Abstract
During metastasis cancer cells disseminate from the primary tumor, invade into surrounding tissues, and spread to distant organs. Metastasis is a complex process that can involve many tissue types, span variable time periods, and often occur deep within organs, making it difficult to investigate and quantify. In addition, the efficacy of the metastatic process is influenced by multiple steps in the metastatic cascade making it difficult to evaluate the contribution of a single aspect of tumor cell behavior. As a consequence, metastasis assays are frequently performed in experimental animals to provide a necessarily realistic context in which to study metastasis. Unfortunately, these models are further complicated by their complex physiology. The chick embryo is a unique in vivo model that overcomes many limitations to studying metastasis, due to the accessibility of the chorioallantoic membrane (CAM), a well-vascularized extra-embryonic tissue located underneath the eggshell that is receptive to the xenografting of tumor cells (figure 1). Moreover, since the chick embryo is naturally immunodeficient, the CAM readily supports the engraftment of both normal and tumor tissues. Most importantly, the avian CAM successfully supports most cancer cell characteristics including growth, invasion, angiogenesis, and remodeling of the microenvironment. This makes the model exceptionally useful for the investigation of the pathways that lead to cancer metastasis and to predict the response of metastatic cancer to new potential therapeutics. The detection of disseminated cells by species-specific Alu PCR makes it possible to quantitatively assess metastasis in organs that are colonized by as few as 25 cells. Using the Human Epidermoid Carcinoma cell line (HEp3) we use this model to analyze spontaneous metastasis of cancer cells to distant organs, including the chick liver and lung. Furthermore, using the Alu-PCR protocol we demonstrate the sensitivity and reproducibility of the assay as a tool to analyze and quantitate intravasation, arrest, extravasation, and colonization as individual elements of metastasis.
Protocol
1. Preparing the eggs for xenografting (spontaneous metastasis)
Freshly laid fertilized chicken eggs are incubated in a rotating incubator for 10 days at 100°F and 60% humidity. The eggs are rotated four times each hour.
On developmental day 10 the eggs are placed on their side in an egg rack. A goose-neck lamp or other suitable light source is used to candle the eggs by shining the light into the eggshell at the blunt end of the egg where the airsac is located.
The chorioallantoic vein is located and marked. This vein is located at the top of the eggshell where it is the junction of several large blood vessels. At this branch point the blood vessel drops down, away from the CAM and attaches to the embryo. A 1cm square box is drawn with pencil on the eggshell approximately 1cm away from the branch point in the vein. The area including and around the square is then cleaned using a cotton swab soaked in iodine.
Using a rotating cutting tool (Dremel) fitted with a silicone carbide grinding stone (Dremel part #84922) a hole is drilled through the blunt end of the egg into the air sac. Using this same tool a hole is drilled within the previously drawn square on top of the egg, stopping short of the eggshell membrane. A 25-gauge syringe needle with a fine bur on the end is used to make a small hole in the eggshell membrane being careful not to tear the underlying CAM. The CAM is subsequently detached from the eggshell membrane in the area of the square using gentle suction created with an automatic pipette aid fitted with a piece of ¼” tygon tubing placed against the hole in the airsac. Upon applying suction, an air pocket should underneath the hole in the square signifying that the CAM has been successfully dropped away from the eggshell. After the CAM is dropped, the hole in the air sac and the hole located in the square near the chorioallantoic vein is sealed with a piece of tape laboratory tape. The eggs are subsequently placed into a stationary incubator at 100°F and 60% humidity in anticipation of grafting the tumor cells. See figure 1 for a sequential depiction.
2. Preparing the eggs for intravenous injection (experimental metastasis)
Freshly laid fertilized chicken eggs are incubated in a rotating incubator for 12 days at 100°F and 60% humidity. The eggs are rotated four times each hour.
On developmental day 12 the eggs are placed on their side in an egg rack. A goose-neck lamp or other suitable light source is used to candle the eggs by shining the light into the eggshell at the blunt end of the egg where the airsac is located.
