Abstract
Myosin-Binding protein-C (MyBP-C) is a family of accessory proteins of striated muscles that contributes to the assembly and stabilization of thick filaments, and regulates the formation of actomyosin cross-bridges, via direct interactions with both thick myosin and thin actin filaments. Three distinct MyBP-C isoforms have been characterized; cardiac, slow skeletal, and fast skeletal. Numerous mutations in the gene for cardiac MyBP-C (cMyBP-C) have been associated with familial hypertrophic cardiomyopathy (FHC) and have led to increased interest in the regulation and roles of the cardiac isoform. This review will summarize our current knowledge on MyBP-C and its role in modulating contractility, focusing on its interactions with both myosin and actin filaments in cardiac and skeletal muscles.
1. Introduction
Striated muscle cells contain a complex, highly ordered cytoskeleton, mainly composed of interdigitating thick myosin and thin actin filaments [1]. Muscle contraction and relaxation occurs by the sliding of myosin filaments past actin filaments under the strict regulation of their accessory proteins, which are responsible for their assembly and maintenance as well as the regulation of contractile activity [1].
Myosin-Binding protein-C (MyBP-C) comprises a family of accessory proteins of the thick myosin filaments that encompasses ~2% of the total myofibrillar protein [2]. To date, two major roles have been attributed to MyBP-C; it contributes to the regular organization and stabilization of thick filaments and modulates the formation of cross-bridges between myosin and actin, via direct interactions with both filamentous systems (as reviewed in [3, 4]). Within the sarcomere, MyBP-C localizes to the C-zone, the cross-bridges containing region of the A-band, in 7–9 transverse stripes that are ~43 nm apart [5–7].
Three MyBP-C isoforms have been identified; cardiac, slow skeletal, and fast skeletal (cMyBP-C, sMyBP-C, and fMyBP-C), encoded by different genes localizing to human chromosomes 11, 12, and 19, respectively [8, 9]. The core structure of MyBP-C is composed of seven immunoglobulin (Ig) domains and three fibronectin type III (Fn-III) repeats, numbered from the NH2-terminus as C1–C10 (Figure 1; [10]). A proline/alanine- (pro/ala-) rich motif and a conserved linker region, termed MyBP-C or M-motif, flank the first Ig domain, C1. Notably, both the cardiac and slow isoforms differ slightly from the core structure. cMyBP-C possesses three unique features, including an additional Ig domain at the extreme NH2-terminus (termed C0), a 9-residue long insertion within the M-motif containing consensus phosphorylation sites and a 28-amino acid long loop in the middle of domain C5 [8, 11]. Conversely, sMyBP-C comprises a subfamily of four alternatively spliced variants (v), v1–v4, differing from one another due to the retention or exclusion of select exons encoding three novel insertions [12]. These are located at the very NH2-terminus within the pro/ala- rich motif, the middle of domain C7, and the very COOH-terminus of the molecule, following the C10 domain (Figure 1; [12]). The presence of these insertions may result in different topographies and possibly functions of the sMyBP-C variants that contain them [12, 13]. Consistent with this, v1, which carries the COOH-terminal insertion preferentially localizes to the periphery of the M-band, rather than the C-zone, where it codistributes with its binding partners obscurin [13] and titin (our unpublished observations) and plays key roles in the structural integrity of M- and A-bands. The last Ig domain (C10) of sMyBP-C v1 interacts with the second Ig domain (Ig2) of obscurin [13]. Although the presence of the novel COOH-terminal insertion further strengthens this interaction, it is not required for binding. However, this novel motif is both necessary and sufficient to support binding of sMyBP-C v1 to the last Ig domain of titin, M10, which is a hotspot for mutations associated with tibial muscular dystrophy [14, 15]. Moreover, our laboratory has recently identified novel phosphorylation sites within the pro/ala- rich motif of the NH2-terminus of sMyBP-C, specifically Ser-59, located in the novel NH2-terminal insertion and Ser-62 are targets of PKA, while Ser-83 and Thr-84 are targets of PKC; Ser-204 is a substrate for both PKA and PKC [16]. Similar to its cardiac counterpart, phosphorylation of these sites may be essential in regulating the activities of sMyBP-C. Nevertheless, despite the differences in the core structure of the cardiac, slow, and fast MyBP-C proteins, they share a ~50% homology and a ~65% identity [12].
Figure 1.
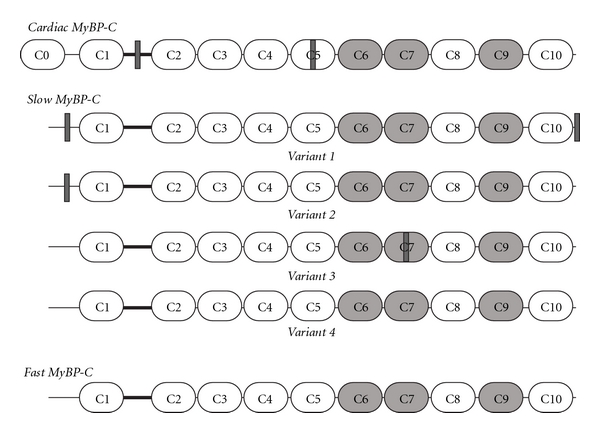
Schematic representation of the three MyBP-C isoforms. White and light grey ovals represent immunoglobulin and fibronectin type-III domains, respectively. The M-motif, flanked by domains C1 and C2 is denoted as a thick black line. Insertions specific to the cardiac isoform and slow variants are shown as dark grey rectangles.
