plates. For each PCR reaction, individual colonies were used to inoculate 12 h LB-Amp cultures in 96-well plates which were submitted to Beckmann-Coulter Genomics for purification and sequencing.
In one of the heterozygous fetuses (2032), the methylation profile of each allele was derived from adrenal medulla/gland and organ of Zuckerkandl (peri-aortic paraganglia), respectively (Fig. 3B). Only one allele was represented in each tissue following multiple bisulfite treatment, PCR, cloning and sequencing experiments. Because genomic DNA was heterozygous in both tissues, skewed allelic representation is the most likely explanation for these results. Nevertheless, we compared the methylation profiles of each allele from two different tissues, because adrenal medulla/gland and organ of Zuckerkandl both belong to the histologically and embryologically related paraganglionic system.
Complementary DNA synthesis was performed using SUPERSCRIPT II (Life Technologies) and oligo(dT) primers; the recommended conditions were used. RT–PCR of UPGL cDNA templates was performed by 30–33 cycles of amplification with primers ProbeF1/Rt-Rb (Supplementary Material, Table S3). PCR products were directly sequenced for allelic expression analyses. Transcriptional activity of the alternative promoter of the UPGL gene was confirmed by RT–PCR using primers Rt-Fa/Rt-Ra.
Bioinformatic analyses
We used two different software programs for robust identification of CGIs, regions >200 bp long that have observed/expected CpG ratio >0.60 and percent C + percent G >50. Initial CpG screening of 80 kb sequence upstream of UPGL major promoter was performed by CpG plot software at http://www.ebi.ac.uk/Tools/emboss/cpgplot/index.html. This software identified a 257 bp long CGI ∼9.5 kb upstream of the major promoter CGI of the UPGL gene. Further analyses of this CpG-rich region, after masking an internal LINE repeat by RepeatMasker (http://www.repeatmasker.org/), revealed two CGIs flanking each side of the repeat by CGI Searcher (http://cpgislands.usc.edu/). Interspecies sequence comparison was performed by ClustalW2 software at http://www.ebi.ac.uk/Tools/clustalw2/. All consensus gene sequences and genomic maps were obtained from UCSC Genome Bioinformatics Web site (http://genome.ucsc.edu/). Similarity between sequences was evaluated by BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Transcription factor-binding sites in the promoters were searched by TFSEARCH software at http://www.cbrc.jp/research/db/TFSEARCH.html.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation was performed as described previously (22). Briefly, sonicated chromatin was incubated with 10 μg of each antibody, coupled to Dynabeads (Invitrogen). The magnetic beads were washed eight times with RIPA buffer containing 50 mm HEPES (pH 8.0), 1 mm EDTA, 1% NP-40, 0.7% DOC and 0.5 m LiCl, supplemented with 1× complete protease inhibitors (Roche Applied Science), and washed once with TE (10 mm Tris at pH 8.0, 1 mm EDTA). After washing, the bound DNA was eluted by heating the beads to 65°C in elution buffer (10 mm Tris at pH 8.0, 1 mm EDTA and 1% SDS). The eluted DNA was incubated at 65°C for 12 h to reverse the cross-links. Following incubation, the immunoprecipitated DNA was treated sequentially with Proteinase K (Roche Applied Science) and RNase A (Sigma), and was desalted using the QIAquick PCR purification kit (Qiagen). The DNA concentration of the recovered ChIP samples was determined using Quant-iT PicoGreen reagent (Invitrogen) using a set of DNA concentration standards generated from the reverse cross-linked total chromatin sample. RAD21 antibody was obtained from Millipore, Billerica, MA.
Primers used for amplification of RAD21 DNA digested by HpaII and MspI (Fig. 5C) were CGCCATTTGAAGCCGCGCAT and CGTTCCTGCGGACCAAGGCA, which spans the UPGL alternative-promoter HpaII sites.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work is supported by National Institutes of Health (R01CA11236 and R01CA140485); Chairman's Challenge Award; Rita Allen Foundation; and Sidney Kimmel Foundation. Portions of this study were also supported by NIH grant R01HL059655 to G.S.S.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs T. MacPherson, T. Parks and R. Dhir for help in obtaining tissues; L. Mock for coordination and anonymization of tissue collection; E. Lawrence in initial sequencing (University of Pittsburgh); Dr P. Hui (Yale University) for help in parental allelic testing; Dr A. Sidow (Stanford University) for information on GABP binding sites. Initial phase of the project is completed at University of Pittsburgh. The remaining data in this paper are reported in the Supplementary Material.
