Abstract
Background
Plant phloem consists of an interdependent cell pair, the sieve element / companion cell complex. Sucrose transporters are localized to enucleate sieve elements (SE), despite being transcribed in companion cells (CC). Due to the high turnover of SUT1, sucrose transporter mRNA or protein must traffic from CC to SE via the plasmodesmata. Localization of SUT mRNA at plasmodesmatal orifices connecting CC and SE suggests RNA transport, potentially mediated by RNA binding proteins. In many organisms, polar RNA transport is mediated through RNA binding proteins interacting with the 3'-UTR and controlling localized protein synthesis. To study mechanisms for trafficking of SUT1, GFP-fusions with and without 3'-UTR were expressed in transgenic plants.
Results
In contrast to plants expressing GFP from the strong SUC2 promoter, in RolC-controlled expression GFP is retained in companion cells. The 3'-UTR of SUT1 affected intracellular distribution of GFP but was insufficient for trafficking of SUT1, GFP or their fusions to SEs. Fusion of GFP to SUT1 did however lead to accumulation of SUT1-GFP in the CC, indicating that trafficking was blocked while translational inhibition of SUT1 mRNA was released in CCs.
Conclusion
A fusion with GFP prevents targeting of the sucrose transporter SUT1 to the SE while leading to accumulation in the CC. The 3'-UTR of SUT1 is insufficient for mobilization of either the fusion or GFP alone. It is conceivable that SUT1-GFP protein transport through PD to SE was blocked due to the presence of GFP, resulting in retention in CC particles. Alternatively, SUT1 mRNA transport through the PD could have been blocked due to insertion of GFP between the SUT1 coding sequence and 3'-UTR.
Background
In plants, sucrose produced in photosynthetic organs is transported through conduits formed by the enucleate but living sieve elements (SEs) [reviewed in [1-4]]. Three sucrose transporter paralogues (SUTs) have been found at the plasma membrane of sieve elements [5-7]. Transgenic plants in which SUT1 had been repressed by cell specific antisense under control of the companion cell (CC)-specific RolC promoter [8] displayed a similar phenotype to plants expressing the same gene in antisense orientation under the CaMV 35S promoter, suggesting that transcription takes place in the CC [8]. The high turnover of SUT1 protein and mRNA, together with the localization of SUT1 mRNA at the plasmodesmatal orifices is consistent with a model according to which SUT mRNA is transcribed in the CC and then moves via plasmodesmata (PDs) to SEs [5]. Alternatively, it is also possible that translation occurs in CC, and the protein is transported to SE e.g. through the ER.
Plasmodesmata (PDs) are complex tubular structures connecting two adjacent cells creating the condition for a plasma membrane continuum [9]. PDs have been shown to be a conduit for transport of both endogenous and foreign mRNAs and proteins. Controlled movement of mRNA is supported by the finding that the RNA-binding protein CmPP16 from phloem sap of Cucurbita maxima is necessary and sufficient to mediate movement of sucrose transporter mRNA between mesophyll cells [10]. This mechanism involving RNA-binding proteins as adaptors for RNA trafficking is similar to that proposed for RNA movement in other organisms [11].
In many organisms (Xenopus, Drosophila or polarized cells in mammals), the 3'-untranslated region (3'-UTR) plays an important role in polar distribution of mRNAs within or between cells [11]. In Drosophila melanogaster, for example, Bicoid and Nanos are transcribed in yolk cells, the RNAs of which are subsequently transported in ribonuclear protein (RNP) complexes to the poles of the embryo [reviewed in [11]]. The specific localization is mediated by RNA binding proteins, such as Staufen and Exuperantia, which bind to specific regions of the 3'-UTR of both Nanos and Bicoid and, via interactions with cytoskeleton-anchoring proteins and motor proteins, are translocated to their respective destination locations [11]. Similarly, trafficking of a fusion of human Staufen with GFP in hippocampal neurons occurs by movement of large ribonucleoprotein (RNP) complexes with an average velocity of 6.4 μm/min [12] to drive localized translation.
In yeast, both Ash1 and Ist2 RNAs traffic during cell division from mother to daughter cells [13]. Similar to Bicoid, Ash1 mRNA has multiple binding sites for RNA binding proteins. She2p binds to Ash1 mRNA and together with other She proteins that jointly mediate cytoskeletal interaction, the entire complex moves from the mother cell to the daughter cell [14,15]. Localization of the membrane protein Ist2 uses the same trafficking mechanism. Moreover, Ist2p is retained in the plasma membrane domain of the daughter cell by a septin barrier localized at the bud neck blocking lateral diffusion of Ist2p back to the mother cell membrane domain [16].
Here we report an analysis of SUT1 and GFP expression in the CC and potential mechanisms for their transport into the SE. RolC-driven GFP expression led to accumulation of GFP in companion cells. The 3'-UTR of the tomato SUT1 gene affected intracellular distribution when fused downstream of the GFP coding region the in CC by aggregation into particle-like structures. The SUT1 3'-UTR did not lead to increased accumulation of GFP in SE.
Also a translational fusion of SUT1 with GFP was retained in CC independent of the presence of the 3'-UTR. Thus additional factors are required to mobilization of SUT1 and GFP between CC and SE.
Results
Differential polyadenylation of SUT1 mRNA
In yeast and animals, the 3'-UTR is often necessary and sufficient as a cis-element for RNA transport. These cis-elements are also implicated in the blocking of translation during transport in cells where the RNA originates. To study a potential role of the SUT1 3'-UTR in SUT1 trafficking, a genomic clone containing a 1.2 kb of downstream region of SUT1 was isolated. The DNA sequence has been deposited in Genbank with the accession number AY380824. To determine the position of transcriptional termination, cDNAs were isolated from a leaf library of Lycopersicum esculentum (tomato). Of 15 cDNAs isolated and representing LeSUT1, twelve had a 3'-UTR of 484 bp, 2 of 294 bp and one of 269 bp; in all cases, the sequences are identical (Fig. 1). The canonical polyadenylation sequence AAUAAA found in animals is much less conserved in plants [17]. Furthermore, upstream sequences are necessary for optimum polyadenylation [17,18]. Sequence analysis of the LeSUT1 clones reveals the presence of polyA signals in the vicinity of the cleavage site in all cDNAs. StSUT1, which is similarly localized to the potato SE (protein) and CC (mRNA) [5], shows a similar number and positions of polyA signals in the mRNA transcript.
Figure 1.
Structural analysis of the genomic sequence of LeSUT1. LeSUT1 genomic sequence contains three introns and has three polyadenylation signals predicted based on sequence comparison with different cDNA clones. The numbers under the introns/exon boxes indicate the size in bp.