Although MyBP-C was identified over forty years ago as a contaminant of purified myosin [17], a better understanding of its physiology has emerged in the last two decades, paralleling the discovery of mutations in cMyBP-C that result in the development of familial hypertrophic cardiomyopathy (FHC; [18, 19]). Consequently, most of our knowledge originates from studies focusing on the cardiac form of the protein. This review will focus on the role of MyBP-C in regulating the formation of actomyosin cross-bridges via its direct interaction with both myosin and actin filaments.
2. Interactions of cMyBP-C with the Myosin Filament
Myosin is a hexameric protein consisting of two heavy chains (MHC) and two light chains (regulatory and essential, RLC and ELC, resp.). A dimer of heavy chains forms a coiled-coil helix that constitutes the rod or light meromyosin (LMM) segment of myosin [20]. Toward its NH2-terminus, the helix unwinds and each MHC gives rise to a catalytic head, referred to as subfragment 1 (S1) that has ATPase activity and participates in the formation of cross-bridges with filamentous actin [21, 22]. The lever arm, referred to as subfragment 2 (S2), separates the S1 segment from the rod portion, and transduces the chemical energy from the hydrolysis of ATP into mechanical movement along the thin filament [23]. A pair of RLC and ELC binds in tandem to each head of the S1 segment modulating the speed and force of contraction [24, 25].
In addition to myosin light chains and actin, myosin is intimately associated with MyBP-C (Table 1 and Figure 2; [26]). The COOH-terminal C10 domain of all three MyBP-C isoforms harbors binding sites for the LMM portion of myosin [27]. Charged residues R1064, E1079, N1083, R1100, and R1101 present on the surface of the C10 domain of MyBP-C support an electrostatic interaction with residues 1554–1581 of the rod domain of myosin that has a modest affinity of ~3.5 μM [28, 29]. Moreover, through domains C8–C10, MyBP-C interacts with titin, a giant sarcomeric protein spanning half a sarcomere, which is also tightly bound to the thick myosin filament [30]. Specifically, MyBP-C binds to the first Ig domain of an 11-domain superrepeat present in the A-band portion of titin [31]. Thus, it has been speculated that the repetitive binding of MyBP-C to the A-band portion of titin likely contributes to its periodicity within the C-zone [31]. Consistent with these findings, a number of studies have further shown that the COOH-terminal C8–C10 domains are necessary and sufficient to target MyBP-C to the A-band [44, 45].
Table 1.
Ligands of the cardiac isoform of MyBP-C.
| Cardiac MyBP-C | ||||
|---|---|---|---|---|
| MyBP-C Domain | Ligand | Method | Affinity | Reference |
| C0 | Myosin RLC | NMR, ITC | 3.2 μM | [32] |
| C1 | Myosin S1-S2 hinge region | NMR | [33] | |
| M-motif | Myosin S2 | Cosedimentation assay, ITC | 4.3 μM | [34] |
| C2 | Myosin S2 | ITC, NMR | 1.1 mM | [35] |
| C10 | Myosin LMM | Cosedimentation | 3.5 μM | [28, 29, 36] |
| C0 | F-actin | Co-IP, cosedimentation | [37] | |
| C0 and C1 | F-actin | Small angle neutron scattering | [38] | |
| C0–C2 | F-actin | Negative staining electron microscopy | [39] | |
| C1-C2 | F-actin | Cosedimentation | 10 μM | [40] |
| M-motif | F-actin | Cosedimentation | [41] | |
| C5–C10 | F-actin | Cosedimentation | 4.3 μM | [42] |
| C5 | MyBP-C C8 | Y2H, SPR | 10 μM | [43] |
| C7 | MyBP-C C10 | Y2H | [43] | |
| C8–C10 | Titin C-Zone 11-repeat super repeat | Dot-blot | [31] | |
Abbreviations used: RLC, regulatory light chain; NMR, nuclear magnetic resonance; ITC, isothermal calorimetry; LMM, light meromyosin; Co-IP, coimmunoprecipitation; Y2H, yeast 2 hybrid; SPR, surface plasmon resonance.
Figure 2.
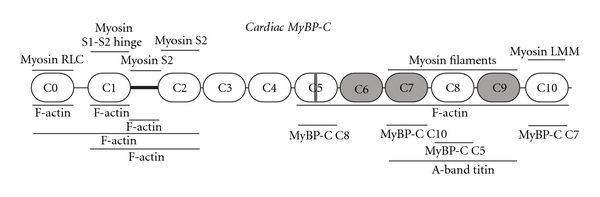
Schematic representation of cardiac MyBP-C, illustrating its domain architecture and known binding partners. Immunoglobulin and fibronectin type III domains are shown as white and grey ovals, respectively. The M-motif is denoted as a dark black line flanked by domains C1 and C2. The cardiac specific insertion within domain C5 is shown as a grey rectangle. Ligands for cMyBP-C are indicated at their sites of interaction.
While binding of the COOH-terminus of MyBP-C to the rod domain of myosin may contribute to the maintenance and stability of the thick filament, (reviewed in [3, 4]), binding of the NH2-terminus of MyBP-C to myosin may mediate contractile regulation [3, 4]. Accordingly, a number of biochemical and structural studies have implicated the NH2-terminal portion of cMyBP-C, containing domains C0–C2, in binding to the S2 region of myosin. Specifically, the M-motif located between the C1 and C2 Ig domains has been shown to bind directly to the NH2-terminal 126 residues of the S2 fragment with an affinity of ~4.3 μM [34]; this interaction is abolished by phosphorylation of specific Ser residues within the M-motif of cMyBP-C via cAMP-dependent protein kinase [46–49]. Similarly, the C2 domain has been also shown to interact with the same 126 residues of the NH2-terminus of the S2 fragment, albeit with considerably lower affinity (~1.1 mM), compared to the M-motif [35]. Interestingly, molecular modeling has suggested that the interaction between the S2 portion of myosin and the C2 domain of cMyBP-C is mediated by polar residues [35]. On the other hand, the C1 domain of cMyBP-C has been reported to interact with the hinge region between the S1 heads and the S2 fragment [33], in close proximity to the myosin light chains, while a recent study reported that the first Ig domain of cMyBP-C, C0, binds to the RLCs with an affinity of ~3.2 μM [32]. In support of this, overexpressed C0 targets to the A-band independently of the rest of the molecule, likely through its interaction with the RLCs [32].
