Abstract
An EtOAc extract from the roots of Sophora flavescens (Kushen) potentiated γ -aminobutyric acid (GABA)-induced chloride influx in Xenopus oocytes transiently expressing GABAA receptors with subunit composition, α1β2γ2S. HPLC-based activity profiling of the extract led to the identification of 8-lavandulyl flavonoids, kushenol I, sophoraflavanone G, (–)-kurarinone, and kuraridine as GABAA receptor modulators. In addition, a series of inactive structurally related flavonoids were characterized. Among these, kushenol Y (4) was identified as a new natural product. The 8-lavandulyl flavonoids are first representatives of a novel scaffold for the target.
Keywords: GABAA receptor modulators, HPLC-based activity profiling, 8-Lavandulyl flavonoids, Sophora flavescens, Fabaceae
Introduction
Gamma-aminobutyric acid type A (GABAA) receptors mediate inhibitory neurotransmission in the brain. The receptors are composed of five subunits forming a central pore that is permeable for chloride ions upon activation by the endogenous ligand γ -aminobutyric acid (GABA). Up to now 19 different subunit isoforms have been identified in the human genome and form GABAA receptors in numerous combinations [1]. The most abundant GABAA receptor subtype is composed of 2α1, 2β2, and 1γ2 subunits. More than 10 subtypes composed of other subunit combinations have been identified [2], which differ in tissue localization, functional characteristics, and pharmacological properties [3,4]. GABAA receptors possess several binding sites for small molecules, and are target for numerous drugs used to treat anxiety, panic, insomnia, and epilepsy [5,6]. However, use of the most frequently prescribed benzodiazepines is associated with undesirable side effects including reduced coordination, cognitive impairment, increased accident proneness, physiological dependence, and withdrawal symptoms [5,7]. Rational lead discovery approaches are presently not possible due to the lack of a high-resolution structure of the GABAA receptor [3,8].
Various natural products modulating GABAA receptors (e.g., flavonoids, monoterpenes, diterpenes, neolignans, β-carbolines, and polyacetylenes) have been identified [9, 10]. Given the diversity of these natural product ligands which structurally differ from synthetic GABAA receptor modulators, we recently initiated a project aiming at the discovery of GABAA receptor agonists with natural product scaffolds new for the target. In a screening of 880 plant and fungal extracts with an automated functional assay using Xenopus oocytes expressing human GABAA receptors of the most abundant subtype composition (α1β2γ2S), an EtOAc extract of the traditional Chinese herbal drug Kushen (roots of Sophora flavescens Aiton, Fabaceae) displayed promising activity.
HPLC-based activity profiling is a miniaturized and highly effective approach for localization, dereplication, and characterization of bioactive natural products in extracts [11]. This approach has been successfully established and used in our labs with various cell-based and biochemical assays [12–14]. We recently developed and validated a profiling approach for the discovery of new GABAA receptor ligands [15], which we subsequently employed to identify scaffolds new for the target [16,17]. We here describe HPLC-based activity profiling of the active S. flavescens extract, identification of the active constituents, and preliminary SAR (structure activity relationship) considerations based on a focused compound library generated from the extract.
Experimental section
General experimental procedures
Optical rotations were measured on a M341 polarimeter (Perkin-Elmer). CD spectra were recorded on a JASCO J-720W spectrophotometer. UV spectra were measured on Hewlett-Packard 8452A diode array spectrophotometer (λmax in nm). IR spectra were measured on a Nicolet Magna-FT-IR-750 spectrometer (νmax in cm−1). NMR spectra were recorded at 291 K with a Bruker Avance III™ spectrometer operating at 500.13 MHz for 1H, and 125.77 MHz for 13C, respectively. 1H-NMR experiments and 2D homonuclear and heteronuclear NMR spectra were measured with a 1 mm TXI probe, and 13C-NMR spectra were obtained in 3 mm tubes (Armar Chemicals) with a 5 mm BBO probe. Spectra were analyzed using Bruker TopSpin 2.1 software. High resolution mass spectra (HPLC–PDA–ESITOFMS) were obtained on a micro TOF ESI–MS system (Bruker Daltonics) connected via T-splitter (1:10) to an HP 1100 series system (Agilent) consisting of a binary pump, autosampler, column oven, and diode array detector (G1315B). Data acquisition and processing was performed using HyStar 3.0 software (Bruker Daltonics). Semi-preparative HPLC separations for activity profiling and offline microprobe NMR was performed with an HP 1100 series system (Agilent) consisting of a quaternary pump, autosampler, column oven, and diode array detector (G1315B). Parallel evaporation of microfractions and semi-preparative HPLC fractions was performed with an EZ-2 plus vacuum centrifuge (Genevac). SunFire C18 (3.5 μm, 3.0×150 mm i.d.) and a SunFire Prep C18 (5 μm, 10 × 150 mm i.d.) columns (Waters) were used for HPLC–PDA–ESITOFMS and semi-preparative HPLC, respectively. HPLC-grade acetonitrile (Scharlau Chemie) and water were used for HPLC separations. Solvents used for extraction and column chromatography were of analytical grade. Sephadex LH-20 gel was obtained from Amersham Biosciences (Uppsala, Sweden). Silica gel 60 F254 pre-coated Al sheets (0.25 mm) and silica gel 60 GF254 glass plates (1.00 mm) (both Merck) were used for TLC and preparative TLC, respectively.
Plant material
Kushen (dried roots of S. flavescens) was purchased from Yong Quan GmbH (Ennepetal, Germany). A voucher specimen (P00414) is deposited at the Institute of Pharmaceutical Biology, University of Basel.
Extraction
The plant material was frozen with liquid nitrogen and ground with a ZM1 ultracentrifugal mill (Retsch). The extract for the screening and HPLC-based activity profiling was prepared with an ASE 200 extraction system with solvent module (Dionex) by extraction with ethyl acetate. Extraction pressure was 120 bar and temperature was set at 70 °C. Extract was obtained by two extraction cycles of 5 min each. For isolation of larger amounts of pure compounds for pharmacological testing, 50 g of ground roots were extracted by maceration at room temperature with petroleum ether (4×0.5 l, 4 h each), followed by ethyl acetate (4 × 0.5 l, 4 h each). The solvents were evaporated at reduced pressure to yield 0.9 and 2.3 g of petroleum ether and ethyl acetate extract, respectively. The extracts were stored at −20 °C until use.
Microfractionation for activity profiling
Microfractionation for GABAA receptor activity profiling was performed as previously described [15], with minor modifications: separation was carried out on a semi-preparative HPLC column with acetonitrile (solvent A) and water containing 0.1% formic acid (solvent B) using the following gradient: 20% A to 100% A for 30 min, hold at 100% A for 5 min. The flow rate was 5 mL/ min, and 50 μL of extract (100 mg/ml in DMSO) were injected. A total of 22 time-based microfractions of 90 s each were collected. Solvent removal of microfractions was carried out with a centrifugal evaporator (Genevac AZ-2), and films reconstituted in DMSO prior to activity testing.
