Abstract
As medicine is currently practiced, doctors send specimens to a central laboratory for testing and thus must wait hours or days to receive the results. Many patients would be better served by rapid, bedside tests. To this end our laboratory and others have developed a versatile, reagentless biosensor platform that supports the quantitative, reagentless, electrochemical detection of nucleic acids (DNA, RNA), proteins (including antibodies) and small molecules analytes directly in unprocessed clinical and environmental samples. In this video, we demonstrate the preparation and use of several biosensors in this "E-DNA" class. In particular, we fabricate and demonstrate sensors for the detection of a target DNA sequence in a polymerase chain reaction mixture, an HIV-specific antibody and the drug cocaine. The preparation procedure requires only three hours of hands-on effort followed by an overnight incubation, and their use requires only minutes.
Keywords: Bioengineering, Issue 52, biosensor, chemistry, detection, electrochemistry, point of care, theranostics, diagnostics, antibody, instrument, electronic
Protocol
1. Setting the Stage
Purchase the relevant, chemically modified probe DNA from a custom oligonucleotide synthesis company such as Biosearch Technologies (Novato, CA) or Midland Certified (Midland, TX). The probe is modified during synthesis by the addition of a C6 thiol at its 3’-end and a redox-active methylene blue at its 5’-end.
Dissolve the probe DNA in phosphate buffered saline pH 7.4 to a concentration of 200 μM and verify its concentration by measuring its absorbance at 260 nm using a spectrophotometer. Due to the methylene blue moiety on the DNA probe the solution should have a visible blue tint.
Freshly prepare 1 mL of a 10 mM solution of tris(2-carboxyethyl)phosphine TCEP in distilled, deionized water (DI-water). In our experience, these TCEP solutions remain fresh for one week when stored in the dark at 4°C.
To reduce any disulfide bonds that might be present in the probe DNA solution, combine 1 μL of the probe DNA stock solution with 2 μL of the TCEP solution and mix gently with a pipette. Incubate the mixture for one hour in a dark, refrigerated container. The initially blue solution should become clear as the TCEP reversibly reduces the methylene blue. If the solution does not become clear, repeat the procedure using a fresh TCEP solution or, possibly, at room temperature.
After one hour dilute the reduced DNA probe solution with 1 mL of buffer; this will dilute it to a concentration of 200nM. Later on, you will incubate a set of gold disk electrodes in 200 μL portions of this diluted probe solution.
Freshly prepare at least 2 mL of 2mM mercaptohexanol in phosphate buffered saline.
2. Sensor Preparation
Combine 0.05 micron alumina powder with water on a fine polishing cloth. Polish a set of gold disk electrodes (CH Instruments, Austin, TX) by pressing the gold surface firmly into the wet cloth, and moving them in a figure eight pattern for approximately three minutes per electrode.
Rinse the polished electrodes with DI-water and immerse them into Eppendorf tubes filled with the same. Sonicate for five minutes to remove any residual alumina powder.
Place the electrodes into a 0.5 M sulfuric acid solution, attach them to a potentiostat along with a platinum counter and silver/silver chloride reference electrode, and run a series of voltammograms to oxidize, reduce, and electrochemically clean their surfaces. Following this perform a second electrochemical cleaning in a solution of 0.01 M KCl in 0.1 M sulfuric acid. The fine details of these cleaning procedures can be found in our Nature Protocols paper1, and in the supplement to this video.
Arrange a set of 2 mL Eppendorf tubes in a rack and fill each with 200 μL of the probe DNA solution. The concentration of the probe DNA in this solution will define the density with which the probe DNAs pack on the sensor surface. Sensor performance is strongly dependent on probe density, with the optimal density varying from one sensor architecture to the next. The probe concentration used at this step should thus be optimized for each new type of sensor2. For the probe architectures we have investigated to date, the probe DNA concentrations we employ in this step range from 15 nM to 2 μm, with 200 nM being a typical value.
Rinse the gold disk electrodes with DI-water, and then immerse them in the relevant probe DNA solution in an Eppendorf tube for one hour. At this point, the probe DNA will attach to the gold electrode surface via the formation of a thiol-on-gold self assembled monolayer.