Taken together, these findings strongly suggest that cMyBP-C may regulate the position and thus proximity of the myosin S1 heads relative to the actin filament, through its interaction with the S2 region and the RLCs, therefore affecting the formation of actomyosin cross-bridges.
3. Interactions of cMyBP-C with the Actin Filament
In addition to the mounting experimental support indicating the intimate association of the NH2-terminus of cMyBP-C with the S2 region of myosin, there is also significant evidence supporting its direct interaction with actin thin filaments. Early studies have shown that purified full-length cMyBP-C binds to filamentous actin (F-actin) [36, 50]. However, reconstituted thin filaments (i.e. preincubated with troponin and tropomyosin) fail to bind purified full length cMyBP-C in the presence of EDTA, a Ca2+ chelating agent, but binding is restored upon addition of Ca2+ [36, 50].
Further studies have recently begun to identify the actin-binding domain(s) of cMyBP-C through biochemical and structural approaches. Neutron scattering experiments have indicated that recombinant C0–C2 binds to F-actin in a repetitive manner, stabilizing it [38], while biochemical studies have shown that recombinant C0 is capable of directly interacting with F-actin [37]. Moreover, bacterially expressed C1-C2 associates with naked F-actin as well as reconstituted thin filaments, likely through the M-motif, exhibiting a binding affinity of ~10 μM to naked F-actin [40, 41]. This finding has been also substantiated by a recent study that used negative stain electron microscopy and three-dimensional reconstruction to show that bacterially expressed C0–C3 is well ordered on actin filaments near subdomain 1 [51].
Noticeably, some of the above studies report linear, nonsaturating binding, suggesting the presence of weak electrostatic interactions, as exemplified by the calculated micromolar affinity [41]. Consistent with this, these binding reactions depend largely on pH and ionic strength and are regulated by phosphorylation of cMyBP-C [42]. Thus, an increase in pH or ionic strength significantly decreases the capacity of the aforementioned cMyBP-C peptides to bind to F-actin [40–42]. Similarly, pretreatment of these recombinant peptides with PKA results in a dramatic reduction of their ability to associate with F-actin [41].
Importantly, a recent study by Rybakova and colleagues investigated the actin-binding capabilities of cMyBP-C across its entire length [42]. In support of previously published studies discussed above, the authors found that the NH2-terminal domains of cMyBP-C bind to F-actin in a linear, nonsaturating manner, likely mediated by weak electrostatic interactions. However, recombinant full-length cMyBP-C, expressed in the baculovirus system, supported a direct and saturating interaction with F-actin with a calculated KD of ~4.3 μM. Moreover, constructs lacking domains C0–C5 exhibited similar binding properties as full-length cMyBP-C [42]; this observation prompted the authors to conclude that the weak actin binding mediated by the NH2-terminus of the molecule does not contribute significantly to the overall affinity of cMyBP-C for F-actin. Moreover, the authors further showed that the actin binding supported by the COOH-terminal C6–C10 domains, unlike the NH2-terminal C0–C2 domains, is independent of the regulatory elements, troponin and tropomyosin, of thin filaments, the levels of Ca2+, and the phosphorylation status of the M-motif [42]. Although the COOH-terminus of cMyBP-C exhibits a higher affinity for F-actin, the minimal binding region and its physiological relevance are still elusive. These open questions are especially important because the COOH-terminal domains C6–C10 also harbor binding sites for myosin and titin filaments (discussed above). Indeed, space constraints would favor dynamic rather than simultaneous interactions of the COOH-terminus of cMyBP-C with the actin, myosin, and titin filaments. However, three-dimensional reconstruction of thick filaments demonstrated that domains C7–C10 of cMyBP-C run along the length of the myosin rod [52], suggesting that its interaction with the LMM portion of myosin is not of dynamic nature.
Collectively these studies suggest the presence of various but weak binding sites for F-actin in the NH2-terminus of cMyBP-C as well as the presence of a high affinity, saturating binding site in the COOH-terminus of the molecule. Obviously, more work is required before we can obtain a clear understanding of the physiological relevance of these interactions during muscle contraction and relaxation.
4. Interactions of Myosin and Actin Filaments with Skeletal Muscle MyBP-C
Early work has shown that the skeletal isoforms of MyBP-C also interact directly with both thick and thin filaments; however, their binding is much less characterized (Figure 3 and Table 2). Similar to cMyBP-C, the skeletal isoforms bind to the rod portion of sarcomeric myosin through their C10 domain and to the S2 region through their NH2-terminus [27]. Further characterization of the NH2-terminal binding has shown that the first two Ig domains of sMyBP-C (C1-C2) bind to the S2 region with an affinity of ~2.2 μM [26, 34]. In addition, native skeletal MyBP-C (presumably containing a mixture of the slow and fast isoforms) coaggregates with F-actin in a Ca2+ sensitive manner [50].
Figure 3.