The chorioallantoic vein is located and marked. A 1cm by 0.5 cm rectangle is drawn with pencil on the eggshell directly above the vein. The area including and around the square is then cleaned using a cotton swab soaked in iodine.
A small window is created in the eggshell in the drawn location using the Dremmel drill bit (#118). The window is removed to expose the underlying eggshell membrane which is subsequently rendered transparent with drop of mineral oil. See figure 5A for a graphical depiction.
At this point the hole in the air sac and the hole located in the square near the chorioallantoic vein is sealed with a piece of tape laboratory tape. The eggs are subsequently placed into a stationary incubator at 100°F and 60% humidity in anticipation of injecting the tumor cells.
3. Preparing tumor cells for grafting or injection.
For xenografting, the tumor cells are detached from from their culture dishes using Trypsin/EDTA and washed with 10 volumes of phosphate buffered saline (PBS) in order to remove any residual media, Trypsin, or EDTA.
For I.V. injection, cultured cells are detached using nonenzymatic cell-dissociation solution, and washed with serum-free DMEM. and)
Cells are counted with a hemacytometer and resuspended in PBS at 10-40 million cells/ml for grafting, or alternatively resuspended in serum-free DMEM at 1 million cells/ml for I.V. injection.
4. Grafting the tumor cells onto the newly exposed CAM (Spontaneous Metastasis)
The eggs are removed from the incubator and a small window is cut in the location of the previously drawn square where the new air pocket has formed. This is most easily done with a rotary tool equipped with a cutting wheel (Dremel tool with the #409 circular cutting wheel). A pair of needle-nose forceps is used to remove the small section of eggshell and expose the underlying CAM.
Using a cotton-tipped applicator (Fisher Brand #23-400-001) the CAM is abraded 5 times. A little blood should be evident on the cotton. Immediately after damaging the CAM, 25μl of the cell suspension is placed onto the damaged area. The optimal number of cells is determined empirically but can range from 0.5X105 to 1X106.The window in the egg is sealed tightly with tape and the chicks are left standing for 10-15 minutes in order to allow the cells to settle. After 15 minutes the eggs are returned to the stationary incubator.
The tumors are allowed to grow for 5-8 days depending on the specific application and the nature of the tumor cell line used. See figure 1 for a sequential depiction.
5. Intravenous injection of tumor cells (Experimental Metastasis)
Using a 30 Gauge insulin syringe 100μl of cell suspension is injected into the allantoic vein of each embryo, the eggs were returned to a stationary incubator and remained there for an additional 1–7 days.
At distinct time points, the lung or lower CAM are harvested from each embryo and processed for the detection of tumor cells, as described below.
6. Harvesting tumors and chick tissues
A dissection area is prepared. The assay detects metastatic cells by quantifying human DNA using a highly sensitive PCR approach. It is therefore very important to prevent contamination with exogenous DNA. Dissections are performed in a dedicated area and throughout the dissections the tools are cleaned in between each animal. In order to prevent cross-contamination of your samples throughout the dissection process 3 separate sets of tools are used: one set for cutting the egg, one set for removing the primary tumor and one set for removing the internal organs. In addition, four wash containers are used for sequential rinsing of tools between each animal. In sequence these washes are: 1) double distilled water, 2) double distilled water, 3) chlorine bleach, and 4) 70% ethanol.
The eggs are removed from the stationary incubator and placed on ice for 15 minutes to anesthetize and euthanize the animals. The windows are opened such that the tumors become visible. Opening the windows bigger by removing some of the eggshell may be necessary. The primary tumors are resected from the CAM, weighed, and processed for histology.