HPLC–PDA–ESITOFMS
The ethyl acetate extract of S. flavescens was analyzed with acetonitrile (solvent A) and water containing 0.1% formic acid (solvent B) using an optimized gradient profile: 20–100% A in 30 min, 100% A, hold at 100% A for 5 min. The flow rate was 0.5 mL/min. The sample was dissolved in DMSO at a concentration of 10 mg/mL, and the injection volume was 10 μL. ESITOFMS spectra were recorded in the range of m/z 100–1500 in positive ion mode. Nitrogen was used as a nebulizing gas at a pressure of 2.0 bar and as a drying gas at a flow rate of 9.0 L/ min (dry gas temperature 240 °C). Capillary voltage was set at 4500 V, hexapole at 230.0 Vpp. Instrument calibration was performed using a reference solution of sodium formiate 0.1% in isopropanol/water (1:1) containing 5 mM sodium hydroxide.
Preparative isolation of compounds for GABAA receptor activity test
A portion (0.8 g) of the ethyl acetate extract was dissolved in 2 mL of DMSO and the solution was filtered. Separation of the ethyl acetate extract was carried out with the same solvent system and gradient elution as for HPLC–PDA–ESITOFMS. The flow rate was set at 5 mL/ min and the injected volume of extract was 200 μL at a concentration of 400 mg/mL in DMSO. A total of 13 peak-based fractions were collected manually. A total of 10 injections were carried out, and corresponding fractions were combined. Final purification were as follows: peaks 2–5, 7, 8, and 10–13 were filtered through a Sephadex LH-20 column (300 × 10 mm, MeOH containing 0.1% formic acid) to yield pure compounds 3 (5.6 mg), 4 (1.8 mg), 5 (2.3 mg), 6 (3.8 mg), 9 (67 mg), 10 (2.9 mg), 13 (4.3 mg), 14 (3.1 mg), 15 (2.1 mg), and 16 (1.2 mg). Peak 1 was separated by preparative TLC (CHCl3:MeOH:formic acid—100:10:0.5) to afford compounds 1 (2.9 mg) and 2 (3.3 mg). Peak 6 was separated by preparative TLC (CHCl3:MeOH:formic acid—150:10:0.5) to give compounds 7 (3.4 mg) and 8 (2.2 mg). Peak 9 was also purified by preparative TLC (CHCl3:MeOH:formic acid—150:10:0.5) to give compounds 11 (2.4 mg) and 12 (3.6 mg).
Microprobe NMR analysis
1 mm NMR tubes (Bruker) were used for 1H detected 1D and 2D NMR experiments, and 0.1 mg of the above 16 pure compounds was dissolved in 10 μl CD3OD. The following settings were used: 128 scans to record 1H-spectra; 8 scans for 1H,1 H-COSY spectra using cosygpqf pulse program; 32 scans and 256 increments to record HSQC experiments using the hsqcedetgp or hsqcetgpsi2 pulse program. HMBC spectra were recorded with 64 scans and 128 increments using the hmbcgplpndqf pulse program.
Kushenol Y (4)
Light yellow amorphous powder; . CD (MeOH) [θ]315 + 5897, [θ]293 − 17061. UV (MeOH) λmax (logε): 287 (4.13), 326 (3.78) nm. IR (film) νmax: 3401 (br), 2968, 2943, 1651, 1610, and 1268 cm−1. 1H-NMR (500.13 MHz, CD3OD) δ: 1.11 (s, 6H, H-6″, 7″), 1.23 (m, 2H, H-3″), 1.35 (m, 2H, H-4″), 1.59 (s, 3H, H-10″), 2.38 (m, 1H, H-2″), 2.42 (m, 2H, H-1″), 4.52 (s, 1H, H-9″), 4.59 (s, 1H, H-9″), 4.74 (d, 1H, J = 11.6 Hz, H-3), 5.39 (d, 1H, J = 11.6 Hz, H-2), 5.96 (s, 1H, H-6), 6.35 (d, 1H, J = 2.1Hz, H-3′), 6.39 (dd, 1H, J = 8.0, 2.1Hz, H-5′), 7.28 (d, 1H, J = 8.0Hz, H-6′); 13C-NMR (125.77 MHz, CD3OD) δ : 196.5 (C-4), 165.7 (C-9), 160.7 (C-7), 158.0 (C-5), 157.9 (C-2′), 155.9 (C-4′), 148.3 (C-8″), 128.8 (C-6′), 115.2 (C-1′), 110.8 (C-9″), 108.8 (8), 107.8 (C-5′), 103.6 (C-10), 100.4 (C-3′), 96.1 (C-6), 78.1 (C-2), 72.7 (C-3), 71.3 (C-5″), 46.9 (C-2″), 41.4 (C-4″), 26.4 (C-3″), 28.6 (C-6″), 27.3 (C-1″), 28.8 (C-7″), 18.7 (C-10″). HREIMS m/z: 481.1846 (calcd. for C25H30O8Na, 481.1838).
Expression of GABAA receptors
Stage V–VI oocytes from Xenopus laevis were prepared and cRNA was injected as previously described by Khom et al. [18]. Female X. laevis (NASCO, Fort Atkinson, WI, USA) were anesthetized by exposing them for 15 min to a 0.2% MS-222 (methanesulfonate salt of 3-aminobenzoic acid ethyl, Sigma, Vienna, Austria) solution before surgically removing parts of the ovaries. Follicle membranes from isolated oocytes were enzymatically digested with 2 mg/mL collagenase (Type 1A, Sigma, Vienna, Austria). Synthesis of capped run-off poly(A+) cRNA transcripts was obtained from linearized cDNA templates (pCMV vector). One day after enzymatic isolation, the oocytes were injected with 50 nL of DEPC-treated water (Sigma) containing different cRNAs at a concentration of approximately 300–3000 pg/nL per subunit. To ensure expression of α1β2γ2S receptors, rat cRNA of α1,β2, and γ2S subunits were mixed in a 1:1:10 ratio. The amount of injected cRNA mixture was determined by means of a NanoDrop ND-1000 (Kisker Biotech, Steinfurt, Germany). Oocytes were then stored at 18 °C in ND96 solution (90mM NaCl, 1mM KCl, 1mM MgCl2, 1 mM CaCl2, and 5 mM HEPES; pH 7.4) [19] Voltage clamp measurements were performed between days 1 and 5 after cRNA injection.
Two-microelectrode voltage clamp studies
Electrophysiological experiments were performed by the two-microelectrode voltage clamp method making use of a TURBO TEC-03X amplifier (npi electronic GmbH, Tamm, Germany) at a holding potential of −70 mV and pCLAMP 10 data acquisition software (Molecular Devices, Sunnyvale, CA, USA). Currents were low-pass-filtered at 1 kHz and sampled at 3 kHz. ND 96 solution was used as bath solution. The electrode filling solution consisted of 2 M KCl. Oocytes with maximal current amplitudes >3 μA were discarded to exclude voltage clamp errors. Before application of test solutions, a dose–response experiment with GABA concentrations ranging from 0.1 to 1 mM was performed to determine GABA EC3–10 (typically between 3 and 10 μM).
