Abstract
Deubiquitinating enzymes (DUBs) regulate diverse cellular functions by their activity of cleaving ubiquitin from specific protein substrates. Ubiquitin-Specific Protease 46 (USP46) has recently been identified as a quantitative trait gene responsible for immobility in the tail suspension test and forced swimming test in mice. Mice with a lysine codon (Lys 92) deletion in USP46 exhibited loss of ‘behavioral despair’ under inescapable stresses in addition to abnormalities in circadian behavioral rhythms and the GABAergic system. However, whether this deletion affects enzyme activity is unknown. Here we show that USP46 has deubiquitinating enzyme activity detected by USP cleavage assay using GST-Ub52 as a model substrate. Interestingly, compared to wild type, the Lys 92 deletion mutant resulted in a decreased deubiquitinating enzyme activity of 27.04%. We also determined the relative expression levels of Usp46 in rat tissues using real-time RT-PCR. Usp46 mRNA was expressed in various tissues examined including brain, with the highest expression in spleen. In addition, like rat USP46, both human and mouse USP46 are active toward to the model substrate, indicating the USP cleavage assay is a simple method for testing the deubiquitinating enzyme activity of USP46. These results suggest that the Lys 92 deletion of USP46 could influence enzyme activity and thereby provide a molecular clue how the enzyme regulating the pathogenesis of mental illnesses.
Introduction
Ubiquitin-specific protease (USP), a subfamily of deubiquitinating enzymes (DUBs) [1], [2], is responsible for the removal of ubiquitin or polyubiquitin from target proteins, the processing of ubiquitin precursors, and the disassembly of unanchored polyubiquitin by catalyzing the hydrolysis of isopeptide bonds in ubiquitin-protein conjugates [3]–[7]. USPs have been implicated in wide variety biological processes [8], [9] and involved in the pathogenesis of numerous diseases, including cancer and neurodegeneration [10]–[14]. Recent studies found that USP have also been associated with neurogenetic disorders, including Parkinson's disease [13], [15] and spinocerebellar ataxia [6].
The tail suspension and forced swimming tests [16] are useful experimental paradigms for assessing antidepressant activity and depression-like behavior. In the tests, animals are subjected to inescapable stress of being suspended by their tail or being forced to swim in a water-filled cylinder. The animals rapidly adopt a characteristic immobile posture that has been named ‘behavioral despair’ on the assumption that the animals have given up hope of escaping. Recent study [17] identified USP46 as a quantitative trait gene responsible for the immobility in the tail suspension and forced swimming tests in mice. In this study, mice with a lysine codon (Lys 92) deletion of USP46 showed loss of ‘behavioral despair’ under inescapable stresses in addition to abnormalities in circadian behavioral rhythms and the GABAergic system. The authors demonstrate that USP46 functions to regulate several behavioral processes, including basal immobility, the anti-immobility effects of imipramine, nest building and the muscimol-induced righting reflex. However, comparing the detailed description of the phenotypes of USP46 mutant mice, the molecular mechanism have not been well documented. In particular, whether the lysine codon (Lys 92) deletion in USP46 affects deubiquitinating enzyme activity is unknown.
Here we report that USP46 has deubiquitinating enzyme activity detected by USP cleavage assay using GST-Ub52 as a model substrate. Notably, the Lys 92 deletion mutant results in a decreased deubiquitinating enzyme activity of 27.04% compared to wild type. We also determined the relative expression levels of Usp46 in rat tissues using real-time RT-PCR. Usp46 mRNA was expressed in various tissues examined including brain, with the highest expression in spleen. In addition, like rat USP46, both human and mouse USP46 are active toward to the model substrate, suggesting that the USP cleavage assay using GST-Ub52 as a model substrate is simple for testing the deubiquitinating enzyme activity of USP46. These results indicate that the Lys 92 deletion of USP46 could influence deubiquitinating enzyme activity and therefore might contribute to the understanding of neural and genetic mechanisms that underlies the mental disorders associated with USP46.