Rinse the electrodes with DI-water, immerse them in 2mM mercaptohexanol in an Eppendorf tube and store them in a dark place for 3 hours to overnight at room temperature to ensure complete formation of the self assembled monolayer. This step incorporates the mercaptohexanol as part of a mixed monolayer to ensure the formation of a stable monolayer. To prevent evaporation you may want to seal the electrode into the Eppendorf tube with parafilm. Sensors can be stored in this solution for several days if necessary.
When you are ready to use the sensor, rinse it with DI-water and then soak it in buffer for at least ten minutes. Sensors aimed at antibody detection must also be immersed in a 100 nM solution of the relevant recognition strand prior to use, followed by an extremely quick rinse with buffer.
3. Sensor Testing, DNA Detection
In this protocol, a 17 nucleotide probe strand is affixed to a gold electrode. It has a methylene blue redox reporter at its 3’-end (Figure 1). When the probe molecule hybridizes with a capture strand, the current through the sensor circuit decreases.
Rinse a fresh sensor with DI-water and immerse it in a blank sample lacking the target in order to record the background signal it produces. Attach the sensor to the working electrode lead of a potentiostat. Place a platinum counter electrode and a silver/silver chloride reference electrode into the solution.
Run a square wave measurement from 0 to -0.6 V with an amplitude of 25 mV and a step size of 1mV. The optimal square wave frequency will depend on the details of the probe architecture2,3; for the probe architectures we have employed the optimal values are typically in the range 60 to 600 Hz. You should see a rounded peak at approximately -0.35 V, the redox potential of methylene blue (the peak potential may shift slightly depending on the precise pH of your test solution). The height from baseline current to this peak is proportional to the efficiency electron transfer between the methylene blue and the gold electrode (Figure2). Save this background measurement.
Move the electrodes to a solution that contains the target DNA molecule of interest, equilibrate (5 to 120 min depending on the size, structure and concentration of the target4,5) and collect a second square wave voltammagram. The height of the peak at -0.35 V will change from the initial, background measurement. The magnitude of this change is related to the concentration of the analyte. It is the main output data of this sensor (Figure 3).
Measuring relative signal change, the percentage decrease or increase in signal relative to the background peak, is often more reproducible than measuring the absolute change in current, as this corrects for variations in the surface area of the electrode. To do this, the difference between the peak current and the background peak current are divided by the background peak current.
4. Sensor Regeneration
Once your measurements are complete, move the sensor into a container filled with DI-water for 30 s, or squirt it with a steady stream of DI-water for 30 s. Repeat this two more times with fresh deionized water. Note: some analytes are resistant to this approach; for them try aggressive rinsing in 6 M guanidine hydrochloride or 70% ethanol.
Put the sensor back into a solution where measurements will be made. Within a minute, the peak height should have returned to its original value. It is worth noting that these sensors often exhibit a slightly larger signal change during their first test and regeneration cycle, and highly consistent results during the subsequent cycles6.
5. Sensor Testing, Antibody Detection
In this protocol, the methylene-blue and thiol-modified DNA probe used in these sensors serves as an "anchor" strand7. It is attached directly to the gold electrode. This is then hybridized with a second, "recognition" DNA strand that has been covalently conjugated to the relevant antigen (Figure 4). We have had good luck with commercial synthesis houses, such as Biosearch Technologies and Panagene, for the synthesis of the needed DNA-antigen chimeras. The hybridization step is carried out by transferring a pre-fabricated sensor into an Eppendorf tube containing 100 nM of the relevant recognition DNA strand in PBS for 1 hour.
Place the sensor in the relevant blank solution. Attach it to the working electrode lead of a potentiostat and place a platinum counter electrode and silver/silver chloride reference electrode into the solution.
Perform square wave voltammetry as described above. For the particular probe architecture we have employed here the optimal square wave frequency is 60 Hz. You should see a rounded peak around -0.35 V. Save this background measurement.
Transfer the electrodes to a solution containing the target analyte, incubate for 5 to 60 min, and collect a second square wave voltammagram. If the target antibody is present the peak at -0.35 V will decrease. The magnitude of this change is related to the antibody concentration.
6. Sensor Testing, Small Molecule Detection
In this case, the probe molecule on the sensor surface is an aptamer, a DNA or RNA molecule that has been selected in vitro to bind a specific molecular analyte, that changes its structure (folds) upon binding to its target analyte8,9 (Figure 5). Here we employ a cocaine-binding, DNA aptamer developed by the Stojanovic10,11 lab.