Test solution application
Test solutions were applied by means of a ScreeningTool (npi electronic GmbH, Tamm, Germany) automated fast perfusion system [20] As previously described, microfractions collected from the semi-preparative HPLC separations were dissolved in 30 μL DMSO and mixed with 2.97 mL of bath solution [15]. After addition of GABA EC3–10, 100 μL of this solution was applied to the oocytes at a perfusion speed of 300 μL/s. Stock solutions of compounds 6–16 (10 mM in DMSO) were diluted to a concentration of 100 μM with bath solution and then mixed with GABA EC3–10. Of each solution, 100 μL was applied to the oocytes at a speed of 300 μL/s.
Data analysis
Enhancement of the chloride current (IGABA) was defined as I(GABA+Comp)/IGABA−1, where I(GABA+Comp) is the current response in the presence of a given compound, and IGABA is the control GABA induced current. Concentration–response curves were generated, and the data were fitted by nonlinear regression analysis using ORIGIN software (OriginLab Corporation, Northampton, MA, USA). Data were fitted to the equation , where EC50 is the concentration of the compound that increases the amplitude of the GABA-evoked current by 50%, and nH is the Hill coefficient. Data are given as mean ± S.E. of at least two oocytes and ≥2 oocyte batches.
Results and discussion
Extracts were screened by means of an automated, fast perfusion system during two-microelectrode voltage clamp measurements in Xenopus oocytes expressing functional GABAA receptors with defined subunit composition (α1β2γ2S) [20]. When tested at 100 μg/mL, the S. flavescens EtOAc extract enhanced GABA induced chloride ion current (IGABA) by 55.0 ± 7.4% (n = 3, Fig. 1).
Fig. 1.
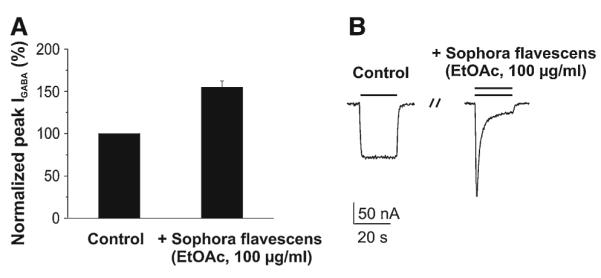
Potentiation of IGABA by Sophora flavescens extract (EtOAc, 100 μg/mL). a The mean IGABA potentiation of 55.0 ± 7.4% (n = 3; compared to 100% control) was observed. b Chloride currents through α1β2γ2S GABAA receptors induced by GABA EC3–10 (control; left column) and currents during co-application of GABA (EC3–10) and 100 μg/mL of the Sophora flavescens extract (right column)
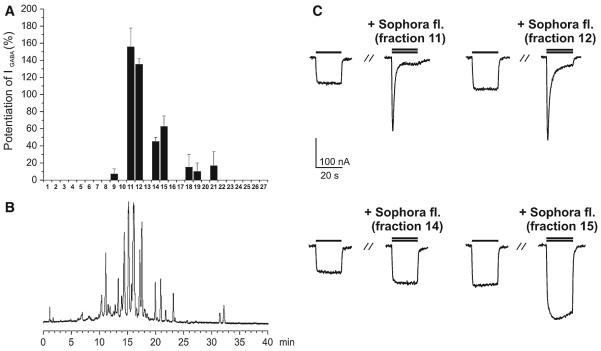
The extract was then submitted to HPLC-based activity profiling using a previously validated protocol [15]. The chromatogram of a semi-preparative separation of 5 mg extract, the corresponding potentiation of IGABA by the time-based fractionations (27 microfractions of 90 s each), and representative currents of the most active fractions are shown in Fig. 2. Fractions 11 and 12 displayed the highest potentiation of IGABA (155.6 ± 22.2%, n = 2 and 135.1 ± 6.6%, n = 2, respectively). These fractions contained the major compounds of the extract. Fractions 14 and 15 potentiated IGABA to a lesser extent (45.0 ± 5.0%, n = 2 and 62.5 ± 12.5%, n = 2, respectively), while the effects of fractions 9, 18, 19, and 21 were not significantly different from zero (P > 0.05, see Fig. 2).
Fig. 2.
HPLC-based activity profiling of Sophora flavescens extract (EtOAc) for GABAA receptor modulating properties. b The HPLC chromatogram (254 nm) of a semi-preparative separation of 5 mg extract is shown. a Potentiation (in %) of IGABA through α1β2γ2S GABAA receptors by 27 fractions of 90 s each fraction is shown above. c Representative currents through α1β2γ2S GABAA receptors in the presence of GABA (EC3–10, single bar, control) and currents recorded during co-application of GABA (EC3–10) and the indicated fraction (double bar). Current recordings for four most active fractions (11, 12, 14, and 15) are shown
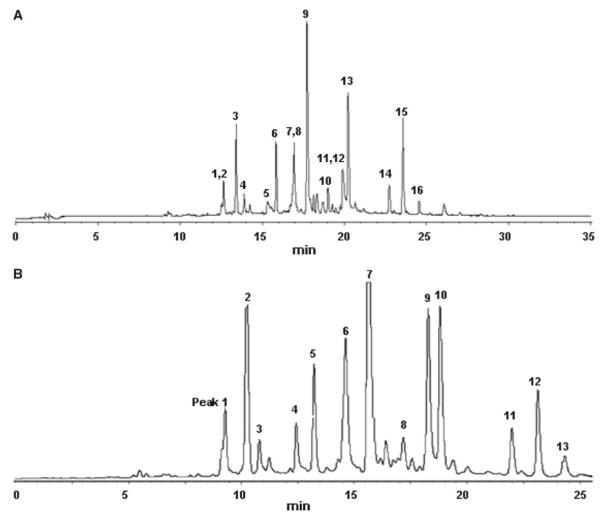
Considering that some HPLC peaks showed partial overlap, separation conditions were optimized for full resolution of the critical HPLC peaks. These conditions were then used to measure high-resolution LC–MS data (Fig. 3a; Table 1). Results of the HPLC–PDA–TOFMS analysis and a database search are summarized in Table 1. The same optimized conditions were also used in preparative isolation of target compounds. A typical semi-preparative HPLC chromatogram obtained with an injection of 80 mg of extract is shown in Fig. 3b. A total of 13 peaks were collected and submitted to further purification with Sephadex LH-20 column and preparative TLC to afford 16 compounds for which 1D and 2D NMR spectra were recorded with a 1 mm TXI probe. The purity was checked by analytical HPLC under conditions as in Fig. 3a.
Fig. 3.
a The optimized HPLC separation for LC–PDA–TOFMS analysis of the EtOAc extract is shown. Peak labeling corresponds to compounds 1–16. 200 μg extract in 20 μL DMSO were injected. The HPLC trace was recorded at 254 nm. b The chromatogram (254 nm) of the optimized, semi-preparative HPLC separation of the EtOAc extract (80 mg in 200 μL DMSO) is shown. Peaks 1–13 were collected for microprobe NMR and further purified if required
Table 1.