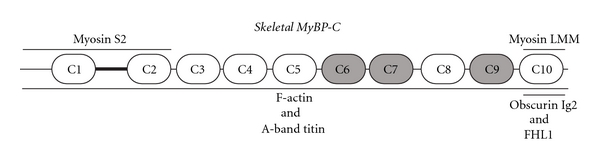
Schematic representation of the core structure of the skeletal isoforms of MyBP-C, depicting their identified binding partners. For ease of representation, the three novel insertions present in MyBP-C slow are not included in the schematic (for detailed description, please see Figure 1 and [12]). Immunoglobulin and fibronectin type III domains are presented as white and grey ovals, respectively. The M-motif is shown as a dark black line flanked by domains C1 and C2. Ligands for skeletal MyBP-C are denoted at their sites of interaction. Notably, obscurin and FHL1 are specific binding partners of v1, since their interactions depend on the presence of the novel COOH-terminal insertion, which is exclusively carried by v1, in addition to the C10 domain.
Table 2.
Ligands of the skeletal isoforms of MyBP-C.
| Skeletal MyBP-C | ||||
|---|---|---|---|---|
| MyBP-C Domain | Ligand | Method | Affinity | Reference |
| C1-C2 | Myosin S2 | Cosedimentation assay, ITC | 2.2 μM | [26, 34] |
| fast C10 | Myosin filaments | Cosedimentation assay | 0.5 μM | [27] |
| Full length | F-actin | Cosedimentation | [36, 50] | |
| Full length | Titin 11-domain super repeat | Dot-blot | [31, 54] | |
| slow variant 1 C10 + insert | Obscurin Ig2 | Y2H, pull down, overlay | [13] | |
| slow variant 1 C10 + insert | FHL1 | Y2H, pull down, Co-IP | [55] | |
Abbreviations used: RLC, regulatory light chain; NMR, nuclear magnetic resonance; ITC, isothermal calorimetry; LMM, light meromyosin; Co-IP, coimmunoprecipitation; Y2H, yeast 2 hybrid; SPR, surface plasmon resonance.
It becomes apparent from the above studies that our knowledge on the interaction of slow or fast MyBP-C with myosin and actin filaments is limited. There are several reasons for this. The identification of multiple mutations in the cardiac isoform that have been causally linked to the development of FHC is a major contributing factor that has shifted the interest of researchers towards cMyBP-C; however, this has recently changed, as mutations in sMyBP-C were identified in patients suffering from distal arthrogryposis type 1 (DA1), a disorder characterized by congenital contractures of the hands and feet ([53]; please see below). More importantly, the molecular diversity of the skeletal isoforms further complicates the relevant studies as there are at least five different skeletal forms of MyBP-C (one fast and four slow variants) that share homologous or common sequences, contain novel insertions in the NH2- and COOH-termini, and may coexist in the same muscle, fiber, or even sarcomere [12]. Further detailed investigation is, therefore, necessary to evaluate the ability of each isoform to bind to myosin and actin filaments as well as the physiological significance of these interactions.
5. MyBP-C in Disease
During the last decade, an overwhelming number of mutations (~200) have been identified in the MYBPC3 gene, which encodes the human cardiac MyBP-C protein. These include missense, nonsense, deletion/insertion, and frame-shift mutations and have been causally linked to the development of modest and late onset familial hypertrophic cardiomyopathy (FCH) [56–58]. The majority of the nonsense and frame-shift mutations result in truncated forms of the protein, that lack the COOH-terminus harboring binding sites for LMM, actin and titin, and thus negate the ability of cMyBP-C to associate with these filaments and regulate contractile activity [59]. Interestingly, these truncated peptides are often undetectable in patient biopsies, possibly due to transcriptional misregulation or rapid degradation [60]. Contrary to non-sense and frame shift mutations, missense mutations are generally associated with a less severe cardiomyopathic phenotype [58] and do not affect the structure or stability of the protein although some of them have been suggested to weaken myosin binding [33].
Moreover, recent genome wide linkage analysis revealed that mutations in the MYBPC1 gene that encodes the human skeletal MyBP-C slow protein lead to the development of distal arthrogryposis type 1, an autosomal dominant disorder characterized by congenital contractures of the hands and feet [53]. Two missense mutations have been identified to date, W236R and Y856H, which are present in the M-motif and C8 domain, respectively [53]. Evaluation of skeletal muscle biopsies obtained from affected individuals revealed that slow twitch fibers were significantly smaller than fast twitch fibers. Importantly, the locations of these two mutations indicate possible alterations in the ability of sMyBP-C to interact with myosin or actin via its NH2-terminus and to associate with the thick and titin filaments through its COOH-terminus.
The causal involvement of the MyBP-C family of proteins in the development of cardiac and skeletal myopathies, as exemplified by the aforementioned studies, clearly indicates that the members of this multifaceted and complex family are essential components and key regulators of muscle structure and function.
