Abstract
The healthy gastrointestinal tract is physiologically colonized by a large variety of commensal microbes that influence the development of the humoral and cellular mucosal immune system1,2.
Microbiota is shielded from the immune system via a strong mucosal barrier. Infections and antibiotics are known to alter both the normal gastrointestinal tract barrier and the composition of resident bacteria, which may result in possible immune abnormalities3.
HIV causes a breach in the gastrointestinal barrier with progressive failure of mucosal immunity and leakage into the systemic circulation of bacterial bioproducts, such as lipopolysaccharide and bacterial DNA fragments, which contribute to systemic immune activation4-7. Microbial translocation is implicated in HIV/AIDS immunopathogenesis and response to therapy 4,8.
We aimed to characterise the composition of bacteria translocating in peripheral blood of HIV-infected patients. To pursue our aim we set up a PCR reaction for the panbacteric 16S ribosomial gene followed by a sequencing analysis.
Briefly, whole blood from both HIV-infected and healthy subjects is used. Given that healthy individuals present normal intestinal homeostasis no translocation of microflora is expected in these patients. Following whole blood collection by venipuncture and plasma separation, DNA is extracted from plasma and used to perform a broad range PCR reaction for the panbacteric 16S ribosomial gene9. Following PCR product purification, cloning and sequencing analyses are performed.
Protocol
Handling of HIV infected blood samples requires some important recommendations. All specimens of blood must be transported in robust leak-proof containers. Care must be taken when collecting the specimen to avoid contamination of the container’s exterior and of any paperwork accompanying the specimen. All persons processing infected blood must wear gloves. Gloves must be changed and hands washed after completion of specimen processing. Processing of HIV-infected blood samples must be done in a class II biohazard cabinet hood. Mechanical pipetting aids should be used. Use of needles or other sharps (including glass e.g. pipettes or capillary tubes) must be limited to situations in which there is no alternative. Laboratory surfaces must be decontaminated with an appropriate chemical disinfectant after a spill of blood and when work activities are completed. Contaminated materials used must be decontaminated before reuse or must be disposed of correctly via the clinical waste route. Every incident of occupational exposure to potentially infectious blood or fluids (i.e., those requiring universal precautions) should be treated as a medical emergency as interventions must be initiated promptly to be effective. The protocol requires 5 days for its completion. The timeframe is detailed in Figure 1. Bacterial species are identified using the methods described in Figure 2.
1. Sample Collection
9 ml of whole blood is drawn into EDTA containing tubes.
Tubes are centrifuged at 2000 rpm for 10 minutes at RT to obtain plasma.
Plasma is collected in a sterile 2 ml Eppendorf-tube.
2. DNA Extraction from Plasma Samples
Adequately disinfect hood, pipettes and material needed for the experiment in order to guarantee sterility.
Position all materials under UV lights for at least 30 minutes.
Wipe gloves with a disinfectant.
Soak a few paper towels in ethanol and place them under the hood. Every time a tip is discarded wipe the pipette on the wet paper towels.
DNA is extracted using a commercial kit following manufacturer’s instructions (Easy-DNA Kit, Invitrogen, Carlsbad CA, USA).
350 μL of plasma are placed in a sterile 2ml eppendorf tube.
350 μL of filtered ultrapure water are used as negative control.
Add 10 μL of lysozyme (1mg/ml) to the samples.
Incubate for 30 minutes at 37°C.
Add 500 μL of Lysis Solution and mix gently to the samples.
Incubate for 7 minutes at 65°C.
Add 900 μL of chloroform to the samples.
Vortex vigorously until the samples are uniformly viscous.
Add 200 μL of Precipitation Solution and vortex vigorously.
Centrifuge samples at 10500 rpm for 10 minutes at RT to separate the phases and form the interface.
Transfer the upper aqueous phase to a fresh microcentrifuge tube containing 1 ml of ethanol 100%.
Centrifuge samples at 10500 rpm for 10 minutes at 4°C.
Remove ethanol.
Add 1ml of ethanol 70%.
Centrifuge samples at maximum speed for 10 minutes at 4°C.
Remove ethanol. The residual ethanol should be removed with a pipettator.
Resuspend the pellet in 50 μL of ultrapure water.
Read DNA concentration with a spectrophotometer.
3. 16S rRNA Gene PCR
PCR amplification is performed as previously described 9.
Amplify DNA in a 100 μL reaction mixture consisting of 10 μL of 10X PCR buffer, 5 μL of 25 mM MgCl2, 5 μL of 2 mM total dNTPs, 1 μL of 50 μM primer RW01 (AACTGGAGGAAGGTGGGGAT), 1 μL of 50 μM primer DG74 (AGGAGGTGATCCAACCGCA), 34 μL of H20 and 0.5 μL of Taq polymerase (AmpliTaq Gold, Applied Biosystem, Foster City, CA, USA).