Key data from HPLC–PDA–ESITOFMS analysis, semi-preparative HPLC, and database search for compounds 1–16
| Cpd | Rt1(min)a | Rt2(min)b | λmax (nm) | Accurate mass [M + Na]+ found |
Accurate mass [M + Na]+ calc. |
Molecular formula | DNP hitsc |
|---|---|---|---|---|---|---|---|
| 1 | 12.6 | 9.4 | 207, 288, 328 | 495.1987 | 495.1995 | C26H32O8 | 2 |
| 2 | 12.6 | 9.4 | 207, 288, 328 | 495.1987 | 495.1995 | C26H32O8 | 2 |
| 3 | 13.4 | 10.3 | 210, 292, 338 | 479.2055 | 479.2046 | C26H32O7 | 3 |
| 4 | 13.9 | 11.7 | 209, 287, 326 | 481.1846 | 481.1838 | C25H30O8 | 1 |
| 5 | 15.3 | 12.5 | 212, 293, 340 | 479.2035 | 479.2046 | C26H32O7 | 3 |
| 6 | 15.8 | 13.3 | 210, 292, 338 | 465.1901 | 465.1889 | C25H30O7 | 2 |
| 7 | 16.9 | 14.2 | 212, 290, 333 | 477.1896 | 477.1889 | C26H30O7 | 5 |
| 8 | 16.9 | 14.2 | 212, 290, 333 | 477.1896 | 477.1889 | C26H30O7 | 5 |
| 9 | 17.8 | 15.6 | 213, 289, 321 | 461.1952 | 461.1940 | C26H30O6 | 16 |
| 10 | 19.1 | 17.2 | 209, 296, 341 | 463.1741 | 463.1733 | C25H28O7 | 8 |
| 11 | 19.9 | 18.3 | nd | 475.2089 | 475.2097 | C27H32O6 | 2 |
| 12 | 19.9 | 18.3 | 273, 304, 363 | 461.1585 | 461.1576 | C25H26O7 | 1 |
| 13 | 20.3 | 19.3 | 208, 292, 336 | 447.1788 | 447.1784 | C25H28O6 | 10 |
| 14 | 22.8 | 22.0 | 213, 294, 318, 340 | 461.1949 | 461.1940 | C26H30O6 | 16 |
| 15 | 23.7 | 23.2 | 246, | 394 461.1937 | 461.1940 | C26H30O6 | 16 |
| 16 | 24.6 | 24.4 | 242, 295, 321 | 431.1839 | 431.1834 | C25H28O5 | 10 |
Retention time in the HPLC–PDA–ESITOFMS analysis,
retention time in the semi-preparative HPLC separation,
hits in the natural products database (Chapman and Hall Dictionary of Natural Products); search query limited by the term “Sophora” and the corresponding molecular formula
Not determined
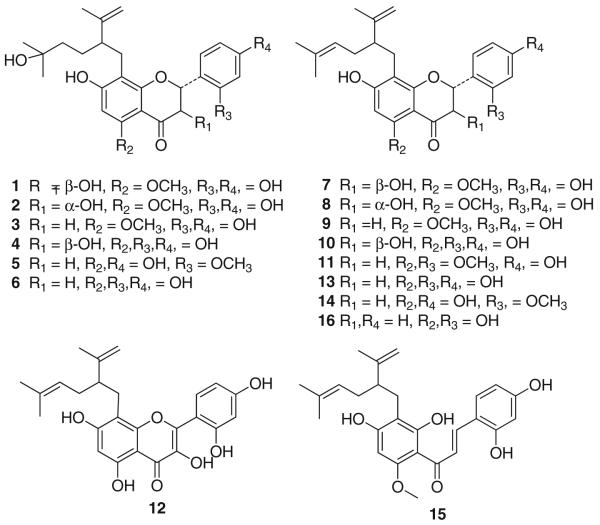
The major peak was attributed to (–)-kurarinone (9), the main lavandulyl flavonoid in S. flavescens. The remaining compounds were all structurally related 8-lavandulyl flavonoids (Fig. 4) which were identified as kushenol H (1), kushenol K (2), kurarinol (3), kushenol P (5), norkurarinol (6), kushenol I (7), kushenol N (8), (–)-kurarinone (9), kushenol X (10), neokurarinol (11), kushenol C (12), sophoraflavanone G (13), leachianone A (14), kuraridine (15), and kushenol A (16) by comparison of their MS, UV, and NMR spectra with published reference data [21–27].
Fig. 4.
Structures of flavonoids 1–16
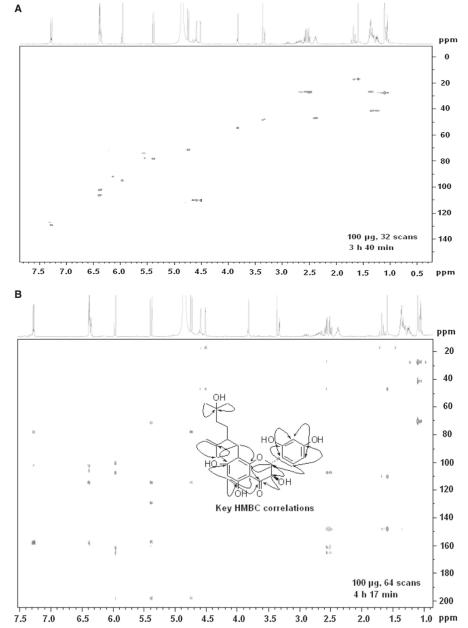
A new but pharmacologically inactive flavonoid 4 was obtained as a light yellow amorphous powder. Its molecular formula C25H30O8 was deduced from HR-ESIMS (M+Na+ found 481.1846, calcd. 481.1838).The UV spectrum showed absorption maxima at 287 and 326 nm. The 1H-NMR spectrum revealed the presence of a 1,2,4-trisubstituted aromatic ring at δH 7.28 (d, 1H, J = 8.0Hz, H-6′), 6.39 (dd, 1H, J = 8.0, 2.1 Hz, H-5′) and 6.35 (d, 1H, J = 2.1Hz, H-3′), an isolated aromatic proton at δH 5.96 (s, 1H, H-6), and two protons at δH 4.74 (d, 1H, J = 11.6 Hz, H-3) and 5.39 (d, 1H, J = 11.6 Hz, H-2) characteristic of 2,3-trans-flavanonol derivatives [24]. Additional signals at δH 1.11 (s, 6H, H-6″, 7″), 1.23 (m, 2H, H-3″), 1.35 (m, 2H, H-4″), 1.59 (s, 3H, H-10″), 2.38 (m, 1H, H-2″), 2.42 (m, 2H, H-1″), 4.52 (s, 1H, H-9″), and 4.59 (s, 1H, H-9″) were attributable to a 5-hydroxy-2-isopropenyl-5-methylhexyl moiety and were virtually superimposable with data reported for lavandulyl moieties of related flavonoids such as kushenol H (1) [21]. Compared to 1 the only difference in the spectrum of 4 observed was a hydroxy group at C-5 instead of a methoxy residue in 1. The 13C-NMR spectrum of 4 contained signals of four oxygenated aromatic carbons and one carbonyl carbon. Chemical shifts and signal patterns in the 1H- and 13C-NMR spectra indicated that the hydroxyl groups were at C-5, C-7, C-2′, and C-4′ .These assignments and the position of the side chain were further confirmed by HSQC and HMBC experiments. Representative spectra obtained with the 1 mm TXI probe after HPLC peak collection are shown in Fig. 5. As in other lavandulyl flavonoids reported from S. flavescens, the absolute configuration at C-2″ has not been determined [24]. The CD spectrum of 4 showed a positive absorption at [θ]315 + 5897 and a negative absorption at [θ]293 − 17061, from which the absolute configuration at C-2 and C-3 were determined as 2R and 3R, respectively [28]. Thus, 4 was (2R,3R)-8-(5-hydroxy-2-isopropenyl-5-methylhexyl)-5,7,2′,4′-tetrahydroxyflavanonol. We named the compound kushenol Y, in accord with trivial names of related lavandulyl flavonoids from S. flavescens.