Analysis of potential RNA structures in the SUT1 3'-UTR
Predicted RNA structures of sucrose transporter genes from tomato (LeSUT1, LeSUT2, LeSUT4) were compared to potato (StSUT1) and Arabidopsis (AtSUC1, AtSUC2) using the Mfold RNA folding software version 3.0 [19] [Additional file 1]. Primary sequence homology of the 3'-UTR is highest between LeSUT1 and StSUT1 at 58%, while the homology between LeSUT1 compared to LeSUT2, LeSUT4, AtSUC1 and AtSUC2 is in the range of 28–45%. Additionally, the predicted secondary structures of LeSUT1 and StSUT1 3'-UTRs share some similarities, while, UTRs from other sucrose transporters analyzed display weak similarity of secondary structures [Additional file 1]. Nevertheless, for tomato, there are regions of high determination, i.e., they routinely appear in a given state (stem or loop). Such regions are of particular interest when they occur within the 3'-UTR, as is the case for LeSUT1 [Additional file 1]. Interestingly, no complex stem-loop structure was found in the 3'-UTR of AtSUC2, a sucrose transporter localized in CC of Arabidopsis [20]. Analysis of cis-elements will be required to determine the function of these predicted structures.
Companion cell expression of GUS fused with the 3'-UTR of SUT1
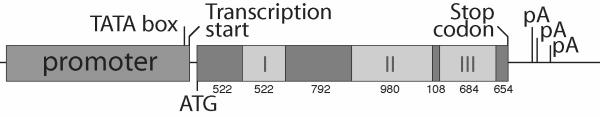
To test whether the LeSUT1 3'-UTR might be able to activate movement of the reporter GUS from the CC to the SE and, a 1.2 kb genomic fragment encompassing the 3'-UTR and subsequent downstream sequences was isolated. This fragment was fused downstream of the β-glucuronidase gene (GUS) behind the CC-specific RolC promoter (Fig. 2A). For each construct (RolC-GUS and RolC-GUS-3'-UTR), ~60 transgenic plants were screened, from which three lines with comparatively high expression levels were selected for further analysis; all lines showed comparable patterns of vascular expression. Histochemical GUS assays were performed with leaves of transgenic plants grown in sterile tissue culture. Both constructs, irrespective of the SUT1 3'-UTR, showed GUS activity in the ab- and adaxial phloem (Fig. 2B,2D). In LR White-embedded tissue, GUS activity was found to be restricted to CCs, again irrespective of the presence of the 3'-UTR (Fig. 2C,2E). Thus the 3'-UTR alone was unable to mobilize GUS RNA or protein movement. The inability of GUS protein to move between CC and SE is consistent with previous studies suggesting that the molecular mass is higher than the size exclusion limit of PDs [21].
Figure 2.
Localization of GUS activity in RolC-GUS transgenic plants. GUS staining was performed on whole tobacco leaves of transgenic plants using X-Gluc as substrate. For higher resolution staining, the leaves were cut to small pieces after the staining, then fixed and embedded in LR White. Thins sections were stained with DAPI for CC nucleus localization and were observed using normal light microscopy coupled to Nomarski imaging and epifluorescence. A) All constructs were prepared in a modified pGPTV-HPT under the control of the CC-specific, RolC, promoter. The asterisk indicates the position of the start codon. T, nos terminator. B) RolC-GUS, C) RolC-GUS-3'-UTR, D) GUS activity localization in leaf thin section of transgenic RolC-GUS, E) GUS activity localization in leaf thin section of transgenic RolC-GUS-3'-UTR. Bar equals to 10 μm.
Effect of SUT1 3'-UTR on GFP localization in companion cells of transgenic plants
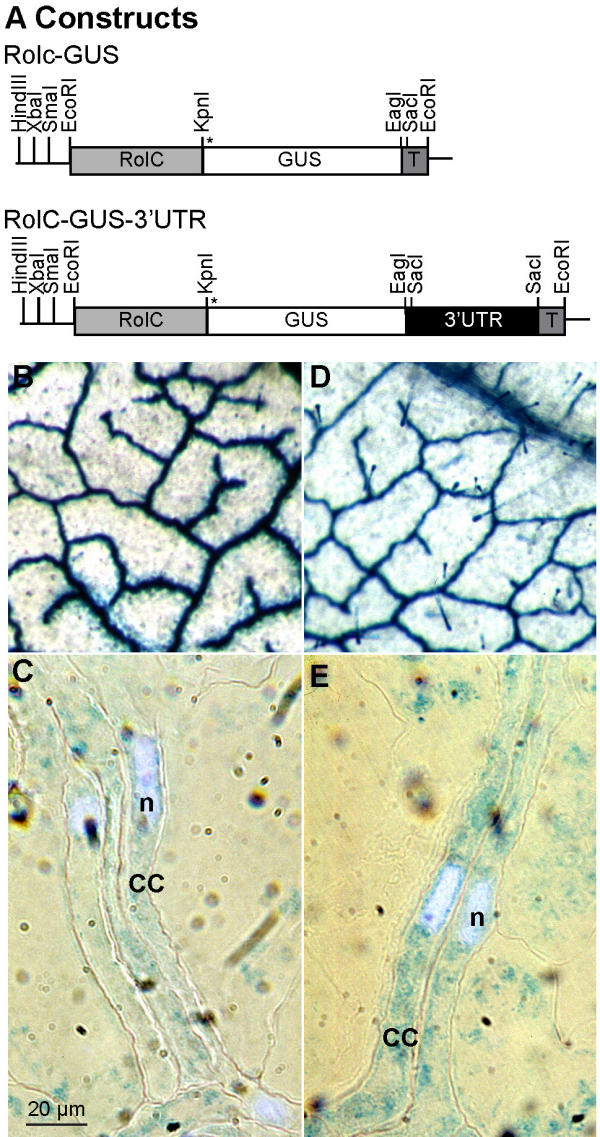
To use a smaller reporter previously shown to move between CC and SE, GFP was fused to the SUT1 3'-UTR under control of the RolC promoter (Fig. 3A). Western analysis was first used to screen for plants expressing high levels of GFP, and selected plants were then analyzed by confocal laser scanning microscopy (CLSM). In both cases (RolC-GFP and RolC-GFP-3'-UTR), ~60 transgenic plants were analyzed, from which three lines with the highest expression levels were selected for further analysis. All lines showed similar expression patterns (data not shown) and none of the transgenic plants analyzed showed obvious phenotypic alterations under tissue culture or greenhouse conditions. Identification of phloem cell types was performed using DAPI staining for CC nuclei and aniline blue staining for the callose of sieve plates. Using CLSM, 3D images could be scanned from which xz or yz 2D images were obtained. In all RolC-GFP plants analyzed, GFP fluorescence was found in the CCs of petiole, leaf midrib, and stem (Fig. 3B,3D,3G). Sink tissues such as sink leaves or unopened flowers did not accumulate GFP (Fig. 3E,3F,3H). In all tissues, GFP protein was found in the cytoplasm and in the proximity of the nucleus. In plants transformed with the GFP-3'-UTR constructs, GFP was also localized in the CC (Fig. 3C), moreover the addition of the 3'-UTR resulted in the formation of particle-like structures and in the loss of the perinuclear localization (Fig. 3C). Thus the 3'-UTR affects GFP distribution in the cell, but is insufficient for trafficking to SE.