6. The Regulatory Activities of MyBP-C-Models and Perspectives
Through its dynamic interactions with myosin and actin filaments, MyBP-C affects the formation and cycling of cross-bridges in three distinct ways: (i) by maintaining the normal structure of myosin and actin filaments, (ii) by regulating the rate at which myosin and actin interact, and (iii) by modulating the ATPase activity of myosin. Consistent with this, the addition of skeletal MyBP-C reduces the critical concentration necessary for myosin polymerization in vitro and results in the formation of longer and more uniform thick filaments [61, 62]. In addition, the NH2-terminal region C0–C2 of cMyBP-C supports actin bundling in vitro, as evidenced by the increased turbidity of F-actin in the solution and the formation of significantly thicker actin filaments, as visualized by electron microscopy [41]. Notably, the effect of cMyBP-C on actin bundling is dependent on pH and regulated by phosphorylation events mediated by PKA [41]. Moreover, in vitro motility assays, examining the sliding of actin filaments over myosin heads in the presence of full-length or truncated forms of cMyBP-C revealed a significant reduction in their sliding velocity [40, 63, 64]. Notably, the degree of reduction was also dependent on the presence of the actin regulatory proteins, troponin, and tropomyosin; in their presence, there was a lesser effect on the decrease of actin motility over full-length myosin [63, 65], HMM [40] or S1 heads [64]. MyBP-C also modulates the ATPase activity of myosin, though in an indirect way, via interactions with both myosin and actin filaments supported by its NH2-terminus. In agreement with this, purified sMyBP-C inhibited the ATPase activity of actin-activated myosin; however, native cMyBP-C moderately enhanced the actin-activated ATPase activity of myosin [66, 67]. Surprisingly though, recombinant cMyBP-C containing the C0–C2 region inhibited the ATPase activity of myosin in the presence of actin [40].
The majority of the aforementioned studies have been performed with native proteins obtained from different species or types of muscles, presumably due to ease of purification. However, Alyonycheva et al. have convincingly shown that the strength of the interaction between myosin and MyBP-C is considerably increased when both proteins are purified from the same source [68]. As such, cMyBP-C and skeletal MyBP-C bound with higher affinity to cardiac and skeletal myosin, respectively [68].
As our knowledge about the molecular and functional properties of MyBP-C broadens, it becomes apparent that through interactions supported by its COOH and NH2 termini, MyBP-C contributes to the maintenance of the normal structure of thick filaments, and the regulation of cross-bridges formation and cycling. It has, therefore, been suggested that MyBP-C may serve as a linker, whereby its COOH-terminus is necessary for anchoring it to LMM in the A-band, leaving the NH2-terminus available to modulate contractility through dynamic interactions with the head region of myosin and actin [4]. Consequently, Calaghan and colleagues proposed the “tether model” to explain how MyBP-C modulates the formation of actomyosin cross-bridges [69]. This model suggests that both myosin binding sites on MyBP-C act together by limiting the movement of the S1 heads relative to LMM and actin, thereby restricting the formation of actomyosin cross-bridges [69]. Phosphorylation of select Ser residues within the M-motif of cMyBP-C, mediated mainly by PKA, regulates the interaction between cMyBP-C and myosin by acting as an “on-off” switch. Consequently, when in the “on” or dephosphorylated state, the NH2-terminus of cMyBP-C binds to myosin and limits its interaction with actin, when in the “off” or phosphorylated state, myosin is free to interact with actin and form cross-bridges [70].
The tether model is centered on the dynamic and highly regulated interaction of MyBP-C with myosin and actin filaments. However, it is necessary to understand the orientation of MyBP-C within the sarcomere, as well. Two proposed models for the topology of MyBP-C have been suggested: a “collar model” in which a trimer of MyBP-C molecules wraps around the rod domain of myosin [43], and an “axial model”, where the COOH-terminus of MyBP-C runs along the length of the rod domain [71]. A stipulation of the collar model is that MyBP-C is required to dimerize [43]. In support of this, it has been shown that MyBP-C dimerizes through interactions mediated by domains C5 and C7 with C8 and C10, respectively [43, 72]. On the other hand, X-ray diffraction in combination with molecular and computational modeling studies support the axial model, suggesting that the last three domains of MyBP-C lay along the length of the myosin rod [71, 73]. Recently, a study using electron microscopy and three-dimensional reconstructions of cardiac thick filaments further demonstrated that the COOH-terminus of cMyBP-C runs along the rod domain surface of thick filaments [52]. Although MyBP-C dimerization is not essential for the arrangement of MyBP-C along the length of the myosin rod, the axial model does not negate its dimerization. Nevertheless, in both models the NH2-terminus of MyBP-C extends into the interfilament space where it is accessible to bind to the S2 portion of myosin and actin. Physiological dynamic interactions of MyBP-C with myosin and actin are possible as the distance between thick and thin filaments is ~9–16 nm [74] and the length of extended MyBP-C is ~40 nm [75]. Therefore, MyBP-C is capable to span the interfilament space, enabling interactions with both actin and myosin filaments.
It is not without precedence that regulation of contractility occurs through dynamic interactions with both actin and myosin filaments, as the essential and regulatory light chains bind tightly to myosin, but also interact with actin [25, 76, 77]. Additionally, twitchin, a protein similar to titin, characterized in C. elegans as a thick filament regulatory protein, interacts with both myosin and actin in a phosphorylation-dependent manner to modulate cross-bridges formation [78, 79]. However, current models (as described above) to explain the mechanism of contractile regulation through dynamic binding to myosin and actin are lacking. Undoubtedly, more work is necessary in order to understand the precise mechanisms by which MyBP-C modulates contractility.
Acknowledgments
Our research has been supported by grants to A. Kontrogianni Konstantopoulos from the National Institutes of Health (R01 AR52768) and to M. A. Ackermann from the National Institutes of Health (F32 AR058079).