Fig. 5.
a HSQC spectrum of kushenol Y (4) recorded with the TXI probe after HPLC peak collection and b HMBC spectrum and key correlations
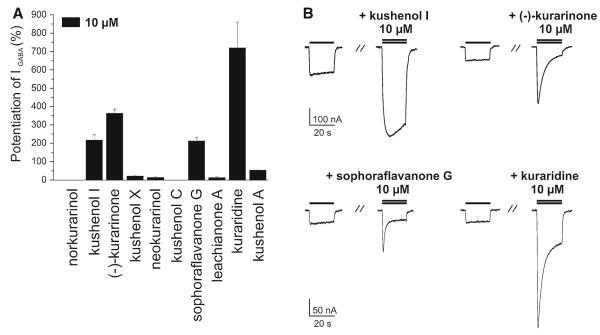
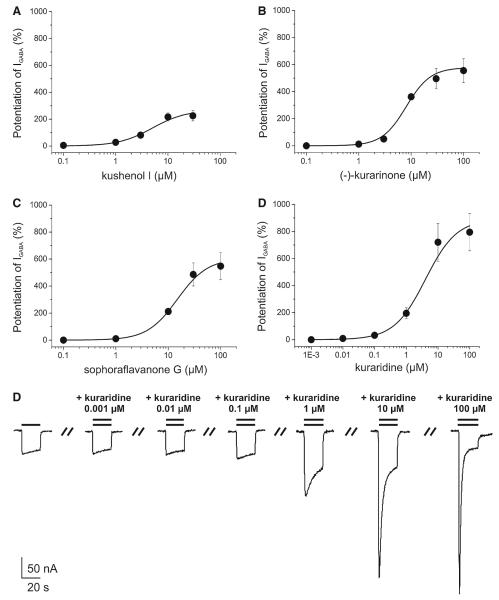
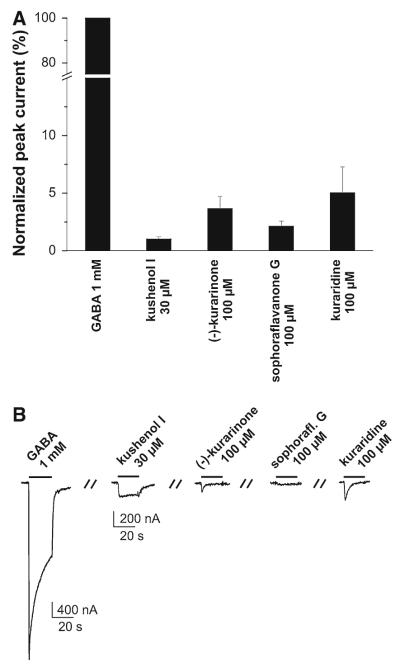
For further evaluation of compounds in the active time windows, 6, 7, and 9–16 (10 μM) were co-applied with a GABA (EC3–10) to oocytes expressing α1β2γ2S receptors. Kushenol I (7), (–)-kurarinone (9), sophoraflavanone G (13), and kuraridine (15) induced the strongest IGABA enhancement (Fig. 6). For these compounds the concentration–response relationships for IGABA enhancement were determined (Fig. 7). Given the lack of activity in other fractions of the activity profile (Fig. 1), the other flavonoids were considered as inactive. Interestingly, application of saturating concentrations of kushenol I (7), (–)-kurarinone (9), sophoraflavanone G (13), and kuraridine (15) induced chloride inward currents in the absence of GABA. These currents did not exceed 10% of the maximal IGABA induced by a saturating GABA concentration (1 mM). This partial agonist activity was most pronounced for kuraridine (15) (Fig. 8).
Fig. 6.
a Potentiation of IGABA through α1β2γ2S GABAA receptors by 10 μM of norkurarinol (0 ± 0%, n = 2), kushenol I (216.5 ± 29.3%, n = 5), (–)-kurarinone (362.3 ± 23.8%, n = 5), kushenol X (21.6 ± 3.4%, n = 2), neokurarinol (13.7±4.6%), n = 2), kushenol C (0±0%, n = 2, sophoraflavanone G (211.6±18.6%, n = 3), leachianone A (13.1±5.1%, n = 2), kuraridine (719.7±140.3%, n = 3), and kushenol A (53.1 ± 3.1%, n = 2). b Representative currents through α1β2γ2S GABAA receptors induced by GABA (EC3–10, single bar, control) and traces recorded during co-application of GABA (EC3–10) and the indicated compound (double bar). Current potentiation induced by the four most active compounds, kushenol I (7), (–)-kurarinone (9), sophoraflavanone G (13), and kuraridine (15) is shown
Fig. 7.
Concentration–response curves for IGABA enhancement by a kushenol I (EC50 = 5.0 ± 2.3 μM, nH = 1.3 ± 0.3, n = 5); b (–)-kurarinone (EC50 = 8.1±1.4 μM, nH = 1.9±0.1, n = 4); c sophoraflavanone G (EC50 = 15.0±3.6 μM, nH = 1.5±0.2, n = 3); and d kuraridine (EC50 = 4.0±2.4 μM, nH = 0.9±0.1) in Xenopus oocytes expressing GABAA receptors composed of α1,β2, and γ2S subunits. A maximum potentiation of IGABA by kushenol I (a, 267.6 ± 56.6%, n = 5), (–)-kurarinone (b, 578.5 ± 68.8%, n = 4), sophoraflavanone G (c, 604.9 ± 108.2%, n = 3), kuraridine (d, 891.5 ± 163.0%, n = 3) was observed. e Representative currents through α1β2γ2S GABAA receptors in the presence of GABA (EC3–10, single bar, control) and currents recorded during co-application of GABA (EC3–10) and 0.001, 0.01, 0.1, 1, 10, and 100 μM of kuraridine (double bar)
Fig. 8.