REFERENCES
- 1.Baysal B.E. Clinical and molecular progress in hereditary paraganglioma. J. Med. Genet. 2008;45:689–694. doi: 10.1136/jmg.2008.058560. [DOI] [PubMed] [Google Scholar]
- 2.Hao H.X., Khalimonchuk O., Schraders M., Dephoure N., Bayley J.P., Kunst H., Devilee P., Cremers C.W., Schiffman J.D., Bentz B.G., et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hensen E.F., Jordanova E.S., van Minderhout I.J., Hogendoorn P.C., Taschner P.E., van der Mey A.G., Devilee P., Cornelisse C.J. Somatic loss of maternal chromosome 11 causes parent-of-origin-dependent inheritance in SDHD-linked paraganglioma and phaeochromocytoma families. Oncogene. 2004;23:4076–4083. doi: 10.1038/sj.onc.1207591. [DOI] [PubMed] [Google Scholar]
- 4.van der Mey A.G., Maaswinkel-Mooy P.D., Cornelisse C.J., Schmidt P.H., van de Kamp J.J. Genomic imprinting in hereditary glomus tumours: evidence for new genetic theory. Lancet. 1989;2:1291–1294. doi: 10.1016/s0140-6736(89)91908-9. [DOI] [PubMed] [Google Scholar]
- 5.Mariman E.C., van Beersum S.E., Cremers C.W., Struycken P.M., Ropers H.H. Fine mapping of a putatively imprinted gene for familial non-chromaffin paragangliomas to chromosome 11q13.1: evidence for genetic heterogeneity. Hum. Genet. 1995;95:56–62. doi: 10.1007/BF00225075. [DOI] [PubMed] [Google Scholar]
- 6.Baysal B.E., Ferrell R.E., Willett-Brozick J.E., Lawrence E.C., Myssiorek D., Bosch A., van der Mey A., Taschner P.E., Rubinstein W.S., Myers E.N., et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 7.Neumann H.P., Erlic Z. Maternal transmission of symptomatic disease with SDHD mutation: fact or fiction? J. Clin. Endocrinol. Metab. 2008;93:1573–1575. doi: 10.1210/jc.2008-0569. [DOI] [PubMed] [Google Scholar]
- 8.Gregg C., Zhang J., Weissbourd B., Luo S., Schroth G.P., Haig D., Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piruat J.I., Pintado C.O., Ortega-Saenz P., Roche M., Lopez-Barneo J. The mitochondrial SDHD gene is required for early embryogenesis, and its partial deficiency results in persistent carotid body glomus cell activation with full responsiveness to hypoxia. Mol. Cell Biol. 2004;24:10933–10940. doi: 10.1128/MCB.24.24.10933-10940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayley J.P., van Minderhout I., Hogendoorn P.C., Cornelisse C.J., van der Wal A., Prins F.A., Teppema L., Dahan A., Devilee P., Taschner P.E. Sdhd and SDHD/H19 knockout mice do not develop paraganglioma or pheochromocytoma. PLoS ONE. 2009;4:e7987. doi: 10.1371/journal.pone.0007987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alimonti A., Carracedo A., Clohessy J.G., Trotman L.C., Nardella C., Egia A., Salmena L., Sampieri K., Haveman W.J., Brogi E., et al. Subtle variations in pten dose determine cancer susceptibility. Nat. Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan L.B., Bartolomei M.S. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv. Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- 13.Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 14.Wei G.H., Badis G., Berger M.F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A.R., et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scarpulla R.C. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 16.Baltzer C., Tiefenbock S.K., Marti M., Frei C. Nutrition controls mitochondrial biogenesis in the drosophila adipose tissue through delg and cyclin D/Cdk4. PLoS ONE. 2009;4:e6935. doi: 10.1371/journal.pone.0006935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valouev A., Johnson D.S., Sundquist A., Medina C., Anton E., Batzoglou S., Myers R.M., Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen E., Horvath S., Tong F., Kraft P., Spiteri E., Riggs A.D., Marahrens Y. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl Acad. Sci. USA. 2003;100:9940–9945. doi: 10.1073/pnas.1737401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen B., Wu H., Bjornsson H., Green R.D., Irizarry R., Feinberg A.P. Overlapping euchromatin/heterochromatin-associated marks are enriched in imprinted gene regions and predict allele-specific modification. Genome Res. 2008;18:1806–1813. doi: 10.1101/gr.067587.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 21.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 22.Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 24.Renfree M.B., Hore T.A., Shaw G., Graves J.A., Pask A.J. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu. Rev. Genomics Hum. Genet. 2009;10:241–262. doi: 10.1146/annurev-genom-082908-150026. [DOI] [PubMed] [Google Scholar]
- 25.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholls R.D., Knepper J.L. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu. Rev. Genomics Hum. Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- 27.Lim D.H., Maher E.R. Genomic imprinting syndromes and cancer. Adv. Genet. 2010;70:145–175. doi: 10.1016/B978-0-12-380866-0.60006-X. [DOI] [PubMed] [Google Scholar]
- 28.Kim Y.J., Cecchini K.R., Kim T.H. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc. Natl Acad. Sci. USA. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottlieb E., Tomlinson I.P. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat. Rev. Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 30.Raimundo N., Baysal B.E., Shadel G.S. Revisiting the TCA cycle: signaling to tumor formation. Trends Mol. Med. 2011 doi: 10.1016/j.molmed.2011.06.001. Epub ahead of print]. doi:10.1016/j.molmed.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung C.Y. Fetal adrenal medulla catecholamine response to hypoxia-direct and neural components. Am. J. Physiol. 1990;258:R1340–R1346. doi: 10.1152/ajpregu.1990.258.6.R1340. [DOI] [PubMed] [Google Scholar]
- 32.Comline R.S., Silver M. The release of adrenaline and noradrenaline from the adrenal glands of the foetal sheep. J. Physiol. 1961;156:424–444. doi: 10.1113/jphysiol.1961.sp006685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slotkin T.A., Seidler F.J. Adrenomedullary catecholamine release in the fetus and newborn: secretory mechanisms and their role in stress and survival. J. Dev. Physiol. 1988;10:1–16. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.