Figure 3.
GFP Localization in RolC-GFP transgenic plants. GFP fluorescence was detected on longitudinal hand sections of transgenic tobacco petiole under CLSM and co-stained with DAPI and aniline blue. A) All constructs were prepared in a modified pGPTV-HPT under the control of the CC-specific, RolC, promoter. The asterisk indicates the position of the start codon. T, nos terminator. B) RolC-GFP petiole, C) RolC-GFP-3'-UTR petiole, D) major vein of mature leaf of RolC-GFP transgenic plant, E) sink leaf (1 cm long) of RolC-GFP transgenic plant, F) leaf petiole of a sink leaf of RolC-GFP transgenic plant, G) Stem of RolC-GFP transgenic plant, H) Veins of an unopened flower of RolC-GFP transgenic plant. I) Untransformed wild type tobacco leaf petiole visualized with the average PMT level used for the above images. CC, companion cell; SE, sieve element; sp, sieve plate; n, nucleus. Bar equals to 10 μm.
The data also suggest that GFP protein is restricted to CCs and did not seem to traffic to SEs or accumulate in sink tissues when expressed from the RolC promoter. The retention of GFP in CC is different from the localization observed when GFP was expressed under control of the SUC2 promoter[21]. The absence of GFP movement in the RolC promoter driven constructs makes this system suitable for the study of cis-elements required for trafficking.
Translational SUT-GFP fusion
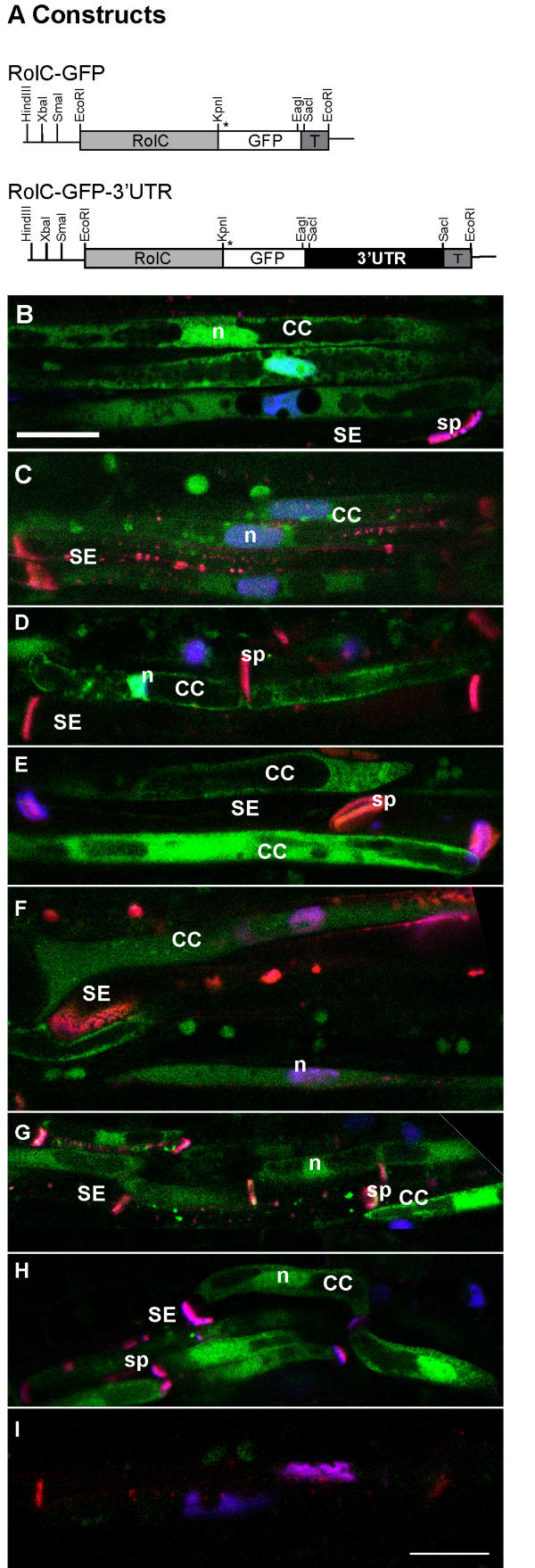
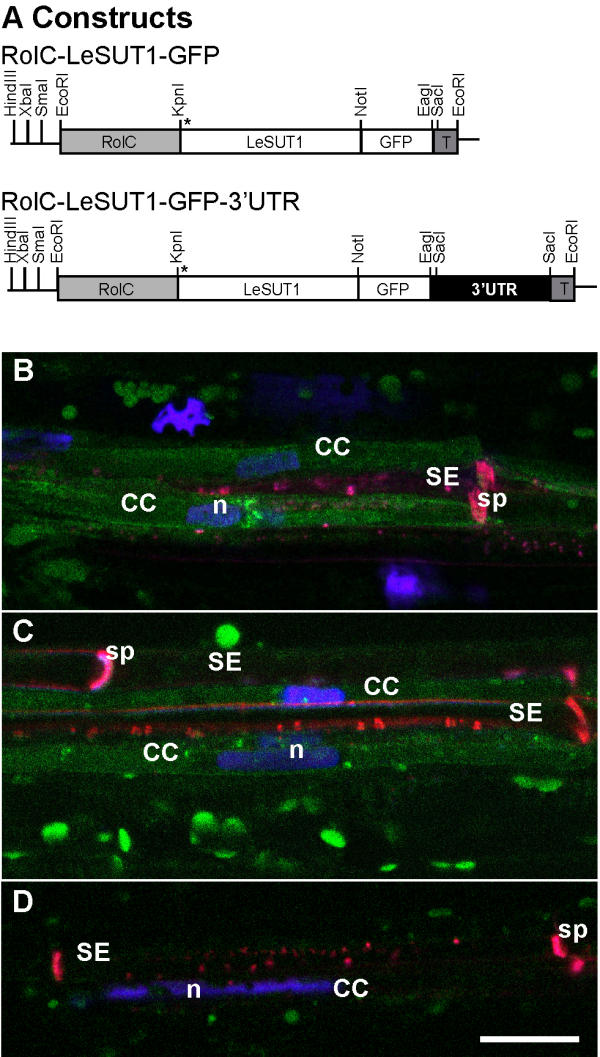
The above results suggested that the 3'-UTR is insufficient by itself to mobilize reporter activity to the SE, indicating that other targeting signals may be required in the LeSUT1 mRNA or protein. To test this hypothesis, GFP was fused translationally to the C-terminus of LeSUT1 (Fig. 4A). Intact folding and targeting of the sucrose transporter-GFP fusion to the plasma membrane was shown by complementation of a sucrose-uptake deficient yeast strain (Fig. 5). Transgenic plants were generated and selected as described above. In lines expressing either LeSUT1-GFP or LeSUT1-GFP-3'-UTR, GFP fluorescence was detected in CC, whereas only very low fluorescence levels were found in the SE (Fig. 4B,4C). As compared to the RolC-GFP plants, the SUT1-GFP fusion constructs showed much lower fluorescence. As a consequence, the sensitivity of the CLSM photomultipliers had to be increased. Thus, the fluorescence seen in the SE is interpreted as an increase in background noise based on similar observations in untransformed plants (compare Fig. 3I with Fig. 4D). The pattern of GFP particles at the cell periphery in LeSUT1-GFP-3'-UTR lines was similar to that described before in