Activation of GABAA receptors by kushenol I, (–)-kurarinone, sophoraflavanone G, and kuraridine. a Bar graphs illustrate the amplitudes of chloride currents induced in the absence of GABA by saturating concentrations of kushenol I (1.0 ± 0.2%, n = 3, (–)-kurarinone (3.7 ± 1.1%, n = 3) sophoraflavanone G (2.1 ± 0.4%, n = 3) and kuraridine 5.0 ± 2.2%, n = 3). 100% correspond to the maximal IGABA induced by a saturating concentration (1 mM) of GABA. b Representative currents illustrating direct activation of GABAA receptors (α1β2γ2S) by kushenol I, (–)-kurarinone, sophoraflavanone G, and kuraridine in comparison to IGABA induced by 1 μM GABA
(–)-Kurarinone (9) and kuraridine (15) are the first representatives of a new scaffold of GABAA receptor modulators with a flavonoid moiety. Interaction with GABAA receptors have been investigated since the 1980s, when first evidence was produced from displacement studies with radiolabelled benzodiazepines [29]. Subsequent binding studies demonstrated that a range of naturally occurring and synthetic flavonoids including the monomeric flavonoids apigenin [30], 6-hydroxyflavone [31], baicalin [32], hesperidin, and 6-methylapigenin [33,34], and biflavonoids such as agathisflavone and amentoflavone [35–37] bind to GABAA receptors with nanomolar affinity. Quercetin, apigenin, morine, and chrysin have been shown to inhibit IGABA through α1β1γ2sGABAA receptors [38]. Simple flavones appear to interact with the benzodiazepine binding site [34,39], whereas amentoflavone is a negative modulator of GABAA receptors whose action appears independent of benzodiazepine modulatory sites on the receptor [36]. Evidence for in vivo activity mediated through GABAA receptor modulation has been reported for selected flavonoids. Simple flavones and flavonols such as chrysin, apigenin, wogonin, 6-methylapigenin, hispidulin, and luteolin-7-O-(2-rhamnosylglucoside) appear to induce pre-dominantly anxiolytic effects and seem less efficient as sedatives, anticonvulsants, and myorelaxants [34,39–42].
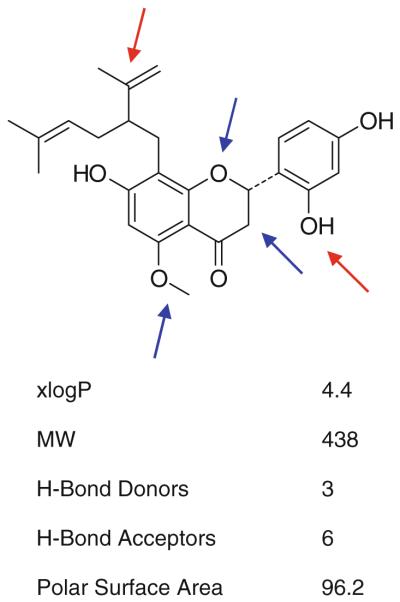
The pharmacology of Sophora flavescens flavonoids has been intensively studied. Neuroprotective [43] and anti-inflammatory [44] properties have been reported, but no pharmacological effects that could be related to modulation of GABAA receptors. Given the close structural relationship of 1–16, some preliminary structure activity considerations can be derived from this focused library via our activity profiling approach (Fig. 9): The compounds differ in some specific attributes, namely in the substitution at C-5″ of the side chain, the substituents at C-5, C-2′, and C-4″ of the flavonoid moiety, and in the nature of the basic flavonoid structure, viz. flavanone, flavanonol or chalcone. Absence of a hydroxy group at C-5″ of the side chain appears essential, as compounds 1-6 bearing a hydroxyl group were all inactive. The methoxy substituent at C-5 contributes to activity, since all compounds with a hydroxy group either showed less (eg. 13) or no activity. Comparison of (–)-kurarinone (9) and kushenol A (16) indicates that a hydroxy group at C-4″ also contributes to activity. Active compounds bear a hydroxy substituent at C-2″ instead of a methoxy group as, for example, in neokurarinol (11). Flavanone (e.g., 9) and chalcone derivatives (e.g., 15) show higher activity than corresponding flavanonol derivatives (7 and 8). In summary, it appears that several structural features contribute to some degree to the GABAA receptor modulating properties of 8-lavandulyl flavonoids and that absence of a hydroxyl group at C-5″ is essential (Fig. 5).
Fig. 9.
Summary of preliminary structure activity information on lavandulyl flavonoids and calculated physicochemical data for (–)-kurarinone (9) (from Pubchem)
The discovery of a new scaffold for potent GABAA modulators (EC50s between 5.0 and 15.0 μM) enlarges the spectrum of natural products acting on this target. The example of 8-lavandulyl flavonoids highlights the advantages of an HPLC-based approach, in particular, the possibility of obtaining valuable preliminary structure–activity information via the rapid characterization of structurally related compound series. (–)-Kurarinone (9) fulfils most criteria of Lipinski’s rule of five [45] and has a polar surface area that may enable CNS penetration [46]. We currently investigate the binding site of the most active compounds 9 and 15, their subunit specificity, and in vivo activity.
Acknowledgments
Financial support from the Swiss National Science Foundation (Projects 31600-113109 and 205321-116157/1), the Steinegg-Stiftung, Herisau, the Fonds zur Förderung von Lehre und Forschung, Basel (M.H.), the FWF Project P19614-B11 (S.H.) is gratefully acknowledged. We thank Jian Gao (Department of Chemistry, University of Basel) for measuring optical rotation, circular dichroism, UV and IR spectra. We thank Dr. Sophia Khom for valuable comments.
Abbreviations
- TOFMS
Time of flight mass spectrometry
- PDA
Photodiode array
- HMBC
Heteronuclear multiple-bond correlation
- HSQC
Heteronuclear single quantum coherence
- CD
Circular dichroism
Contributor Information
Xinzhou Yang, Institute of Pharmaceutical Biology, University of Basel, Klingelbergstrasse 50, 4056 Basel, Switzerland.
Igor Baburin, Institute of Pharmacology and Toxicology, University of Vienna, Althanstrasse 14, 1090 Vienna, Austria.
Inken Plitzko, Institute of Pharmaceutical Biology, University of Basel, Klingelbergstrasse 50, 4056 Basel, Switzerland.
Steffen Hering, Institute of Pharmacology and Toxicology, University of Vienna, Althanstrasse 14, 1090 Vienna, Austria.
Matthias Hamburger, Institute of Pharmaceutical Biology, University of Basel, Klingelbergstrasse 50, 4056 Basel, Switzerland, e-mail: Matthias.Hamburger@unibas.ch.