Rinse a fresh sensor with DI-water and immerse it in a blank sample lacking the target in order to record the background signal it produces. Attach the sensor to the working electrode lead of a potentiostat. Place a platinum counter and a silver/silver chloride reference into the solution.
Perform square wave voltammetry as described above. For the particular probe architecture we have employed here the optimal square wave frequency is 200Hz (But 60 Hz also works). You should see a rounded peak around -0.35 V. Save this background measurement.
Transfer the electrodes to a solution containing the target analyte, incubate for ˜5 min, and collect a second square wave voltammagram. The height of the peak at -0.35 V will change. The magnitude of this change is related to the concentration of the target analyte. If you cannot obtain a cocaine sample, procaine, the use of which is unregulated, can be used as a substitute.
7. Representative Results:
When used to detect DNA using the first architecture, the signal should decrease by at least 60% when equilibrated at 200 nM target. After three brief rinses in deionized water, the signal should return very close (within 0.1-5%) to its original value. Antibody detection sensors should undergo a signal decrease of 40 to 80%. Aptamer-based sensors for the detection of cocaine exhibit a signal increase of up to 200% depending on the frequency and surface coverage at which they operate. For the cocaine sensor, a low surface coverage is best3.
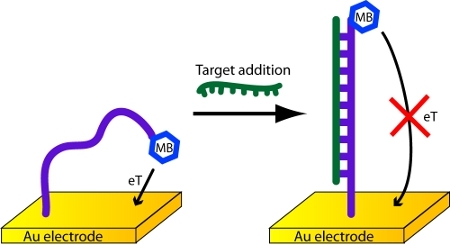 Figure 1. Detection of DNA with an electrochemical DNA biosensor.
Figure 1. Detection of DNA with an electrochemical DNA biosensor.
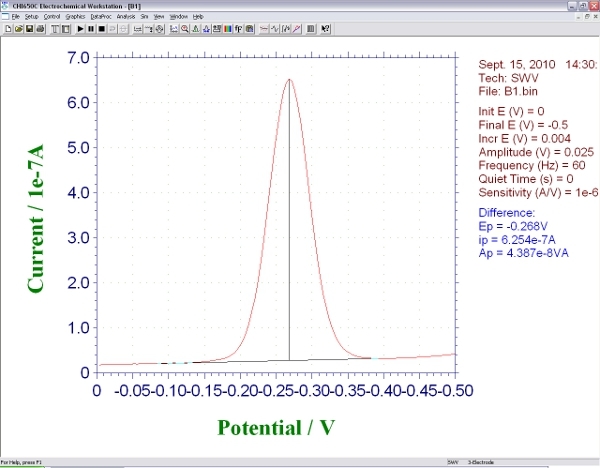 Figure 2. Screen shot showing the signal produced by an E-DNA biosensor during square wave voltammetry.
Figure 2. Screen shot showing the signal produced by an E-DNA biosensor during square wave voltammetry.
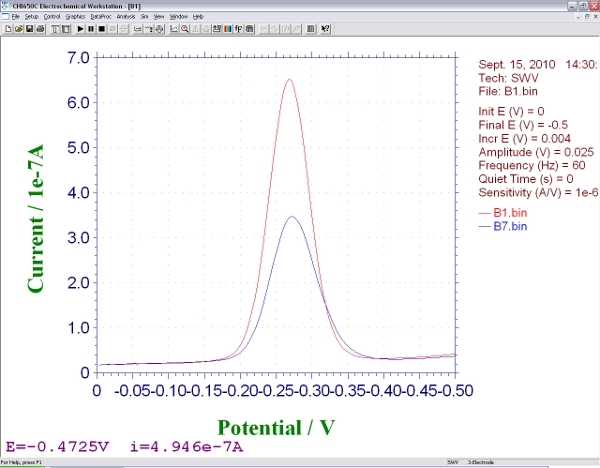 Figure 3. Screen shot showing the signals produced by an E-DNA biosensor during square wave voltammetry, before and after hybridization with an analyte.
Figure 3. Screen shot showing the signals produced by an E-DNA biosensor during square wave voltammetry, before and after hybridization with an analyte.