The chick is removed from the eggshell by cutting the shell radially into equal halves. The embryo is decanted from the cut shell and transferred to a clean weigh boat breast side up. The animal is dissected with using a fresh, clean set of tools. The animal is opened by cutting through the sternum. Once the embryo is open, a piece of the liver from either the right or left lobe is collected. The internal organs are subsequently removed by pulling gently on the esophagus attached to the gizzard and cutting the visera that connect the organs. In order to collect lungs cut the rib cage is opened the breast plate is separated and the heart is removed. At this point it is possible to cut around the four sides of the lung (the top closest to the head the left side along the rib cage, the bottom near the diaphragm and the right side along the trachea). For the sake of consistency, the same lung is collected in each animal. Harvested tissue samples are stored at -20°C for later use or can be processed immediately for DNA isolation.
7. Genomic DNA isolation and quantitative PCR Analysis
Genomic DNA is extracted from tissues using the Sybr Green Extract–N-Amp Tissue PCR Kit (Sigma-Aldrich Catalog #XNATRG). The extracted DNA can be transferred to a clean eppendorf tube and stored at -20°C or used immediately. It is important to dilute the DNA 1:50 in nuclease free water before using it in the subsequent PCR reaction. Alternatively, DNA can be extracted using any number of standard DNA extraction protocols (Zijlstra et al., 2002).
Human DNA is detected in the chick tissues using quantitative PCR specific for human Alu sequences. Quantitative PCR using Alu-specific primers is performed using the SYBR green amplification from the Exract-N-Amp kit. The PCR kit comes with a 2x reaction mix containing SYBR green, buffer, salts dNTPS, Taq polymerase and the Jumpstart Taq Antibody. The reaction mix is made up in a final volume of 15μl, with 0.4μM of each primer. For most instances using the extracted DNA diluted 1:50 will provide quantitative data, however, the optimal amount of template DNA may need to be optimized empirically. Chicken GAPDH primers are used as an internal control The PCR is run under the following conditions for Alu sequences: polymerase activation at 95°C for 2 min followed by 30 cycles at 95°C for 30 s., 63°C for 30 s., 72°C for 30 s. The reaction conditions for chicken GAPDH are identical to those described for Alu however it may be necessary to extend the PCR to 40 cycles.
Quantitative analysis of metastasis is performed using the ΔΔCt method comparing experimental groups to a control group (Schmittgen and Livak, 2008; Zijlstra et al., 2002). Alternatively a standard curve can be generated using a dilution series of human tumor cells extracted using the Extract-N-Amp kit (Zijlstra et al., 2002). Statistical significance is evaluated using Student T-test or Anova analysis of control and experimental groups.
8. Representative Results:
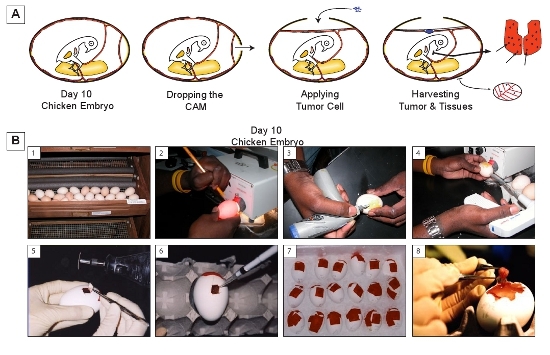 Figure 1. The chicken embryo metastasis model. A) A four panel diagram illustrating the appearance of the model in cross section at key steps: 1. After 10 days of incubation. The CAM has reached near-maximum size. The arteries and veins are distinctly visible with the Allantoic vein positioned against the top of the egg. 2. The egg immediately after drilling two holes in the eggshell: one into the airsac and one adjacent to the attachment point for the allantoic vein. 3. The appearance after a mild vacuum has been used to evacuate the airsac and drop the CAM. Tumor cells (blue) are applied to the dropped CAM. 4. After allowing the grafted tumor cell to grow, the tumor along with tissues from the chicken embryo are harvested for analysis. B) A serial depiction of the model: 1. Incubating the eggs., 2. Marking the location of the allantoic vein., 3. Drilling holes in the eggshell., 4. Dropping the CAM., 5. Cutting an opening for tumor cell grafting., 6. Applying the tumor cells., 7. Cooling the eggs on ice prior to collecting tissue., 8. Harvesting the tumor after 7 days of growth.