the GFP-3'-UTR lines (Fig. 4C, and 3C). Thus in contrast to the native SUT1 protein, which was found exclusively in SE, the SUT1-GFP fusion protein is found almost exclusively in CC. To confirm the presence of SUT1 in the CC, LeSUT1 was also immunolocalized using a SUT1-specific antibody affinity-purified against solanaceous SUT1 proteins [5]. As shown previously, SUT1 protein localizes to the sieve elements in wild type tobacco and tomato plants, (Fig. 6A,6B and [5]). In contrast, SUT1 protein was detected in CC of both the RolC-LeSUT1-GFP and RolC-LeSUT1-GFP-3'-UTR transgenic lines (Fig. 6C,6D). The cross-reactivity detected in the SE can be attributed to the presence of NtSUT1 (Fig. 6A and 6B). Consistent with the GFP localization, a LeSUT1-GUS-3'-UTR construct (including a 2.3 kb LeSUT1 promoter; Fig. 7A) displayed GUS activity localized to the CC (Fig. 7B), suggesting that the fusion protein is produced in the CC.
Figure 4.
GFP fluorescence localization in RolC-SUT-GFP fusion. GFP fluorescence was detected on longitudinal hand sections of tobacco transgenic petiole under CLSM and co-stained with DAPI and aniline blue. In both cases, GFP fluorescence is localized to companion cells. A) All constructs were prepared in a modified pGPTV-HPT under the control of the CC-specific, RolC, promoter. The asterisk indicates the position of the start codon. T, nos terminator. B) RolC-LeSUT1-GFP petiole, C) RolC-LeSUT1-GFP-3'-UTR petiole. D) Untransformed wild type tobacco leaf petiole visualized with the average PMT level used for the above images CC, companion cell; SE, sieve element; sp, sieve plate; n, nucleus. Bar equals to 10 μm.
Figure 5.
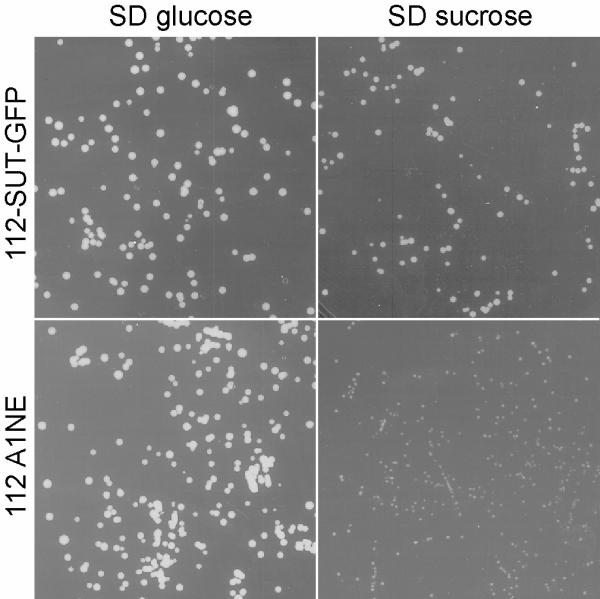
Yeast heterologous expression of SUT1-GFP. The functionality of the SUT1 in the SUT1-GFP fusion is shown by complementation of a yeast strain deficient in sucrose uptake mechanism. SUSY7 yeast strain was transformed with the plasmid containing the fusion LeSUT1-GFP under the Adh1 promoter and the empty vector (112A1NE). A similar amount of cells were plated on 2% sucrose or 2% glucose containing media.
Figure 6.
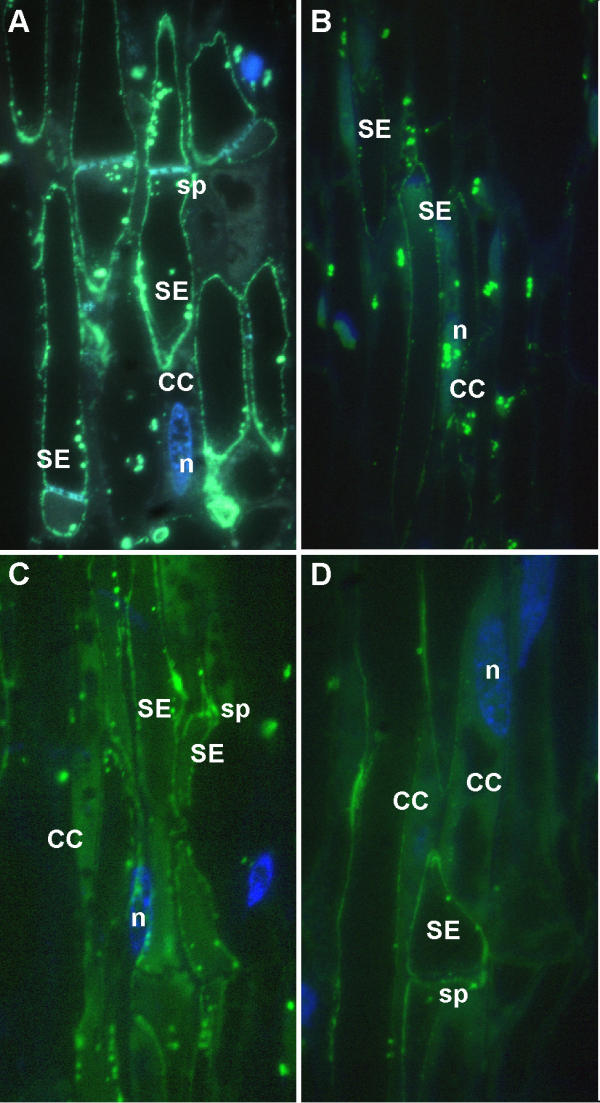
LeSUT1 immunolocalization. SUT1 was immunolocalized in different transgenic tobacco lines using antibody against StSUT1 [5] and detected using a secondary antibody conjugated to FITC. SUT1 localized to sieve element in wild type tobacco and tomato; but appears present in companion cells of transgenic plants. A) Wild type tobacco, B) Wild type tomato, C) RolC-LeSUT1-GFP, D) RolC-LeSUT1-GFP-3'-UTR. CC, companion cell; SE, sieve element; sp, sieve plate, nucleus. Bar indicates 10 μm.