References
- 1.Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annual Review of Cell and Developmental Biology. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 2.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extraction, purification and characterization. Journal of Molecular Biology. 1973;74(4):653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 3.Flashman E, Redwood C, Moolman-Smook J, Watkins H. Cardiac myosin binding protein C: its role in physiology and disease. Circulation Research. 2004;94(10):1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 4.Oakley CE, Chamoun J, Brown LJ, Hambly BD. Myosin binding protein-C: enigmatic regulator of cardiac contraction. International Journal of Biochemistry and Cell Biology. 2007;39(12):2161–2166. doi: 10.1016/j.biocel.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Bennett P, Craig R, Starr R, Offer G. The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. Journal of Muscle Research and Cell Motility. 1986;7(6):550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 6.Pepe FA, Ashton FT, Street C, Weisel J. The myosin filament. X. Observation of nine subfilaments in transverse sections. Tissue and Cell. 1986;18(4):499–508. doi: 10.1016/0040-8166(86)90016-9. [DOI] [PubMed] [Google Scholar]
- 7.Pepe FA, Drucker B. The myosin filament. III. C protein. Journal of Molecular Biology. 1975;99(4):609–617. doi: 10.1016/s0022-2836(75)80175-6. [DOI] [PubMed] [Google Scholar]
- 8.Gautel M, Zuffardi O, Freiburg P, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO Journal. 1995;14(9):1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber FE, Vaughan KT, Reinach FC, Fischman DA. Complete sequence of human fast-type and slow-type muscle myosin-binding-protein C (MyBP-C). Differential expression, conserved domain structure and chromosome assignment. European Journal of Biochemistry. 1993;216(2):661–669. doi: 10.1111/j.1432-1033.1993.tb18186.x. [DOI] [PubMed] [Google Scholar]
- 10.Einheber S, Fischman DA. Isolation and characterization of a cDNA clone encoding avian skeletal muscle C-protein: an intracellular member of the immunoglobulin superfamily. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(6):2157–2161. doi: 10.1073/pnas.87.6.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasuda M, Koshida S, Sato N, Obinata T. Complete primary structure of chicken cardiac C-protein (MyBP-C) and its expression in developing striated muscles. Journal of Molecular and Cellular Cardiology. 1995;27(10):2275–2286. doi: 10.1016/s0022-2828(95)91731-4. [DOI] [PubMed] [Google Scholar]
- 12.Ackermann MA, Kontrogianni-Konstantopoulos A. Myosin binding protein-C slow: an intricate subfamily of proteins. Journal of Biomedicine and Biotechnology. 2010;2010:10 pages. doi: 10.1155/2010/652065. Article ID 652065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ackermann MA, Hu LYR, Bowman AL, Bloch RJ, Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Molecular Biology of the Cell. 2009;20(12):2963–2978. doi: 10.1091/mbc.E08-12-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Udd B. Third filament diseases. Advances in Experimental Medicine and Biology. 2008;642:99–115. doi: 10.1007/978-0-387-84847-1_9. [DOI] [PubMed] [Google Scholar]
- 15.Udd B, Haravuori H, Kalimo H, et al. Tibial muscular dystrophy - from clinical description to linkage on chromosome 2q31. Neuromuscular Disorders. 1998;8(5):327–332. doi: 10.1016/s0960-8966(98)00024-8. [DOI] [PubMed] [Google Scholar]
- 16.Ackermann MA, Kontrogianni-Konstantopoulos A. Myosin Binding Protein-C Slow is a Novel Substrate for Protein Kinase A (PKA) and C (PKC) in Skeletal Muscle. Journal of proteome research. 2011 ;10(10):4547–4555. doi: 10.1021/pr200355w. Article ID 652065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starr R, Offer G. Polypeptide chains of intermediate molecular weight in myosin preparations. FEBS Letters. 1971;15(1):40–44. doi: 10.1016/0014-5793(71)80075-3. [DOI] [PubMed] [Google Scholar]
- 18.Bonne G, Carrier L, Bercovici J, et al. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nature Genetics. 1995;11(4):438–440. doi: 10.1038/ng1295-438. [DOI] [PubMed] [Google Scholar]
- 19.Watkins H, Conner D, Thierfelder L, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nature Genetics. 1995;11(4):434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 20.Chakrabarty T, Xiao M, Cooke R, Selvin PR. Holding two heads together: stability of the myosin II rod measured by resonance energy transfer between the heads. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(9):6011–6016. doi: 10.1073/pnas.082024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayment I, Rypniewski WR, Schmidt-Base K, et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 22.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padrón R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436(7054):1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 23.Rayment I, Holden HM, Whittaker M, et al. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261(5117):58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 24.Colson BA, Locher MR, Bekyarova T, et al. Differential roles of regulatory light chain and myosin binding protein-C phosphorylations in the modulation of cardiac force development. Journal of Physiology. 2010;588(6):981–993. doi: 10.1113/jphysiol.2009.183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morano I. Tuning the human heart molecular motors by myosin light chairs. Journal of Molecular Medicine. 1999;77(7):544–555. doi: 10.1007/s001099900031. [DOI] [PubMed] [Google Scholar]
- 26.Starr R, Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. Biochemical Journal. 1978;171(3):813–816. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okagaki T, Weber FE, Fischman DA, Vaughan KT, Mikawa T, Reinach FC. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. Journal of Cell Biology. 1993;123(3):619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flashman E, Watkins H, Redwood C. Localization of the binding site of the C-terminal domain of cardiac myosin-binding protein-C on the myosin rod. Biochemical Journal. 2007;401(1):97–102. doi: 10.1042/BJ20060500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto CA, Fischman DA, Reinach FC. The interface between MyBP-C and myosin: site-directed mutagenesis of the CX myosin-binding domain of MyBP-C. Journal of Muscle Research and Cell Motility. 1999;20(7):703–715. doi: 10.1023/a:1005513312939. [DOI] [PubMed] [Google Scholar]