References
- 1.Simon J, Wakimoto H, Fujita N, Lalande M, Barnard EA. Analysis of the set of GABAA receptor genes in the human genome. J Biol Chem. 2004;279:41422–41435. doi: 10.1074/jbc.M401354200. doi:10.1074/jbc.M401354200. [DOI] [PubMed] [Google Scholar]
- 2.Olsen RW, Sieghart W. International union of pharmacology. LXX. Subtypes of γ -aminobutyric acidA receptors: classification on the basis of subunit composition, pharmacology, and function. Update. Pharmacol Rev. 2008;60:243–260. doi: 10.1124/pr.108.00505. doi:10.1124/pr.108.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrera NP, Edwardson JM. The subunit arrangement and assembly of ionotropic receptors. Trends Neurosci. 2008;31:569–576. doi: 10.1016/j.tins.2008.08.001. doi:10.1016/j.tins.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Sieghart W, Sperk G. Subunit composition, distribution and function of GABAA receptor subtypes. Curr Top Med Chem. 2002;2:795–816. doi: 10.2174/1568026023393507. [DOI] [PubMed] [Google Scholar]
- 5.Whiting PJ. GABA-A receptors: a viable target for novel anxiolytics? Curr Opin Pharmacol. 2006;6:24–29. doi: 10.1016/j.coph.2005.08.005. doi:10.1016/j.coph.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Riss J, Cloyd J, Gates J, Collins S. Benzodiazepines in epilepsy: pharmacology and pharmacokinetics. Acta Neurol Scand. 2008;118:69–86. doi: 10.1111/j.1600-0404.2008.01004.x. doi:10.1111/j.1600-0404.2008.01004.x. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan EM, DuPont RL. Benzodiazepines and anxiety disorders: a review for the practicing physician. Curr Med Res Opin. 2005;21:941–950. doi: 10.1185/030079905X48401. doi:10.1185/030079905X48401. [DOI] [PubMed] [Google Scholar]
- 8.Rupprecht R, Eser D, Zwanzger P, Moeller HJ. GABAA receptors as targets for novel anxiolytic drug. World J Biol Psychiatry. 2006;7:231–237. doi: 10.1080/15622970600868525. doi:10.1080/15622970600868525. [DOI] [PubMed] [Google Scholar]
- 9.Johnston GAR, Hanrahan JR, Chebib M, Duke RK, Mewett KN. Modulation of ionotropic GABA receptors by natural products of plant origin. Adv Pharmacol. 2006;54:285–316. doi: 10.1016/s1054-3589(06)54012-8. doi:10.1016/S1054-3589(06)54012-8. [DOI] [PubMed] [Google Scholar]
- 10.Tsang SY, Xue H. Development of effective therapeutics targeting the GABAA receptor: naturally occurring alternatives. Curr Pharm Design. 2004;10:1035–1044. doi: 10.2174/1381612043452767. doi:10.2174/1381612043452767. [DOI] [PubMed] [Google Scholar]
- 11.Potterat O, Hamburger M. Natural products in drug discovery—concepts and approaches for tracking bioactivity. Curr Org Chem. 2006;10:899–920. doi:10.2174/138527206776894401. [Google Scholar]
- 12.Potterat O, Wagner K, Gemmecker G, Mack J, Puder C, Vettermann R, Streicher R. BI-32169, a bicyclic 19-peptide with strong glucagon receptor antagonist activity from Streptomyces sp. J Nat Prod. 2004;67:1528–1531. doi: 10.1021/np040093o. doi:10.1021/np040093o. [DOI] [PubMed] [Google Scholar]
- 13.Danz H, Stoyanova S, Wippich P, Brattstroem A, Hamburger M. Identification and isolation of the cyclooxygenase-2 inhibitory principle in Isatis tinctoria. Planta Med. 2001;67:411–416. doi: 10.1055/s-2001-15805. doi:10.1055/s-2001-15805. [DOI] [PubMed] [Google Scholar]
- 14.Dittmann K, Gerhaeuser C, Klimo K, Hamburger M. HPLC-based activity profiling of Salvia miltiorrhiza for MAO-A and iNOS inhibitory activities. Planta Med. 2004;70:909–913. doi: 10.1055/s-2004-832615. doi:10.1055/s-2004-8326156. [DOI] [PubMed] [Google Scholar]
- 15.Kim HJ, Baburin I, Khom S, Hering S, Hamburger M. HPLC-based activity profiling approach for the discovery of GABAA receptor ligands using an automated two microelectrode voltage clamp assay on Xenopus oocytes. Planta Med. 2008;74:521–526. doi: 10.1055/s-2008-1074491. doi:10.1055/s-2008-1074491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaugg J, Baburin I, Strommer B, Kim HJ, Hering S, Hamburger M. HPLC-based activity profiling: discovery of piperine as a positive GABAA receptor modulator targeting a benzodiazepine-independent binding site. J Nat Prod. 2010;73:185–191. doi: 10.1021/np900656g. doi:10.1021/np900656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Plitzko I, Zaugg J, Hering S, Hamburger M. HPLC-based activity profiling for GABAA receptor modulators: a new dihydroisocoumarin from Haloxylon scoparium. J Nat Prod. 2010;73:768–770. doi: 10.1021/np900803w. doi:10.1021/np900803w. [DOI] [PubMed] [Google Scholar]
- 18.Khom S, Baburin I, Timin EN, Hohaus A, Sieghart W, Hering S. Pharmacological properties of GABAA receptors containing γ 1 subunits. Mol Pharmacol. 2006;69:640–649. doi: 10.1124/mol.105.017236. doi:10.1124/mol.105.017236. [DOI] [PubMed] [Google Scholar]
- 19.Methfessel C, Witzemann V, Takahashi T, Mishina M, Numa S, Sakmann B. Patch clamp measurements on Xenopus laevis oocytes: currents through endogenous channels and implanted acetylcholine receptor and sodium channels. Pflug Arch Eur J Phy. 1986;407:577–588. doi: 10.1007/BF00582635. [DOI] [PubMed] [Google Scholar]
- 20.Baburin I, Beyl S, Hering S. Automated fast perfusion of Xenopus oocytes for drug screening. Pflug Arch Eur J Phys. 2006;453:117–123. doi: 10.1007/s00424-006-0125-y. doi:10.1007/s00424-006-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu LJ, Miyase T, Ueno A, Kuroyanagi M, Noko T, Fukushima S. Studies on the constituents of Sophora flavescens Ait. III. Yakugaku Zasshi. 1985;105:736–741. [Google Scholar]
- 22.Sato S, Takeo J, Aoyama C, Kawahara H. Na+-glucose co-transporter (SGLT) inhibitory flavonoids from the roots of Sophora flavescens. Bioorg Med Chem. 2007;15:3445–3449. doi: 10.1016/j.bmc.2007.03.011. doi:10.1016/j.bmc.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Kim JH, Ryu YB, Kang NS, Lee BW, Heo JS, Jeong IY, Park KH. Glycosidase inhibitory flavonoids from Sophora flavescens. Biol Pharm Bull. 2006;29:302–305. doi: 10.1248/bpb.29.302. doi:10.1248/bpb.29.302. [DOI] [PubMed] [Google Scholar]
- 24.Kuroyanagi M, Arakawa T, Hirayama Y, Hayashi T. Anti-bacterial and antiandrogen flavonoids from Sophora flavescens. J Nat Prod. 1999;62:1595–1599. doi: 10.1021/np990051d. doi:10.1021/np99000051d. [DOI] [PubMed] [Google Scholar]