Figure 7.
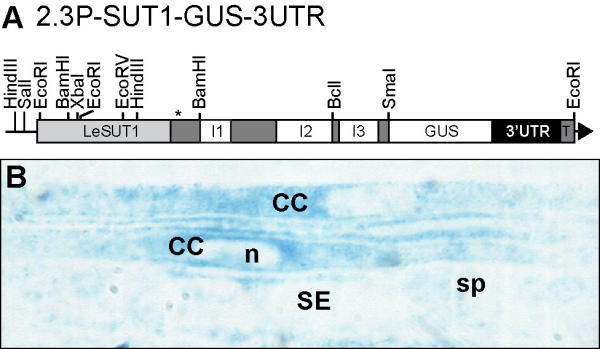
GUS localization in 2.3P-SUT-GUS-3'-UTR. The construct was prepared in pGPTV-HPT binary vector under the control of a 2.3 kb fragment of LeSUT1 promoter fused to the genomic sequence of LeSUT1, to GUS gene and to the 3'-UTR. The asterisk indicates the position of the start codon. I, intron; T, nos terminator.
Discussion
SEs are formed by asymmetric cell division of a mother cell producing two cell types, a CC and a SE. Upon differentiation, the SE loses its nucleus and most of its organelles, while the CC differentiates into a nurse cell for the SE [reviewed in [22]]. Previous experiments indicated that SUT1 mRNA is expressed in the CC [8], while the protein resides in the SE [5]. The short half-life of SUT1, which is in the range of a few hours, suggests that new protein must be synthesized continuously [5,23]. Furthermore, the localization of SUT1 mRNA at the orifices of plasmodesmata of both the CC and SE is compatible with an RNA trafficking mechanism. Thus one of the questions arising was how SUT1 protein can be produced in the SE in the absence of a nucleus. Two alternative hypotheses have been put forward: (i) the mRNA produced in the CC is translated in the CC itself, and the protein is targeted and to the SE via PD, or (ii) the mRNA produced in CC is targeted to CC/SE PDs, moves through the PDs and is then translated in the SE.
In organisms such as Drosophila and Xenopus embryos or mammalian nerve cells, asymmetric distribution of mRNA is important for localized translation and for polarity. This asymmetry is, in most cases, attributed to RNP complexes that target, move and anchor specific RNAs via cis-sequences present in the 3'-UTRs. Considering the role of the 3'-UTR in Drosophila and other organisms, we used a transgenic approach to study whether the 3'-UTR of SUT1 mRNA may play a role in targeting from CC to SE. We used two different reporter genes, GUS and GFP, alone or as C-terminal fusions to SUT1, and this with or without a 1.2 kb fragment corresponding to the 3'-UTR. The constructs were expressed under the control of the CC-specific RolC promoter [8,24], allowing expression of the reporter genes in the CC and analysis of the role of the 3'-UTR.
Reporter Genes Are Localized to Companion Cells
In plants expressing the RolC-GFP construct, GFP fluorescence was detected only in the CC in both source and sink tissues (Fig. 3), with no detectable signal in other cell types above background. Restriction of GFP protein to CC is in contrast with earlier observations where GFP expressed in the CC under the Arabidopsis AtSUC2 promoter moves into SEs and unloads in sink tissues [21]. The only obvious difference between the two experimental setups are the promoters and may thus be explained by differences in promoter strength in the CC. Although no direct comparison of the relative activity of the two promoters is available, Imlau et al. [21], reported that GFP fluorescence was detected using a fluorescence stereo microscope, whereas in this study, even the highest expressing plants described showed no visible fluorescence when analyzed under these conditions. If the differences are due to relative promoter strength, the movement of GFP in the SUC2 promoter-driven expression may be explained by overloading of a receptor-coupled trafficking system, rather than non-selective movement of polypeptides smaller than the SEL. The absence of movement of GFP in case of RolC-GFP will allow us to use this system for studying the presence of cis-element in the targeting, anchoring and/or movement of mRNA from CC to SE.
Role of the 3'-UTR
To test the hypothesis that cis-elements in SUT1 mRNA are required for trafficking, we used different reporter genes fused to the tomato SUT1 3'-UTR. Our results suggest that the 3'-UTR has at least two roles: (i) release of a potential block in translation in the CC since the reporter was translated and (ii) association with particles.
Translational fusions of SUT1 with either GUS or GFP (with or without 3'-UTR) led to an accumulation of the reporter in the CC. Thus one potential interpretation would be that signals for mRNA trafficking overlap the coding region and the 3'-UTR.
Addition of other upstream elements such as the native SUT1 promoter (2.3 kb) and the introns did not have any effect on localization (Fig. 7). The common feature between all constructs is the fusion of either the Nos terminator (constructs without LeSUT1 3'-UTR) or the LeSUT1 3'-UTR behind the stop codon of the reporter genes. None of the constructs had a continuity SUT1 coding region with its 3'-UTR.
In the case of the yeast Ash1 mRNA, localization to the daughter cell depends on a signal of 7 nucleotides overlapping the stop codon [25,26]. Insertion of GFP at the stop codon disrupts the signal and results in a loss of asymmetric localization. This region of the 3'-UTR of Ash1 contains secondary structures recognized specifically by RNA-binding proteins. The similarity of Ash1 to the SE-localized sugar transporters is further reinforced by an analysis of the 3'-UTR of SUT1 RNAs (tomato, potato and tobacco), which reveals more complex secondary structures as compared to other plant RNAs [Additional file 1]. The absence of complex structures in the 3'-UTR of the Arabidopsis SUC1 and SUC2 mRNAs is consistent with the lack of trafficking to be expected for the pollen SUC1 and for SUC2, which cannot leave the CC.