Transfer the PCR mix in filters in order to avoid possible contamination by bacterial Taq polymerase.
Centrifuge filters (Microcon, Millipore, Billerica, MA, USA) for 30 minutes at 500 rcf at 4°C.
Aliquot the PCR mix according to the number of samples which need to be amplified (positive and negative PCR controls, samples and extracted water).
Use the following thermal cycler conditions: 94°C for 10 minutes; 40 times for 1 minute each time at 94°C, 55°C and 72°C, and 10 min at 72°C.
Visualize the PCR product on a 2% agarose gel. Use 100 bp DNA ladder. PCR product size is about 360 bp (Figure 3).
4. PCR Product Purification
Only PCR positive samples should be purified. Purification is performed using a commercial kit following manufacturer’s instructions (PureLink PCR microkit, Invitrogen, Carlsbad CA, USA).
Add 4 volumes of Binding Buffer containing isopropanol per 1 volume of PCR product.
Transfer PCR product with Binding Buffer to a column.
Centrifuge for 1 minute at 10000 rcf at RT.
Wash column with Wash Buffer containing ethanol.
Centrifuge for 1 minute at 10000 rcf at RT.
Centrifuge at maximum speed for 1 minute at RT to dry the silica membrane and remove any residual Wash Buffer with ethanol.
Add 10 μL of ultrapure water.
Incubate for 1 minute at RT.
Centrifuge at maximum speed for 2 minutes at RT to collect the purified DNA.
5. Cloning
Lysogeny Broth (LB) Plate Preparation
Dissolve 25 gr of LB mix in approximately 800 ml of water.
Bring the final volume to 1 L.
Add 15 gr of Bactoagar.
Autoclave for 20 minutes.
After autoclaving, vigorously swirl the solution in the flask to mix molten agar.
Cool the solution to 50°C.
Add Ampicillin (50 μg/ml) and swirl until it dissolves.
Pour plates to a depth of approximately 3mm.
Leave the plates out at room temperature.
Spread X-Gal (40mg/ml) and IPTG (100 mM) on each LB plate and incubate at 37°C until ready for use.
Competent Cells Transformation
Transformation is performed using a commercial kit following manufacturer’s instructions (Topo TA cloning kit, Invitrogen, Carlsbad CA, USA).
Prepare the cloning reaction mix (1-4 μL of fresh PCR product, 1 μL of salt solution, 1 μL of vector and water if necessary).
Mix reaction gently and incubate for 5-10 minutes at RT.
Place the reaction on ice.
Add 2 μL of the cloning reaction into a vial of competent cells and mix gently.
Incubate on ice for 30 minutes.
Heat-shock the cells for 30 seconds at 42°C.
Immediately transfer the tube into ice.
Add 250 μL of medium.
Cap the tube tighly and shake the tube horizontally at 37°C for 1 hour.
Spread 50 μL from each transformation on a prewarmed selective plate.
Incubate overnight at 37°C.
Following incubation, white and blue colonies develop on plates. White colonies are positive for PCR product insertion and blue colonies are negative for PCR product insertion.
6. Sequencing Analysis
Sequencing Reaction
A mix is made using these reagents: 2,5 μl DNA, 1 μl primer (5pmol/ μl) 1,1 μl BigDye (Applied Biosystem, Foster City, CA, USA ).
Thermal cycler conditions: 96°C for 1 minutes; 25 times for 96°C for 10 sec, 55°C for 15 sec and 60°C for 4 min.
Column Purification
For each reaction, prepare one MicroSpin Column (Qiagen, , Milan, Italy).
Invert column and vortex to mix resin.
Snap off the bottom of column, loosen lid ¼ turn, and place column in a microcentrifuge tube.
Centrifuge 3200 rpm for 1 minute.
Transfer column to a clean microcentrifuge tube.
Carefully pipet the entire PCR sequencing reaction into the center of the column.
Centrifuge 3200 rpm for 1 minute.
Purified DNA will eluite into tube (20 μl of ultrapure water).
Sequencing Analysis
Load the purified sample (20 μl) into 96-multiwell plate.
Load the plate into the sequencer. Automated DNA Sequencers generate a four-color chromatogram showing the results of the sequencing run.
Input the nucleotide sequence as a query against the public sequence databases. The search is performed on the NCBI databases and servers, with Basic Local Alignment Search Tool (BLAST).
Consider only bacteria with 98-100% homologies.