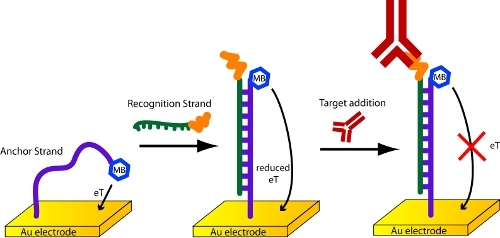 Figure 4. Detection of antibodies with a scaffold biosensor.
Figure 4. Detection of antibodies with a scaffold biosensor.
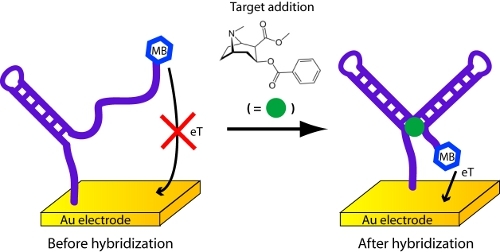 Figure 5. Detection of cocaine or procaine with an electrochemical aptamer biosensor.
Figure 5. Detection of cocaine or procaine with an electrochemical aptamer biosensor.
| Custom Oligo | Sequence | Comments |
| Linear Probe DNA (LP17) | 5′-HS−(CH2)6−TGGATCGGCGTTTTATT−(CH2)7−NH−MB-3′ | HPLC Purified, can be ordered with S-S |
| Target Analyte DNA | AATAAAACGCCGATCCA | Unmodified |
| Recognition Strand | 5'-Antigen-TEG- CAGTGGCGTTTTATTCTTGTTACTG-3' | |
| Scaffold Anchor | 5'-HS-(CH2)6-GCAGTAACAAGAATAAAACGC CACTGC-(CH2)7-MB | HPLC Purified, can be ordered with S-S |
| A4 Cocaine Aptamer | 5'-HS-AGACAAGGAAAATCCTTCAATGAAGTGGGTCG-MethyleneBlue-3' | HPLC Purified, can be ordered with S-S |
Table 1. Probe and Target DNA Sequences.
Discussion
An important note is that none of the experiments described above will work properly unless the electrodes have been properly cleaned. Here is a guide to our electrochemical cleaning procedure. When working with CH Instruments potentiostats, we run these cleaning steps using a set of three macro programs.
Phase Zero (E-clean O) Immerse the electrodes in 0.5M H2SO4 and connect them to the working electrodes of a potentiostat. Also attach and immerse an Ag/AgCl reference and platinum counter electrode. Start with an oxidation step (2 V for 5 s) and then a reduction step (0.35 V for 10 s).
Phase One (E-clean 1) Initiate oxidation and reduction scans under the same acidic conditions (0.5M H2SO4) from 0.35 to 1.5 V (20 scans at a scan rate of 4 V/s and a sample interval of 0.01 V, followed by four scans at a scan rate of 0.1 V/s and a sample interval of 0.01 V).
Phase Two (E-clean 2) Conduct another set of electrochemical oxidation and reduction scans under acidic conditions (0.01 M KCl/0.1 MH2SO4) covering four different potential ranges (all performed for 10 segments at a scan rate of 0.1 V s 1 and a sample interval of 0.01 V): (i) potential range from 0.2 to 0.75 V; (ii) potential range from 0.2 to 1.0 V; (iii) potential range from 0.2 to 1.25 V; (iv) potential range from 0.2 to 1.5 V.
Many types of gold electrodes can be used to conduct these experiments. In addition to gold disk electrodes such as those employed here, we have had success with microfabricated gold surfaces, gold wire, and gold on printed circuit boards.
Along with the sensors described in this paper, many other electrochemical DNA biosensor architectures have been reported. This includes sensors with a pseudoknot12, triple strand13, sandwich14, super sandwich15, or triplex16 architecture.
In the future, we expect that these sensors will be used in point of care medical diagnostics. They have been successfully integrated into several microfluidic devices17,18, and offer many advantages over optical analyte detection systems. In particular, these sensors can function in turbid, optically dense and highly auto-fluorescent samples.
Disclosures
There is nothing to disclose.
Acknowledgments
This work was funded by a grant (OPP1015402) from the Bill and Melinda Gates Foundation through the Grand Challenges Explorations Initiative, and by the NIH through grants GM062958-01 and 2R01EB002046. This work was performed in part under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344.