Similar to other organisms, the association of GFP fluorescence with particles (Figs. 3C and 4C) might correspond to RNP complexes as found in neuronal cells, where polarized localization of Staufen occurs in granules [12]. These particles are thought to contain all the machinery necessary for the translation of the RNA at its destination point, as well as microtubule anchoring proteins for the movement of the particles themselves [27,28]. The particles observed here might thus represent an intermediary complex, which plays a role in SUT1 trafficking to PD. SUT1 translation is expected to occur on the ER in CC, SE or associated with the PD connecting these cells. SUT1 3'-UTR resulted in GFP and SUT1-GFP association with particles. Presumably, information in the 3'-UTR targeted the mRNA to the particles, where translation occurred and where the protein was retained. It is possible that SUT1-GFP protein transport through PD to SE was blocked due to the presence of GFP, resulting in retention in CC particles. Alternatively, SUT1 mRNA transport through the PD could have been blocked due to insertion of GFP between the SUT1 coding sequence and 3'-UTR.
Conclusions
An unexpected finding of the analysis was that when driven from the RolC promoter, GFP remained in CC, a result that is in seeming contradiction with GFP trafficking when GFP was expressed under control of the SUC2 promoter [21]. The difference in GFP distribution may be explained if trafficking of proteins through PDs is receptor-coupled and SUC2 promoter driven expression leads to overloading of receptors. Similarly, SUT1-GFP fusion protein was found in CC, suggesting that the fusion led to accumulation of SUT1 in CC and inhibited trafficking to SE. Accumulation of the fusion was confirmed by immunolocalization. However, addition of the SUT1 3'-UTR to either GFP or the SUT1-GFP fusion was insufficient to mobilize the RNA in either case, suggesting that additional signals are required.
Localization of SUT1 in the SE suggests a barrier in the plasma membrane preventing trafficking through the continuous plasma membrane between CC and SE. Therefore an RNA-based trafficking of SUT1 mRNA is feasible and supported by mRNA localization to SE PDs as well as by CmPP16-mediated intercellular mobilization of SUT1 mRNA [10]. Therefore, we propose that one possibility to explain the accumulation of SUT1 fusion proteins in the CC is the effect of an interruption of cis-elements, potentially encoded in RNA structures, at the junction between the SUT1 coding region and the 3'-UTR. These cis-elements seem to be involved both in trafficking and a block of translation in the CC. This is consistent with the ability of the 3'-UTR alone to localize GFP to particle-like structures in the cytosol of the CC. However the 3'-UTR alone seems insufficient for movement of reporter mRNAs into the CC when expressed from the weak RolC promoter.
Alternatively, the results are also compatible with a model in which the SUT1 protein moves between CC and SE. In this case, the 3'-UTR could have a role in targeting SUT1 mRNA to particles within the CC where translation of SE-destined membrane proteins occurs, presumably part of the ER. The translational fusion of SUT1 to GFP may have inhibited trafficking. Further experiments will be required to clarify the mechanism leading to accumulation of SUT1 and other transporter proteins in SE.
Methods
Plant material, growth and transformation
Tobacco plants (Nicotiana tabacum var. SNN) were transformed with an Agrobacterium-mediated gene transfer method using leaf disks [29]. Regenerated plants were selected on 50 mg/mL hygromycin containing media and were maintained in sterile conditions on 2MS media [30] at 21°C with a 16/8 h light/dark cycle. Selection of expressing plants was performed either using a confocal laser scanning microscope (CLSM; see below) for plants containing GFP, or by staining for GUS activity (see below). A minimum of three independent lines per constructs was analyzed.
Constructs
All constructs were made in the plant binary vector pGPTV-HPT in which a KpnI restriction site was added in the existing SmaI restriction site [31]. An EcoRI-KpnI fragment of the RolC promoter [32] was inserted in the modified pGPTV-HPT and a SacI restriction site was removed by treating with the T4 DNA polymerase [33]. The reporter genes β-glucuronidase (GenBank Acc. No. M14641) and mGFP5 (GenBank Acc. No. U87974) [34], LeSUT1 (GenBank Acc. No. X82275) and LeSUT1 3'-UTR (GenBank Acc. No. AY380824) were amplified by PCR using gene specific primers supplemented with restriction (Table 1) sites and the Pfu polymerase (Stratagene). Each PCR product was individually cloned into pBluescript SK+ (Stratagene) or pDR195 [35], sequenced and tested for functionality in yeast. LeSUT1 cDNA was isolated by screening a flower cDNA library from Lycopersicon esculentum cv UC82b using NtSUT3 [36] as a probe. For mGFP5, the original start and stop codons of wild type GFP were used; hence removing the chitinase leader peptide and the ER retention signal. For the isolation of the 3'-UTR, a genomic LeSUT1 clone isolated from a tomato library [37,38] was used as template for the PCR reaction generating a fragment of 1.2 kb. GFP, GUS and LeSUT1 were digested by KpnI and EagI and ligated into the plasmid containing the 3'-UTR. The whole cassette was partially digested with SacI and the fragments corresponding to the gene and the 3'-UTR were ligated into the modified binary vector containing the RolC promoter. To create the constructs without the 3'-UTR, the vector was digested by SacI and re-ligated in a larger volume. For the LeSUT1-GFP fusion, specific primers with restriction sites were used to create a NotI fusion between the two genes (Table 1).
Table 1.
List of cloning primers
| Primer Name | Sequencea |
| KpnI-mGFP5 | TCG GGTACC ATG AGT AAA GGA GAA GAA CTT TTC |
| SacI/EagI-mGFP5 | GTGT CGGCCGGAGCTC TTA TTT GTA TAG TTC ATC CAT GCC |
| KpnI-GUS | CTCG GGTACC ATG TTA CTT CCT GTA GAA ACC |
| SacI/EagI-GUS | GTGT CGGCCGGAGCTC TCA TTG TTT GCC TCC CTG C |
| KpnI-LeSUT1 | GAA GGTACC CAA ATG GAG AAT GGT ACA AAA GGG |
| SacI/EagI-LeSUT1 | CGTCTT CGGCCGGAGCTC TTA ATG GAA ACC GCC CAT GG |
| EagI-3'-UTR | TTC CGGCCG AAA AAA TTA CAA AAG ACG AGG AAG |
| SacI-3'-UTR | TACC GAGCTCCTAGG CGA GGT CGA CGG TAT CG |
| NotI-LeSUT1 | CGTCTT GCGGCCGC TTA ATG GAA ACC GCC CAT GG |
| NotI-GFP | TCG GCGGCCGC TTA TTT GTA TAG TTC ATC CAT GCC |
a Restriction sites are underlined
Cytochemistry
GFP localization
Longitudinal hand sections of petiole of sterile cultured plants were double stained with DAPI (4',6-diamidino-2-phenylindole; 2.5 μg/mL in water) and aniline blue (0.05% (w/v) in potassium phosphate buffer, pH 8.5) and observed under a microscope (Leica DMR fitted with a large numerical aperture objective and water immersion lens) equipped with a confocal laser scanning head (Leica, TCS SP). GFP was detected using an Ar/Kr mixed gas laser with an excitation line at 488 nm and emission was recorded between 495–525 nm. A UV laser (50 mW – Coherent Inc.) with an excitation 350–365 nm was used to excite aniline blue and DAPI, emission was recorded in the range of 510–540 nm for aniline Blue and 435–485 nm for DAPI. Scanning was done sequentially between the UV and Ar/Kr laser to avoid cross talk in the excitation of the different fluorescent compounds. To increase the specificity of the GFP signal, several emission channels were simultaneously recorded and overlaid (chlorophyll, 625–690 nm, lignified compounds, 570–600 nm). Images were processed and assembled using Photoshop® 7.0 and Illustrator® 10.0.