7. Representative Results
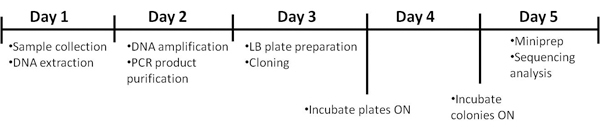 Figure 1. Timeline of the procedure.
Figure 1. Timeline of the procedure.
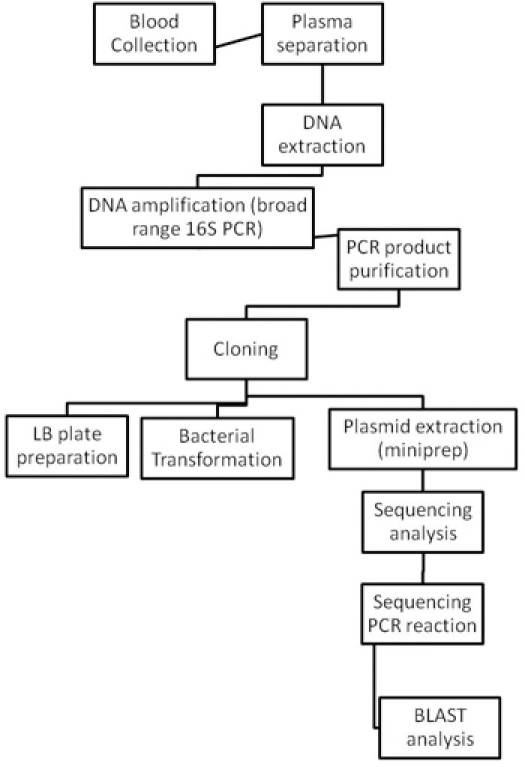 Figure 2. Flow charts of the entire bacterial identification procedure.
Figure 2. Flow charts of the entire bacterial identification procedure.
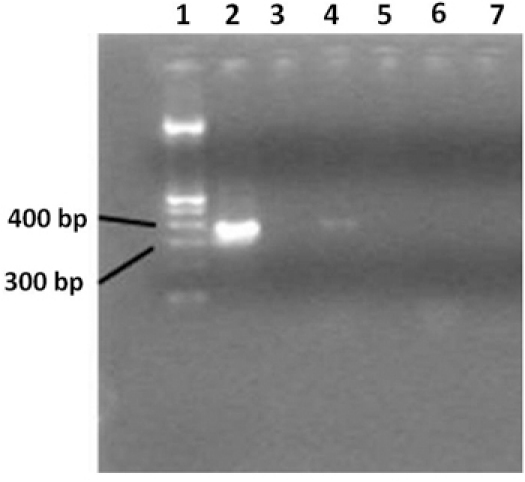 Figure 3. 2% agarose gel showing broad range 16S rRNA gene PCR products. Lane 1 contains a 100bp DNA ladder, lane 2 contains
the PCR positive control, lane 3 shows the negative PCR control. Lane 4 shows samples from an HIV-positive patient, and lane 5 contains water.
Lane 6 shows samples from a healthy individual and a negative PCR reaction; lane 7 contains water. Only HIV positive patients displays a
positive PCR amplification. Ultrapure water used as a negative control during the extraction step, indicates that no contamination occurred.
Figure 3. 2% agarose gel showing broad range 16S rRNA gene PCR products. Lane 1 contains a 100bp DNA ladder, lane 2 contains
the PCR positive control, lane 3 shows the negative PCR control. Lane 4 shows samples from an HIV-positive patient, and lane 5 contains water.
Lane 6 shows samples from a healthy individual and a negative PCR reaction; lane 7 contains water. Only HIV positive patients displays a
positive PCR amplification. Ultrapure water used as a negative control during the extraction step, indicates that no contamination occurred.
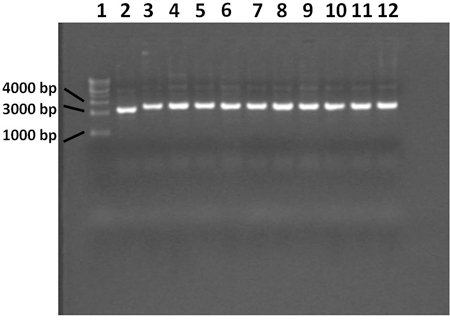 Figure 4. 0.7% agarose gel showing plasmid extracted with miniprep procedure. Lane 1 contains a 1 Kilobase DNA ladder, lane 2
contains the blue control colony, lanes 3 through 12 contain white colonies. The plasmid from the blue colony does not contain the PCR product
insert. All 10 white colonies contain plasmid with the correct insert.