References
- Xiao Y, Lai RY, Plaxco KW. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat. Protocols. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- Ricci F, Lai RY, Heeger AJ, Plaxco KW, Sumner JJ. Effect of Molecular Crowding on the Response of an Electrochemical DNA Sensor. Langmuir. 2007;23:6827–6834. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Phares N, Lubin AA, Xiao Y, Plaxco KW. Optimization of Electrochemical Aptamer-Based Sensors via Optimization of Probe Packing Density and Surface Chemistry. Langmuir. 2008;24:10513–10518. doi: 10.1021/la800801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin AA, Vander Stoep Hunt B, White RJ, Plaxco KW. Effects of Probe Length, Probe Geometry, and Redox-Tag Placement on the Performance of the Electrochemical E-DNA Sensor. Analytical Chemistry. 2009;81:2150–2158. doi: 10.1021/ac802317k. [DOI] [PubMed] [Google Scholar]
- Lubin AA, Plaxco KW. Folding-Based Electrochemical Biosensors: The Case for Responsive Nucleic Acid Architectures. Accounts of Chemical Research. 2010;43:496–505. doi: 10.1021/ar900165x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin AA, Lai RY, Baker BR, Heeger AJ, Plaxco KW. Sequence-Specific, Electronic Detection of Oligonucleotides in Blood, Soil, and Foodstuffs with the Reagentless, Reusable E-DNA Sensor. Analytical Chemistry. 2006;78:5671–5677. doi: 10.1021/ac0601819. [DOI] [PubMed] [Google Scholar]
- Cash KJ, Ricci F, Plaxco KW. An Electrochemical Sensor for the Detection of Protein?Small Molecule Interactions Directly in Serum and Other Complex Matrices. Journal of the American Chemical Society. 2009;131:6955–6957. doi: 10.1021/ja9011595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BR. An Electronic, Aptamer-Based Small-Molecule Sensor for the Rapid, Label-Free Detection of Cocaine in Adulterated Samples and Biological Fluids. Journal of the American Chemical Society. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- Ferapontova EE, Olsen EM, Gothelf KV. An RNA Aptamer-Based Electrochemical Biosensor for Detection of Theophylline in Serum. Journal of the American Chemical Society. 2008;130:4256–4258. doi: 10.1021/ja711326b. [DOI] [PubMed] [Google Scholar]
- Stojanovic MN, Landry DW. Aptamer-Based Colorimetric Probe for Cocaine. Journal of the American Chemical Society. 2002;124:9678–9679. doi: 10.1021/ja0259483. [DOI] [PubMed] [Google Scholar]
- Stojanovic MN, Prada Pde, Landry DW. Aptamer-Based Folding Fluorescent Sensor for Cocaine. Journal of the American Chemical Society. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- Cash KJ, Heeger AJ, Plaxco KW, Xiao Y. Optimization of a Reusable, DNA Pseudoknot-Based Electrochemical Sensor for Sequence-Specific DNA Detection in Blood Serum. Analytical Chemistry. 2009;81:656–661. doi: 10.1021/ac802011d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y. An Electrochemical Sensor for Single Nucleotide Polymorphism Detection in Serum Based on a Triple-Stem DNA Probe. Journal of the American Chemical Society. 2009;131:15311–15316. doi: 10.1021/ja905068s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Xiao Y, Plaxco KW. High Specificity, Electrochemical Sandwich Assays Based on Single Aptamer Sequences and Suitable for the Direct Detection of Small-Molecule Targets in Blood and Other Complex Matrices. Journal of the American Chemical Society. 2009;131:6944–6945. doi: 10.1021/ja901315w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia F. An Electrochemical Supersandwich Assay for Sensitive and Selective DNA Detection in Complex Matrices. Journal of the American Chemical Society. 2010;132:14346–14348. doi: 10.1021/ja104998m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson A. Using Triplex-Forming Oligonucleotide Probes for the Reagentless, Electrochemical Detection of Double-Stranded DNA. Analytical Chemistry. 2010;82:9109–9115. doi: 10.1021/ac1024528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BS. Integrated Microfluidic Electrochemical DNA Sensor. Analytical Chemistry. 2009;81:6503–6508. doi: 10.1021/ac900923e. [DOI] [PubMed] [Google Scholar]
- Swensen JS. Continuous, Real-Time Monitoring of Cocaine in Undiluted Blood Serum via a Microfluidic, Electrochemical Aptamer-Based Sensor. Journal of the American Chemical Society. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]