Figure 1. The chicken embryo metastasis model. A) A four panel diagram illustrating the appearance of the model in cross section at key steps: 1. After 10 days of incubation. The CAM has reached near-maximum size. The arteries and veins are distinctly visible with the Allantoic vein positioned against the top of the egg. 2. The egg immediately after drilling two holes in the eggshell: one into the airsac and one adjacent to the attachment point for the allantoic vein. 3. The appearance after a mild vacuum has been used to evacuate the airsac and drop the CAM. Tumor cells (blue) are applied to the dropped CAM. 4. After allowing the grafted tumor cell to grow, the tumor along with tissues from the chicken embryo are harvested for analysis. B) A serial depiction of the model: 1. Incubating the eggs., 2. Marking the location of the allantoic vein., 3. Drilling holes in the eggshell., 4. Dropping the CAM., 5. Cutting an opening for tumor cell grafting., 6. Applying the tumor cells., 7. Cooling the eggs on ice prior to collecting tissue., 8. Harvesting the tumor after 7 days of growth.
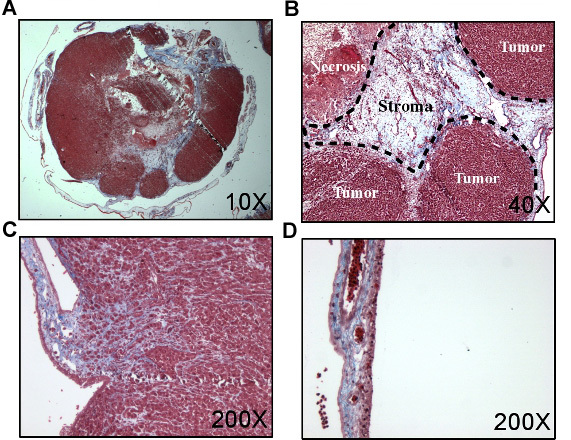 Figure 2. Histology of CAM tumors. Tissues taken from tumor bearing chorioallantoic membrane was fixed in formalin, sectioned and processed with a trichrome stain. A) HEp3 tumor after seven days of growth on the CAM. B and C show magnified regions from A showing both tumor and stroma. D) normals CAM.
Figure 2. Histology of CAM tumors. Tissues taken from tumor bearing chorioallantoic membrane was fixed in formalin, sectioned and processed with a trichrome stain. A) HEp3 tumor after seven days of growth on the CAM. B and C show magnified regions from A showing both tumor and stroma. D) normals CAM.
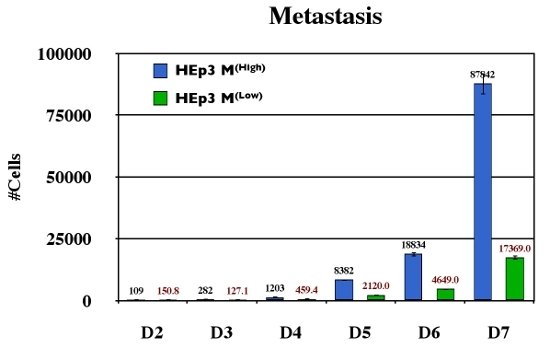 Figure 3. Quantitative longitudinal analysis of metastasis. Variants for the head and neck carcinoma HEp3 with high or low metastatic ability (HEp3 M(High) and HEp3 M(Low) respectively) were analyzed for their ability to disseminate using Alu PCR. Ten animals were analyzed at each time point for the duration of 7 days. These samples were quantified against a standard curve and subsequently expressed as #cells/50mg tissue.
Figure 3. Quantitative longitudinal analysis of metastasis. Variants for the head and neck carcinoma HEp3 with high or low metastatic ability (HEp3 M(High) and HEp3 M(Low) respectively) were analyzed for their ability to disseminate using Alu PCR. Ten animals were analyzed at each time point for the duration of 7 days. These samples were quantified against a standard curve and subsequently expressed as #cells/50mg tissue.