- 30.Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiological Reviews. 2009;89(4):1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. European Journal of Biochemistry. 1996;235(1-2):317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 32.Ratti J. Structure and interactions of myosin-binding protein c domain c0: cardiac-specific regulation of myosin at its neck? Journal of Biological Chemistry. 2011;286(14):12650–12658. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ababou A, Rostkova E, Mistry S, Masurier CL, Gautel M, Pfuhl M. Myosin binding protein C positioned to play a key role in regulation of muscle contraction: structure and interactions of domain C1. Journal of Molecular Biology. 2008;384(3):615–630. doi: 10.1016/j.jmb.2008.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruen M, Gautel M. Mutations in β-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. Journal of Molecular Biology. 1999;286(3):933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 35.Ababou A, Gautel M, Pfuhl M. Dissecting the N-terminal myosin binding site of human cardiac myosin-binding protein C: structure and myosin binding of domain C2. Journal of Biological Chemistry. 2007;282(12):9204–9215. doi: 10.1074/jbc.M610899200. [DOI] [PubMed] [Google Scholar]
- 36.Moos C, Mason CM, Besterman JM. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. Journal of Molecular Biology. 1978;124(5):571–586. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 37.Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. Effect of MyBP-C binding to actin on contractility in heart muscle. Journal of General Physiology. 2003;122(6):761–774. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: implications for cardiac function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(47):18360–18365. doi: 10.1073/pnas.0808903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kensler RW, Shaffer JF, Harris SP. Binding of the N-terminal fragment C0-C2 of cardiac MyBP-C to cardiac F-actin. Journal of Structural Biology. 2011;174(1):44–51. doi: 10.1016/j.jsb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Razumova MV, Shaffer JF, Tu AY, Flint GV, Regnier M, Harris SP. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: evidence for long-lived cross-bridges. Journal of Biological Chemistry. 2006;281(47):35846–35854. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 41.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. Journal of Biological Chemistry. 2009;284(18):12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rybakova IN, Greaser ML, Moss RL. Myosin binding protein C interaction with actin: characterization and mapping of the binding site. The Journal of Biological Chemistry. 2011;286(3):2008–2016. doi: 10.1074/jbc.M110.170605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moolman-Smook J, Flashman E, De Lange W, et al. Identification of novel interactions between domains of myosin binding protein-C that are modulated by hypertrophic cardiomyopathy missense mutations. Circulation Research. 2002;91(8):704–711. doi: 10.1161/01.res.0000036750.81083.83. [DOI] [PubMed] [Google Scholar]
- 44.Gilbert R, Cohen JA, Pardo S, Basu A, Fischman DA. Identification of the A-band localization domain of myosin binding proteins C and H (MyBP-C, MyBP-H) in skeletal muscle. Journal of Cell Science. 1999;112(1):69–79. doi: 10.1242/jcs.112.1.69. [DOI] [PubMed] [Google Scholar]
- 45.Gilbert R, Kelly MG, Mikawa T, Fischman DA. The carboxyl terminus of myosin binding protein C (MyBP-C, C-protein) specifies incorporation into the A-band of striated muscle. Journal of Cell Science. 1996;109(1):101–111. doi: 10.1242/jcs.109.1.101. [DOI] [PubMed] [Google Scholar]
- 46.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Letters. 1999;453(3):254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 47.Sadayappan S, Gulick J, Klevitsky R, et al. Cardiac myosin binding protein-C phosphorylation in a β-myosin heavy chain background. Circulation. 2009;119(9):1253–1262. doi: 10.1161/CIRCULATIONAHA.108.798983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadayappan S, Gulick J, Osinska H, et al. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circulation Research. 2005;97(11):1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadayappan S, Osinska H, Klevitsky R, et al. Cardiac myosin binding protein c phosphorylation is cardioprotective. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(45):16918–16923. doi: 10.1073/pnas.0607069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto K. The binding of skeletal muscle C-protein to regulated actin. FEBS Letters. 1986;208(1):123–127. doi: 10.1016/0014-5793(86)81545-9. [DOI] [PubMed] [Google Scholar]
- 51.Mun JY, et al. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C) Journal of Molecular Biology. 2011;410(2):214–225. doi: 10.1016/j.jmb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(7):2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurnett CA, Desruisseau DM, McCall K, et al. Myosin binding protein C1: a novel gene for autosomal dominant distal arthrogryposis type 1. Human Molecular Genetics. 2010;19(7):1165–1173. doi: 10.1093/hmg/ddp587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fürst DO, Vinkemeier U, Weber K. Mammalian skeletal muscle C-protein: purification from bovine muscle, binding to titin and the characterization of a full-length human cDNA. Journal of Cell Science. 1992;102(4):769–778. doi: 10.1242/jcs.102.4.769. [DOI] [PubMed] [Google Scholar]
- 55.McGrath MJ, Cottle DL, Nguyen MA, et al. Four and a half LIM protein 1 binds myosin-binding protein C and regulates myosin filament formation and sarcomere assembly. Journal of Biological Chemistry. 2006;281(11):7666–7683. doi: 10.1074/jbc.M512552200. [DOI] [PubMed] [Google Scholar]