Histochemical localization of β-glucuronidase activity
For the localization of the β-glucuronidase activity transgenic plants or parts of transgenic plants were infiltrated with 1 mM 5-bromo-4-chloro-3-indolyl-glucuronide (X-Gluc) in 50 mM sodium phosphate buffer pH 7.2 containing 0.5 % Triton X-100 and incubated overnight at 37°C [39,40]. Addition of 0.5 mM potassium ferri-/ ferrocyanide was needed to increase the specificity of the staining. After incubation, plant material was cleared with 70 % ethanol.
For the cellular GUS localization, plant material was first incubated in X-Gluc solution as described above. After staining, plant leaves were cut into small pieces (2 mm2) and fixed overnight under light vacuum in 100 mM PIPES buffer containing 1.6% (v/v) glutaraldehyde, 2 mM MgCl2, and 5 mM EDTA. Following fixation, material was washed in 50 mM PIPES buffer, dehydrated by successive passage through increasing ethanol concentration, gradually infiltrated and embedded in LR White. Thin sections (4 μm) were prepared using an ultramicrotome (Leica, Germany). Sections were stained with DAPI for nuclei identification.
Immunolocalization
Immunolocalization of LeSUT1 was performed with modifications according to Barker et al. [6]. Briefly, hand-cut pieces (1 mm2) from tobacco leaves, and petioles were fixed in 100 mM PIPES buffer pH 7.2, 5 mM EDTA, 2 mM MgCl2 containing 0.1% (v/v) glutaraldehyde and 4% (w/v) formaldehyde overnight under light vacuum at 4°C. The material was dehydrated by incubation in ascending ethanol concentration, followed by gradual infiltration in LR White (London Resin Company Ltd, Reading, UK). Polymerization was performed at 58°C for 24 h. Semi-thin sections (1 μm) were mounted and SUT1 was immunolocalized as in Barker et al. [6]. For triple staining of the transporter protein with that of nuclei and sieve plate, sections were stained with DAPI (0.5 μg/mL in water) and aniline blue. DAPI and aniline blue fluorescence was detected with an excitation light of 365 nm. Photographs were taken on Kodak Chrome 400, slides were scanned, processed and assembled using Photoshop® 7.0 and Illustrator® 10.0.
Abbreviations
3'-UTR 3'-untranslated region
CC companion cell
CLSM confocal laser scanning microscope
DAPI 4',6-diamidino-2-phenylindole
FITC fluorescein isothioacetate
PD(s) plasmodesma (plasmodesmata)
RNP ribonucleoprotein
SE sieve element
SUT sucrose transporter
TEM transmission electron microscope
X-Gluc 5-bromo-4-chloro-3-indolyl-glucuronide
Authors' contributions
SL carried out the molecular studies, made the constructs, plant transformations and analysis of the transgenic plants (except for construct 2.3P-SUT-GUS-3'-UTR and its analysis) and drafted the manuscript. AW carried out molecular studies, made the construct 2.3P-SUT-GUS-3'-UTR, plant transformation and analysis of the transgenic plants. RPW carried out the bioinformatic 3'UTR studies. JMW participated in the design of the study. WBF conceived the study, participated in its design and drafted the manuscript together with SL. All authors read and approved the final manuscript.
Supplementary Material
Secondary structures predictions of the 3'-UTR of different sucrose transporters. From left to right, LeSUT1 (Acc No. X82275), LeSUT2 (Acc. No. AF166598), LeSUT4 (Acc. No. AF176950), and StSUT1 (Acc. No. X69165), on the bottom, AtSUC1 (Acc. No. X75365) and AtSUC2 (NM_102118). The secondary structures were predicted using the MFold 3.0 server at [http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi[19]]. The color code indicates the "determination" of the bases. Regions depicted as yellow, orange or red dots indicate a high degree of determination (with red being the highest degree, followed by orange, then yellow). Regions that are highly indeterminate (are not routinely predicted to appear in a given state) are depicted with green, blue, and black dots (black being the highest state of indetermination). Stars indicate the beginning of the 3'-UTR.
Acknowledgments
Acknowledgements
We would like to acknowledge Sabine Hummel and Volker Pott for outstanding technical assistance, Nicole Thiele for LeSUT1 clone isolation, Brigitte Hirner for the LeSUT1 genomic clone isolation, and Michael Burnet for reading the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 446) and Körber Stiftung to WBF and the Alexander von Humboldt Stiftung to SL.
Contributor Information
Sylvie Lalonde, Email: sylvie.lalonde@zmbp.uni-tuebingen.de.
Andreas Weise, Email: aweise@greenovation.com.
Rama Panford Walsh, Email: rpanford@zmbp.uni-tuebingen.de.
John M Ward, Email: jward@tc.umn.edu.
Wolf B Frommer, Email: frommer@andrew2.stanford.edu.