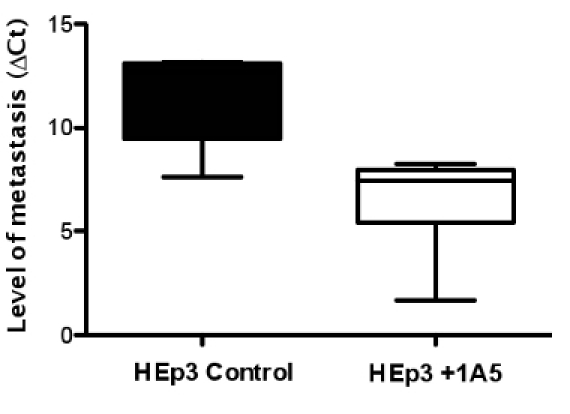 Figure 4. The inhibition of spontaneous metastasis cells in animals treated with the metastasis inhibiting antibody 1A5. Tumors were formed using 5X105 HEp3 cells applied to the CAM of day-10 embryos. Three days after applying the tumor cells half the animals were treated with the anti-CD151 antibody 1A5. The lungs from tumor-bearing animals were harvested two days later and subjected to quantitative real-time alu PCR. Conversely quantitative real-time PCR of chGAPDH was used to confirm the presence of equivalent quantities of host genomic DNA (The ΔΔCt method was used to compare the two groups quantitatively (B).
Figure 4. The inhibition of spontaneous metastasis cells in animals treated with the metastasis inhibiting antibody 1A5. Tumors were formed using 5X105 HEp3 cells applied to the CAM of day-10 embryos. Three days after applying the tumor cells half the animals were treated with the anti-CD151 antibody 1A5. The lungs from tumor-bearing animals were harvested two days later and subjected to quantitative real-time alu PCR. Conversely quantitative real-time PCR of chGAPDH was used to confirm the presence of equivalent quantities of host genomic DNA (The ΔΔCt method was used to compare the two groups quantitatively (B).
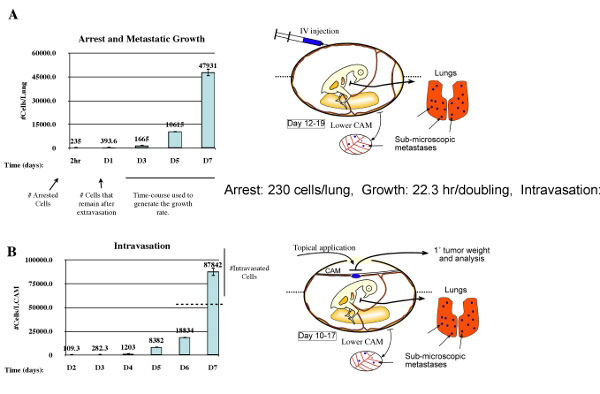 Figure 5. Evaluating arrest, growth, and intravasation in the metastatic cascade. Fundamentally, metastasis to a secondary organ is defined by the contributions of three aspects: arrest, rate of growth, and rate of intravasation. These can be quantified using a combination of experimental metastasis (A) and spontaneous metastasis (B). Using experimental metastasis (A) arrest is defined as the number of cell detected 2hr post injection. Survival and growth of arrested cells is determined as the change in cell number 24hr post injection. Growth is defined as the rate of proliferation during days 3-7. Intravasation is determined using spontaneous metastasis (B) where the number intravasated cells is defined as the metastatic burden present on day 7 that exceeds the value predicted by the growth of the metastatic burden present on day 6.