Figure 4. 0.7% agarose gel showing plasmid extracted with miniprep procedure. Lane 1 contains a 1 Kilobase DNA ladder, lane 2
contains the blue control colony, lanes 3 through 12 contain white colonies. The plasmid from the blue colony does not contain the PCR product
insert. All 10 white colonies contain plasmid with the correct insert.
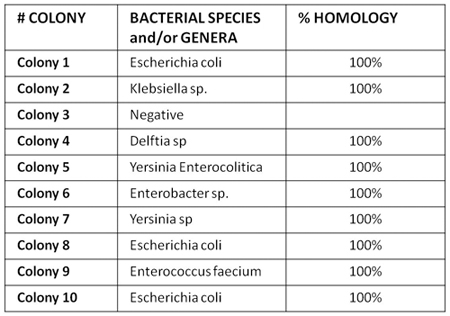 Figure 5. Shows an example of bacterial sequencing analysis in plasma from one HIV-positive individual. Our results show that microbial translocation in HIV disease involves a polimicrobic flora, which is not seen in HIV-negative subjects, suggesting substantial failure of gut immunity in controlling bacteria translocation.
Figure 5. Shows an example of bacterial sequencing analysis in plasma from one HIV-positive individual. Our results show that microbial translocation in HIV disease involves a polimicrobic flora, which is not seen in HIV-negative subjects, suggesting substantial failure of gut immunity in controlling bacteria translocation.
Discussion
We hereby show a PCR/sequencing protocol to characterise the translocation of bacteria in peripheral blood of HIV-infected individuals.
Most reliable results are obtained when using plasma collected in EDTA-containing tubes. After blood sampling, plasma should be separated by centrifugation within 2/3 hours, to avoid haemolysis and DNA fragment degradation. Samples are stored first at -20°C for approximately 5 days and then at -80°C. Multiple freeze and thaw cycles which may cause loss of bacterial DNA should be avoided.
Plasma samples should be used within 12 hours after thawing.
Optimization of a DNA extraction protocol was performed in order to obtain the maximum quantity of DNA from donors’ peripheral blood. More specifically, plasma instead of whole blood is used , because the latter contains low amounts of bacterial DNA which is also under-represented by the great amount of DNA derived from peripheral blood cells.
We deliberately chose to exclude from our study subjects with signs and/or symptoms of acute infection. Nonetheless, given that subjects with HIV-related immunedepression present impairment of the gastro-intestinal barrier and translocation of bacterial products, we could not exclude translocation of entire microorganisms. Thus the rationale behind the use of lysozyme: given that this enzyme damages the bacterial wall, its use allows for the release of bacterial DNA from extremely low numbers of circulating microorganism which do not necessarily account for overt clinical manifestations.
The major issue in the present experiment is the risk of contamination. Given the high risk of PCR contamination from environment- and reagent-borne bacteria, we optimized laboratory procedures to avoid contamination in each step of the experiment, starting from sample collection.
In particular, for best sample preparation, it is essential to avoid contamination of plasma samples and surfaces of plastic consumables and containers. This is achieved by using disinfectants, powder free gloves and by respecting standard laboratory procedures. Within the described protocol the most critical steps in terms of contamination risk are DNA extraction and amplification, due to the possible presence of contaminating bacterial DNA, deriving, for example, from Taq polymerase.
Using this approach, we demonstrated that HIV-infected patients display translocation of polymicrobic bacteria in peripheral blood. No microbes are detected in the peripheral blood of healthy individuals, possibly reflecting the integrity of the gastrointestinal barrier. Future research will focus on the investigation of the role of peripheral blood microbiota in the pathogenesis of HIV/AIDS and response to anti-retroviral therapy.
Disclosures
No conflicts of interest declared.
Acknowledgments
We are grateful to Gianni Scimone for excellent assistance with video making.
References
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Kanauchi O, Mitsuyama K, Araki Y, Andoh A. Modification of intestinal flora in the treatment of inflammatory bowel disease. Curr Pharm Des. 2003;9:333–346. doi: 10.2174/1381612033391883. [DOI] [PubMed] [Google Scholar]
- Brenchley JM. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Douek DC. HIV disease: fallout from a mucosal catastrophe. Nat Immunol. 2006;7:235–239. doi: 10.1038/ni1316. [DOI] [PubMed] [Google Scholar]
- Jiang W. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti G. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS. 2008;22:2035–2038. doi: 10.1097/QAD.0b013e3283112d29. [DOI] [PubMed] [Google Scholar]
- Marchetti G. Role of Microbial Translocation and Immune Hyperactivation in Disease Progression of HIV+ Patients with Preserved CD4 Count in the Absence of ART. The 17th Conference on Reteroviruses and Opportunistic Infections (CROI); San Francisco, CA, USA. 2010. [Google Scholar]
- Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]