References
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM. The dual function of sugar carriers: transport and sugar sensing. Plant Cell. 1999;11:707–726. doi: 10.1105/tpc.11.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Barker L, Bürkle L, Frommer WB. Update on sucrose transport in higher plants. Journal of Experimental Botany. 1999;50:935–953. doi: 10.1093/jexbot/50.suppl_1.935. [DOI] [Google Scholar]
- Sjölund RD. The phloem sieve element: A river runs through it. Plant Cell. 1997;9:1137–1146. doi: 10.1105/tpc.9.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJE, Ehlers K, Knoblauch M. Sieve elements caught in the act. Trends in Plant Science. 2002;7:126–132. doi: 10.1016/S1360-1385(01)02225-7. [DOI] [PubMed] [Google Scholar]
- Kühn C, Franceschi VR, Schulz A, Lemoine R, Frommer WB. Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science. 1997;275:1298–1300. doi: 10.1126/science.275.5304.1298. [DOI] [PubMed] [Google Scholar]
- Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB. SUT2, a putative sucrose sensor in sieve elements. Plant Cell. 2000;12:1153–1164. doi: 10.1105/tpc.12.7.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Schulze W, Kühn C, Barker L, Schulz A, Ward JM, Frommer WB. Protein-protein interactions between sucrose transporters of different affinities co-localized in the same enucleate sieve element. Plant Cell. 2002;14:1567–1577. doi: 10.1105/tpc.002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Quick WP, Schulz A, Riesmeier JW, Sonnewald U, Frommer WB. Companion cell-specific inhibition of the potato sucrose transporter SUT1. Plant, Cell and Environment. 1996;19:1115–1123. [Google Scholar]
- Zambryski P, Crawford K. Plasmodesmata: gatekeepers for cell-to-cell transport of developmental signals in plants. Annual Review of Cell and Developmental Biology. 2000;16:393–421. doi: 10.1146/annurev.cellbio.16.1.393. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares B, Xiang Y, Ruiz-Medrano R, Wang HL, Monzer J, Yoo BC, McFarland KC, Franceschi VR, Lucas WJ. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science. 1999;283:94–98. doi: 10.1126/science.283.5398.94. [DOI] [PubMed] [Google Scholar]
- Palacios IM, St. Johnston D. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annual Review of Cell and Developmental Biology. 2001;17:569–614. doi: 10.1146/annurev.cellbio.17.1.569. [DOI] [PubMed] [Google Scholar]
- Köhrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Molecular Biology of the Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen R-P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Böhl F, Kruse C, Frank A, Ferring D, Jansen R-P. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO Journal. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-Myo3p complex to ASH1 mRNA. EMBO Journal. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for the maintenance of cell polarity in yeast. Molecular Cell. 2000;5:841–851. doi: 10.1016/S1097-2765(00)80324-X. [DOI] [PubMed] [Google Scholar]
- Graber JH, Cantor CR, Mohr C, Smith TF. In silico detection of control signals: mRNA 3'-end-processing sequences in diverse species. Proceedings of the National Academy of Sciences of the USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Hunt AG. The polyadenylation of RNA in plants. Plant Physiology. 1997;115:321–325. doi: 10.1104/pp.115.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Mathews DH, Turner DH. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J and Clark BFC, editor. RNA Biochemistry and Biotechnology. Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]
- Stadler R, Sauer N. The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Botanica Acta. 1996;109:299–308. [Google Scholar]
- Imlau A, Truernit E,, Sauer N. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 1999;11:309–322. doi: 10.1105/tpc.11.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJE, Knoblauch M. Sieve element and companion cell: The story of the comatose patient and the hyperactive nurse. Australian Journal of Plant Physiology. 2000;27:477–487. doi: 10.1071/PP99172. [DOI] [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR. Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proceedings of the National Academy of Sciences of the USA. 2002;99:10876–10880. doi: 10.1073/pnas.172198599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guivarc'h A, Caissard JC, Azmi A, Elmayan T, Chriqui D, Tepfer M. In situ detection of expression of the gus reporter gene in transgenic plants: ten years of blue genes. Transgenic research. 1996;5:281–288. [Google Scholar]
- Gonzales I, Buonomo SBC, Nasmyth K, von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Current Biology. 1999;9:337–340. doi: 10.1016/S0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- Chartrand P, Meng X-H, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of green fluorescent protein reporter particle in vivo. Current Biology. 1999;9:333–336. doi: 10.1016/S0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, DesGroseillers L. Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron. 2000;25:19–28. doi: 10.1016/s0896-6273(00)80868-5. [DOI] [PubMed] [Google Scholar]
- Mallardo M, Deitinghoff A, Müller J, Goetze B, Macchi P, Peters C, Kiebler MA. Isolation and characterization of staufen-containing ribonucleoprotein particles from rat brain. Proceedings of the National Academy of Sciences of the USA. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, van Montagu M, Leemans J. Efficient octopine Ti-plasmid derived vector for Agrobacterium-mediated gene transfer. Nucleic Acids Research. 1985;13:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Molecular Biology. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Lerchl J, Geigenberger P, Stitt M, Sonnewald U. Impaired photoassimilate partitioning caused by phloem-specific removal of pyrophosphate can be complemented by phloem-specific cytosolic yeast-derived invertase in transgenic plants. Plant Cell. 1995;7:259–270. doi: 10.1105/tpc.7.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. 2nd. Cold Spring Harbor, USA, Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences of the USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB. NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Letters. 1995;370:264–268. doi: 10.1016/0014-5793(95)00853-2. [DOI] [PubMed] [Google Scholar]
- Bürkle L, Hibberd JM, Quick WP, Kühn C, Hirner B, Frommer WB. The H+-sucrose co-transporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiology. 1998;118:59–68. doi: 10.1104/pp.118.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A. Biology. Tübingen, Germany, Eberhard-Karls-Universität; 2000. Expression analysis of sucrose transporter genes from Lycopersicon esculentum and Arabidopsis thaliana. [Google Scholar]
- Hirner B. Biology. Berlin, Germany, Freien Universität Berlin; 1996. Molekular Characterisierung der Be- und Entladungsvorgänge beim Transport von Saccharose und Aminosäure in Solanum tuberosum, Lycopersicon esculentum und Arabidopsis thaliana. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Molecular Biology Report. 1987;5:387–405. [Google Scholar]
- Martin T, Hellman H, Schmidt R, Willmitzer L, Frommer WB. Identification of mutants in metabolically regulated gene expression. Plant Journal. 1997;11:53–62. doi: 10.1046/j.1365-313X.1997.11010053.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Secondary structures predictions of the 3'-UTR of different sucrose transporters. From left to right, LeSUT1 (Acc No. X82275), LeSUT2 (Acc. No. AF166598), LeSUT4 (Acc. No. AF176950), and StSUT1 (Acc. No. X69165), on the bottom, AtSUC1 (Acc. No. X75365) and AtSUC2 (NM_102118). The secondary structures were predicted using the MFold 3.0 server at [http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi[19]]. The color code indicates the "determination" of the bases. Regions depicted as yellow, orange or red dots indicate a high degree of determination (with red being the highest degree, followed by orange, then yellow). Regions that are highly indeterminate (are not routinely predicted to appear in a given state) are depicted with green, blue, and black dots (black being the highest state of indetermination). Stars indicate the beginning of the 3'-UTR.