- 56.Harris SP, Lyons RG, Bezold KL. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circulation Research. 2011;108(6):751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konno T, Chang S, Seidman JG, Seidman CE. Genetics of hypertrophic cardiomyopathy. Current Opinion in Cardiology. 2010;25(3):205–209. doi: 10.1097/HCO.0b013e3283375698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Q, Dewey S, Nguyen S, Gomes AV. Malignant and benign mutations in familial cardiomyopathies: insights into mutations linked to complex cardiovascular phenotypes. Journal of Molecular and Cellular Cardiology. 2010;48(5):899–909. doi: 10.1016/j.yjmcc.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 59.Oakley CE, Hambly BD, Curmi PM, Brown LJ. Myosin binding protein C: structural abnormalities in familial hypertrophic cardiomyopathy. Cell Research. 2004;14(2):95–110. doi: 10.1038/sj.cr.7290208. [DOI] [PubMed] [Google Scholar]
- 60.Rottbauer W, Gautel M, Zehelein J, et al. Novel splice donor site mutation in the cardiac myosin-binding protein- C gene in familial hypertrophic cardiomyopathy. Characterization of cardiac transcript and protein. Journal of Clinical Investigation. 1997;100(2):475–482. doi: 10.1172/JCI119555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davis JS. Interaction of C-protein with pH 8.0 synthetic thick filaments prepared from the myosin of vertebrate skeletal muscle. Journal of Muscle Research and Cell Motility. 1988;9(2):174–183. doi: 10.1007/BF01773739. [DOI] [PubMed] [Google Scholar]
- 62.Koretz JF. Effects of C-protein on synthetic myosin filament structure. Biophysical Journal. 1979;27(3):433–446. doi: 10.1016/S0006-3495(79)85227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saber W, Begin KJ, Warshaw DM, VanBuren P. Cardiac myosin binding protein-C modulates actomyosin binding and kinetics in the in vitro motility assay. Journal of Molecular and Cellular Cardiology. 2008;44(6):1053–1061. doi: 10.1016/j.yjmcc.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaffer JF, Razumova MV, Tu AY, Regnier M, Harris SP. Myosin S2 is not required for effects of myosin binding protein-C on motility. FEBS Letters. 2007;581(7):1501–1504. doi: 10.1016/j.febslet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Shchepkin DV, Kopylova GV, Nikitina LV, Katsnelson LB, Bershitsky SY. Effects of cardiac myosin binding protein-C on the regulation of interaction of cardiac myosin with thin filament in an in vitro motility assay. Biochemical and Biophysical Research Communications. 2010;401(1):159–163. doi: 10.1016/j.bbrc.2010.09.040. [DOI] [PubMed] [Google Scholar]
- 66.Yamamoto K, Moos C. The C-proteins of rabbit red, white, and cardiac muscles. Journal of Biological Chemistry. 1983;258(13):8395–8401. [PubMed] [Google Scholar]
- 67.Margossian SS. Reversible dissociation of dog cardiac myosin regulatory light chain 2 and its influence on ATP hydrolysis. Journal of Biological Chemistry. 1985;260(25):13747–13754. [PubMed] [Google Scholar]
- 68.Alyonycheva TN, Mikawa T, Reinach FC, Fischman DA. Isoform-specific interaction of the myosin-binding proteins (MyBPs) with skeletal and cardiac myosin is a property of the C-terminal immunoglobulin domain. Journal of Biological Chemistry. 1997;272(33):20866–20872. doi: 10.1074/jbc.272.33.20866. [DOI] [PubMed] [Google Scholar]
- 69.Calaghan SC, Trinick J, Knight PJ, White E. A role for C-protein in the regulation of contraction and intracellular Ca2+ in intact rat ventricular myocytes. Journal of Physiology. 2000;528(1):151–156. doi: 10.1111/j.1469-7793.2000.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carrier L. Cardiac myosin-binding protein C in the heart. Archives des Maladies du Coeur et des Vaisseaux. 2007;100(3):238–243. [PubMed] [Google Scholar]
- 71.Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. Journal of Molecular Biology. 2003;331(3):713–724. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 72.Flashman E, Korkie L, Watkins H, Redwood C, Moolman-Smook JC. Support for a trimeric collar of myosin binding protein C in cardiac and fast skeletal muscle, but not in slow skeletal muscle. FEBS Letters. 2008;582(3):434–438. doi: 10.1016/j.febslet.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 73.Rome E, Offer G, Pepe FA. X ray diffraction of muscle labelled with antibody to C protein. Nature New Biology. 1973;244(135):152–154. doi: 10.1038/newbio244152a0. [DOI] [PubMed] [Google Scholar]
- 74.Julian FJ, Moss RL, Waller GS. Mechanical properties and myosin light chain composition of skinned muscle fibres from adult and new-born rabbits. Journal of Physiology. 1981;311:201–218. doi: 10.1113/jphysiol.1981.sp013581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hartzell HC, Sale WS. Structure of C protein purified from cardiac muscle. Journal of Cell Biology. 1985;100(1):208–215. doi: 10.1083/jcb.100.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Timson DJ. Fine tuning the myosin motor: the role of the essential light chain in striated muscle myosin. Biochimie. 2003;85(7):639–645. doi: 10.1016/s0300-9084(03)00131-7. [DOI] [PubMed] [Google Scholar]
- 77.Farman GP, Miller MS, Reedy MC, et al. Phosphorylation and the N-terminal extension of the regulatory light chain help orient and align the myosin heads in Drosophila flight muscle. Journal of Structural Biology. 2009;168(2):240–249. doi: 10.1016/j.jsb.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Butler TM, Mooers SU, Narayan SR, Siegman MJ. The N-terminal region of twitchin binds thick and thin contractile filaments: redundant mechanisms of catch force maintenance. Journal of Biological Chemistry. 2010;285(52):40654–40665. doi: 10.1074/jbc.M110.166041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Funabara D, Hamamoto C, Yamamoto K, et al. Unphosphorylated twitchin forms a complex with actin and myosin that may contribute to tension maintenance in catch. Journal of Experimental Biology. 2007;210(24):4399–4410. doi: 10.1242/jeb.008722. [DOI] [PMC free article] [PubMed] [Google Scholar]