Materials and Methods
Molecular cloning of Usp46
Total RNA was isolated using TRIzol reagent (Tiangen, Beijing, China) from Wistar rat brain (7 weeks old), by the Experimental Animal Center of Hebei Medical University, and human esophageal squamous cell carcinoma cell line TE-1 (Cell Bank of Type Culture Collection of Chinese Academy of Sciences, Shanghai, China), respectively. The cDNA was synthesized by reverse transcription-PCR according to the instructions of Quantscript RT Kit (Tiangen, Beijing, China). Forward primer 5′-GGATCCATGACTGTCCGAAACATCGC-3′ (introducing a BamHIsite) and reverse primer 5′-GTCGACCACGCTCCACCAAGTTATTC-3′ (introducing a SalI site) were used to amplify rat USP46 cDNA. Similarly, human USP46 was amplified using forward primer 5′-GGATCCATGACTGTCCGAAACATCGC-3′ (introducing a BamHIsite) and reverse primer 5′-GTCGACCGCAGGTCTTTCAGTTACTC-3′ (introducing a SalI site). The PCR amplified fragment with a predicted size was subcloned into pGEM-T Easy vector (Promega). Escherichia coli DH5α harboring pGEM-Usp46 clone was cultured to prepare supercoiled recombinant plasmid DNA using a Mini Prep kit (QIAGEN). The DNA sequences were determined by automated sequencing (ABI PRISM).
Expression GST-USP46 fusion protein in Escherichia coli DH5α
Plasmid construction
To construct a expression vector suitable for production of glutathione S-transferase (GST) fusion protein (GST-USP46) in Escherichia coli, we first generated a 1.1-kb DNA fragment containing the coding sequence for rat USP46, and subcloned in-frame with GST into the BamHI and SalIsites of the expression vector pGEX-6p-1 (Amersham Pharmacia Biotech) to generate pGEX-USP46. The recombined plasmid was then used to transform Escherichia coli DH5α.
Expression in Escherichia coli DH5α
Escherichia coli DH5α harboring pGEX-USP46 was incubated in LB medium containing 100 µg of ampicillin per ml at 37°C overnight. The pre culture was diluted 1/10 into fresh LB medium containing 100 µg of ampicillin per ml and incubated at 37°C until an OD 600 of 0.5 is reached, then added IPTG at a final concentration of 1 mM and continued the incubation for 4 h. After the incubation, cells were collected by centrifugation at 15,000× g for 10 min at 4°C, and resuspended in buffer PBS (pH 7.4) containing 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4. Cells were disrupted by sonication (six times for 10 s each), then added equal value of 2× SDS sample buffer, boiled for 2 min. The expressed GST-USP46 fusion protein was detected by 10% SDS PAGE.
USP cleavage assay
The deubiquitinating enzyme activity was detected by USP cleavage assay using GST-Ub52 fusion protein as a model substrate, which has been previously described [18]. The model substrate (GST-Ub52) comprising the natural human Ub52 fusion protein precursor linked to GST, such that the ubiquitin sequences comprise the middle portion of the hybrid protein, was cleaved efficiently by UBP2 (positive control) to yield a product of the expected size. USP46 enzyme activity was assayed basically following the method described by Tian et al. [19]. Briefly, a pAC-T7 plasmid was produced by inserting a T7 promoter from BglII and HindIII fragment of pET-3d (a pBR322 Ampr replicon) into pACYC184 (Cmr replicon) at its BamHI and HindIII site. Then, a pAC-T7-Usp46 plasmid was produced by inserting the complete coding sequence of Usp46 into pAC-T7 plasmid at the BamHI site. The plasmid pACYC-UBP2 (pACYC184 Cmr replicon) containing yeast UBP2 co-transformed with pGEX-Ub52 as a positive control for USP cleavage assay. For cleavage of the GST-Ub52 substrate, E. coli strain BL21 (DE3) cells harboring pGEX-Ub52 were further transformed with either pAC-T7-Usp46 or pACYC-UBP2, and colonies resistant to both ampicillin and chloramphenicol were grown-up. Protein expression was induced by IPTG and the cells were further incubated for 3 hours, when the USPs cleaved the substrate. Soluble protein extracts were prepared after sonication of the cell lysate. GST fusion proteins and their cleavage products were purified by GSH-Sepharose™ Resin (Sangon Biotech, Shanghai, China) and detected by 10% SDS PAGE. The relative intensity of the resulting bands was analyzed by Odyssey V3.0 software. The arbitrary unit calculated by the ratio of 36 KDa to 45 KDa protein bands was used to measure the relative deubiquitinating enzyme activity of wild type and mutant of USP46.