Figure 5. Evaluating arrest, growth, and intravasation in the metastatic cascade. Fundamentally, metastasis to a secondary organ is defined by the contributions of three aspects: arrest, rate of growth, and rate of intravasation. These can be quantified using a combination of experimental metastasis (A) and spontaneous metastasis (B). Using experimental metastasis (A) arrest is defined as the number of cell detected 2hr post injection. Survival and growth of arrested cells is determined as the change in cell number 24hr post injection. Growth is defined as the rate of proliferation during days 3-7. Intravasation is determined using spontaneous metastasis (B) where the number intravasated cells is defined as the metastatic burden present on day 7 that exceeds the value predicted by the growth of the metastatic burden present on day 6.
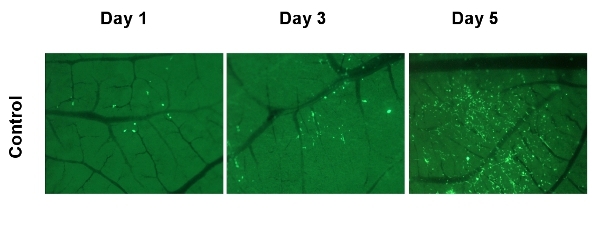 Figure 6. Visualizing expansion of the metastatic in the CAM. HEp3 cells expressing GFP were injected intravenously on developmental day 12. On day 1, 3, and 5 an egg was sacrificed and a section of CAM was imaged with a fluorescent stereo microscope. The dark green background is created by the eggshell, the vasculature is visible as black lines, and the tumor cells are visible as bright green spots.
Figure 6. Visualizing expansion of the metastatic in the CAM. HEp3 cells expressing GFP were injected intravenously on developmental day 12. On day 1, 3, and 5 an egg was sacrificed and a section of CAM was imaged with a fluorescent stereo microscope. The dark green background is created by the eggshell, the vasculature is visible as black lines, and the tumor cells are visible as bright green spots.
Discussion
The chick CAM has been used for decades as a model system to study various aspects of cancer biology and metastatic progression. Principle evaluation of metastatic abilities have been performed in this model by a variety of groups including the analysis of dormancy (Aguirre Ghiso et al., 1999; Ossowski and Reich, 1983), tissue remodeling (Deryugina et al., 2005; Hahn-Dantona et al., 1999), and metastasis (Zijlstra et al., 2008; Zijlstra et al., 2002). It continues to be a very attractive model for cancer biology because of its ease of use, its ability to accept xenografts, and its low cost (Chambers et al., 1982; Chambers et al., 1990). Using human specific Alu-based quantitative PCR (Kim et al., 1998; Zijlstra et al., 2002) we demonstrate the ability to analyze metastatic dissemination of human cpidermoid carcinoma cell line HEp3 (Figure 3). In addition we demonstrate the utility of this technique in the evaluation of therapeutic inhibition using the metastasis inhibitory antibody 1A5 (figure 4, (Testa et al., 1999; Zijlstra et al., 2008)).
In addition to being able to evaluating metastatic ability and treatment response, this model also can be used to quantitatively study individual components of the metastatic cascade (Zijlstra et al., 2002). Using experimental metastasis by intravenous injection of tumor cells, this model allows for the analysis of cancer cell arrest in the secondary organs and their subsequent growth (figure 5A). This rate of growth can be used to predict the metastatic burden in a longitudinal spontaneous metastasis assay (figure 5B). Intravasation is determined using spontaneous metastasis (B) where the number intravasated cells is defined as the metastatic burden present on day 7 minus the value predicted by the growth of the metastatic burden present on day 6.
As an extension of the techniques reported here, we recently developed novel strategies to visualize tumor cell behavior in vivo (Zijlstra et al., 2008), which use fluorescently labeled nanoparticles to target the tumor microenvironment (Leong et al., 2010; Lewis et al., 2006). Through these advancing methods, the utility of this model can be exploited to visualize the complex and dynamic changes that support the survival, establishment and growth of tumor cells. These technologies, coupled with the relatively low cost, immunodeficient nature, ease of use, and adaptability of the chick CAM make this model valuable for imaging and quantitative analysis of cancer metastasis.
Disclosures
No conflicts of interest declared.