- 25.Hatayama K, Komatsu M. Studies on the constituents of Sophora species. V. Constituents of the root of Sophora angustifolia SIEB. et. ZUCC. Chem Pharm Bull. 1971;19:2126–2131. doi: 10.1248/yakushi1947.90.4_463. [DOI] [PubMed] [Google Scholar]
- 26.Hillerns PI, Wink M. Binding of flavonoids from Sophora flavescens to the rat uterine estrogen receptor. Planta Med. 2005;71:1065–1068. doi: 10.1055/s-2005-871302. doi:10.1055/s-2005-871302. [DOI] [PubMed] [Google Scholar]
- 27.Kyogoku K, Hatayama K, Komatsu M. Constituents of Chinese drug, Kushen (the root of Sophora flavescens Arr.). Isolation of five new flavonoids and formononetin. Chem Pharm Bull. 1973;21:2733–2738. doi: 10.1248/cpb.21.2733. [DOI] [PubMed] [Google Scholar]
- 28.Gaffield W. Circular dichroism, optical rotatory dispersion and absolute configuration of flavanones, 3-hydroxyflavanones and their glycosides: determination of aglycone chirality in flavanone glycosides. Tetrahedron. 1970;26:4093–4108. doi:10.1016/S0040-4020(01)93050-9. [Google Scholar]
- 29.Luk KC, Stern L, Weigele M, O’Brien RA, Spirt N. Isolation and identification of “diazepam-like” compounds from bovine urine. J Nat Prod. 1983;46:852–861. doi: 10.1021/np50030a005. doi:10.1021/np50030a005. [DOI] [PubMed] [Google Scholar]
- 30.Avallone R, Zanoli P, Puia G, Kleinschnitz M, Schreier P, Baraldi M. Pharmacological profile of apigenin, a flavonoid isolated from Matricaria chamomilla. Biochem Pharmacol. 2000;59:1387–1394. doi: 10.1016/s0006-2952(00)00264-1. doi:10.1016/S0006-2952(00)00264-1. [DOI] [PubMed] [Google Scholar]
- 31.Ren L, Wang F, Xu Z, Chan WM, Zhao C, Xue H. GABAA receptor subtype selectivity underlying anxiolytic effect of 6-hydroxyflavone. Biochem Pharmacol. 2010;79:1337–1344. doi: 10.1016/j.bcp.2009.12.024. doi:10.1016/j.bcp.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 32.Wang F, Xu Z, Ren L, Tsang SY, Xue H. GABAA receptor subtype selectivity underlying selective anxiolytic effect of baicalin. Neuropharmacology. 2008;55:1231–1237. doi: 10.1016/j.neuropharm.2008.07.040. doi:10.1016/j.neuropharm.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 33.Wasowski C, Marder M, Viola H, Medina JH, Paladini AC. Isolation and identification of 6-methylapigenin, a competitive ligand for the brain GABAA receptors, from Valeriana wallichii D. C. Planta Med. 2002;68:934–936. doi: 10.1055/s-2002-34936. [DOI] [PubMed] [Google Scholar]
- 34.Marder M, Viola H, Wasowski C, Fernandez S, Medina JH, Paladini AC. 6-Methylapigenin and hesperidin: new valeriana flavonoids with activity on the CNS. Pharmacol Biochem Behav. 2003;75:537–545. doi: 10.1016/s0091-3057(03)00121-7. doi:10.1016/S0091-3057(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 35.Svenningsen AB, Madsen KD, Liljefors T, Stafford GI, van Staden J, Jäger AK. Biflavones from Rhus species with affinity for the GABAA/benzodiazepine receptor. J Ethnopharmacol. 2006;103:276–280. doi: 10.1016/j.jep.2005.08.012. doi:10.1016/j.jep.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Hanrahan JR, Chebib M, Davucheron NLM, Hall BJ, Johnston GAR. Semisynthetic preparation of amentoflavone: a negative modulator at GABAA receptors. Bioorg Med Chem Lett. 2003;13:2281–2284. doi: 10.1016/s0960-894x(03)00434-7. doi:10.1016/S0960-894X(03)00434-7. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen M, Frøkjaer S, Braestrup C. High affinity of the naturally-occurring biflavonoid, amentoflavon, to brain benzodiazepine receptors in vitro. Biochem Pharmacol. 1988;37:3285–3287. doi: 10.1016/0006-2952(88)90640-5. doi:10.1016/0006-2952(88)90640-5. [DOI] [PubMed] [Google Scholar]
- 38.Goutman JD, Waxemberg MD, Doñate-Oliver F, Pomata PE, Calvo DJ. Flavonoid modulation of ionic currents mediated by GABAA and GABAC receptors. Eur J Pharmacol. 2003;461:79–87. doi: 10.1016/s0014-2999(03)01309-8. doi:10.1016/S0014-2999(03)01309-8. [DOI] [PubMed] [Google Scholar]
- 39.Hui KM, Huen MSY, Wang HY, Zheng H, Sigel E, Baur R, Ren H, Li ZW, Wong JTF, Xue H. Anxiolytic effect of wogonin, a benzodiazepine receptor ligand isolated from Scutellaria baicalensis Georgi. Biochem Pharmacol. 2002;64:1415–1424. doi: 10.1016/s0006-2952(02)01347-3. doi:10.1016/S0006-2952(02)01347-3. [DOI] [PubMed] [Google Scholar]
- 40.Coleta M, Batista MT, Campos MG, Carvalho R, Cotrim MD, Lima TC, Cunha AP. Neuropharmacological evaluation of the putative anxiolytic effects of Passiflora edulis Sims, its subfractions and flavonoid constituents. Phytother Res. 2006;20:1067–1073. doi: 10.1002/ptr.1997. doi:10.1002/ptr.1997. [DOI] [PubMed] [Google Scholar]
- 41.Medina JH, Viola H, Wolfman C, Marder M, Wasowski C, Calvo D, Paladini AC. Neuroactive flavonoids: new ligands for the benzodiazepine receptors. Phytomedicine. 1998;5:235–243. doi: 10.1016/S0944-7113(98)80034-2. [DOI] [PubMed] [Google Scholar]
- 42.Kavvadias D, Sand P, Youdim KA, Qaiser MZ, Rice-Evans C, Baur R, Sigel E, Rausch WD, Riederer P, Schreier P. he flavone hispidulin, a benzodiazepine receptor ligand with positive allosteric properties, traverses the blood-brain barrier and exhibits anticonvulsive effects. Br J Pharmacol. 2004;142:811–820. doi: 10.1038/sj.bjp.0705828. doi:10.1038/sj.bjp.0705828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SJ, Nam KW, Lee HJ, Cho EY, Koo U, Mar W. Neuroprotective effects of an alkaloid-free ethyl acetate extract from the root of Sophora flavescens Ait. against focal cerebral ischemia in rats. Phytomedicine. 2010;16:1042–1051. doi: 10.1016/j.phymed.2009.03.017. doi:10.1016/j.phymed.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Jin JH, Kim JS, Kang SS, Son KH, Chang HW, Kim HP. Anti-inflammatory and anti-arthritic activity of total flavonoids of the roots of Sophora flavescens. J Ethnopharmacol. 2009;127:589–595. doi: 10.1016/j.jep.2009.12.020. doi:10.1016/j.jep.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 45.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. doi:10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 46.Pajouhesh H, Lenz GR. Medicinal chemical properties of successful central nervous system drugs. NeuroRX. 2005;2:541–553. doi: 10.1602/neurorx.2.4.541. doi:10.1602/neurorx.2.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]