Acknowledgments
We want to recognize Tyson Foods Inc. for generously providing the fertilized eggs. Shanna Arnold was instrumental during the filming of this protocol. Trenis Palmer was supported by the National Institutes of Health under the Ruth L. Kirschstein National Research Service Award (F31 National Cancer Institute). This work was partially supported by CCSRI Grant #700537 to JDL and NIH/NCI grant #CA120711-01A1 and CA120711-01A1 to AZ.
References
- Ghiso Aguirre, Kovalski JA, K , Ossowski L. Tumor dormancy induced by downregulation of urokinase receptor in human carcinoma involves integrin and MAPK signaling. J Cell Biol. 1999;147:89–104. doi: 10.1083/jcb.147.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers AF, Shafir R, Ling V. A model system for studying metastasis using the embryonic chick. Cancer research. 1982;42:4018–4025. [PubMed] [Google Scholar]
- Chambers AF, Wilson SM, Tuck AB, Denhardt GH, Cairncross JG. Comparison of metastatic properties of a variety of mouse, rat, and human cells in assays in nude mice and chick embryos. In Vivo. 1990;4:215–219. [PubMed] [Google Scholar]
- Deryugina EI, Zijlstra A, Partridge JJ, Kupriyanova TA, Madsen MA, Papagiannakopoulos T, Quigley JP. Unexpected effect of matrix metalloproteinase down-regulation on vascular intravasation and metastasis of human fibrosarcoma cells selected in vivo for high rates of dissemination. Cancer Res. 2005;65:10959–10969. doi: 10.1158/0008-5472.CAN-05-2228. [DOI] [PubMed] [Google Scholar]
- Hahn-Dantona E, Ramos-DeSimone N, Sipley J, Nagase H, French DL, Quigley JP. Activation of proMMP-9 by a plasmin/MMP-3 cascade in a tumor cell model. Regulation by tissue inhibitors of metalloproteinases. Ann N Y Acad Sci. 1999;878:372–387. doi: 10.1111/j.1749-6632.1999.tb07696.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Yu W, Kovalski K, Ossowski L. Requirement for specific proteases in cancer cell intravasation as revealed by a novel semiquantitative PCR-based assay. Cell. 1998;94:353–362. doi: 10.1016/s0092-8674(00)81478-6. [DOI] [PubMed] [Google Scholar]
- Leong HS, Steinmetz NF, Ablack A, Destito G, Zijlstra A, Stuhlmann H, Manchester M, Lewis JD. Intravital imaging of embryonic and tumor neovasculature using viral nanoparticles. Nat Protoc. 2010;5:1406–1417. doi: 10.1038/nprot.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. Viral nanoparticles as tools for intravital vascular imaging. Nat Med. 2006;12:354–360. doi: 10.1038/nm1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski L, Reich E. Changes in malignant phenotype of a human carcinoma conditioned by growth environment. Cell. 1983;33:323–333. doi: 10.1016/0092-8674(83)90414-2. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Testa JE, Brooks PC, Lin JM, Quigley JP. Eukaryotic expression cloning with an antimetastatic monoclonal antibody identifies a tetraspanin (PETA-3/CD151) as an effector of human tumor cell migration and metastasis. Cancer Res. 1999;59:3812–3820. [PubMed] [Google Scholar]
- Zijlstra A, Lewis J, Degryse B, Stuhlmann H, Quigley JP. The Inhibition of Tumor Cell Intravasation and Subsequent Metastasis via Regulation of In Vivo Tumor Cell Motility by the Tetraspanin CD151. Cancer Cell. 2008;13:221–234. doi: 10.1016/j.ccr.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra A, Mellor R, Panzarella G, Aimes RT, Hooper JD, Marchenko ND, Quigley JP. A quantitative analysis of rate-limiting steps in the metastatic cascade using human-specific real-time polymerase chain reaction. Cancer Res. 2002;62:7083–7092. [PubMed] [Google Scholar]