The USP cleavage assay using ubiquitin-β-galactosidase (Ub-Met-β-gal) as a model substrate has been previously described [18]. The substrate is a fusion protein in which the ubiquitin was fused to the N-terminus of β-galactosidase. Upon meeting the active deubiquitinating enzyme in the cell, the substrate was cleaved efficiently and yields a product of the expected size. Plasmid pAC-M-β-gal expresses the Ub-Met-β-gal fusion protein substrate in a pACYC184 Cmr replicon. E. coli BL21 (DE3) bacteria harboring pGEX-6P-1-USP46 were transformed with pAC-M-β-gal, Ampr Cmr colonies were grown and induced with IPTG, and total protein extracts were analyzed by Western blotting with anti-β-galactosidase purified monoclonal antibody (Promega) and anti-mouse IgG (H&L) (GOAT) antibody IRDye800CW® Conjugated (Rockland) and detected by the Bio Imaging Navigator FSX100 system (Olympus). The relative intensity of the resulting bands was analyzed by Odyssey V3.0 software.
Real-time RT-PCR analysis
Real-time RT-PCR analysis (qRT-PCR) was used to measure the expression levels of Usp46 in Wistar rat tissues (Experimental Animal Center of Hebei Medical University). Reactions utilizing equal amounts of total RNA from each sample were carried out on Rotor Gene 6000 real-time detection system (Bio-Rad) using the Quant one step SYBR Green RT-PCR reagent kit (Tiangen, Beijing, China) according to the instructions. All qRT-PCR were carried out for 40 cycles under the standard PCR conditions (one cycle at 50°C for 30 min, one cycle at 95°C for 2 min and 40 cycles of 94°C for 20 s, 55°C for 20 sec, 68°C for 20 min, and 72°C∼95°C for Melt Curve Analysis). The results were analyzed based on cycle threshold (Ct) values of a method called 2−ΔΔCt method using GAPDH as an internal control. Briefly, a ΔCt was first calculated by subtracting the averaged Ct value of GAPDH from the averaged Ct value of Usp46 of each tissue, and then a ΔΔCt value was calculated by subtracting the ΔCt values of Usp46 derived from brain. Fold differences were determined by 2 to the power of minus ΔΔCt. The primers used were: Forward primer 5′-CCGATGTGATTAGCTCATG-3′ and reverse primer 5′- GCCACACACATACAGTAAC-3′.
Site-directed mutagenesis
The Cys 44 within the Usp46 conserved Cys-box was mutated to serine and the Lys 92 was deleted using deoxyoligonucleotides and the Stratagene Quik-Change™ site-directed mutagenesis kit according to the manufacturer's instructions using pAC-T7-Usp46 as a template. The resulting plasmids were pAC-T7-Usp46 (C44S) and pAC-T7-Usp46 (ΔK92), respectively. Mutations were confirmed by DNA sequencing.
Other methods
Animals were handled in accordance with the National Institute of Health Guide for Care and Use of Laboratory Animals (NIH Publication no. 80–23). The use of the animals was in agreement with the Animal Care and Use Committee of the Hebei Medical University (Permit Number: 1106110).
Results
Molecular cloning of the open reading frame of Usp46 and expression of GST-USP46 fusion protein in Escherichia coli DH5α
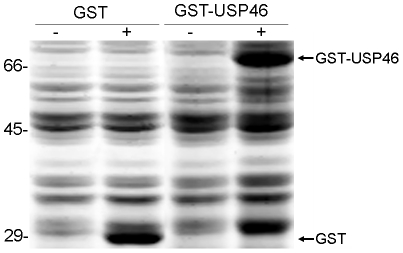
In order to investigate the deubiquitinating enzyme activity of Usp46, we isolated the open reading frame of Usp46 from rat brain by RT-PCR using a pair of primers designed with reference to sequences of Rattus norvegicus (GenBank accession number XM214034). The open reading frame of Usp46 consists of 1101 bp encoding a polypeptide of 366 amino acid residues, and contains highly conserved Cys, Asp and His domains, which is the characteristic of the ubiquitin-specific proteases [1]. The nucleotide sequence of Usp46 has been submitted to the GenBank under accession number GU455413. Similarly, we also cloned human Usp46 from TE-1 cell line (Genbank accession number GU455414). Comparison of the sequence of USP46 of human, rat and mouse revealed 99.7% homology between human and rat and 100% between human and mouse at the amino acid level (Fig. 1). The calculated molecular mass of USP46 is about 42 KDa. As expected, GST-USP46 expressed in the transformants with approximate 68 KDa detected by 10% SDS-PAGE (Fig. 2).
Figure 1. Multi-alignment of amino acid sequences of Homo sapiens, Mus musculus, and Rattus norvegicus of USP46.
Conserved Cys-, Asp- and His- domains of deubiquitinating enzyme are boxed. The amino acid sequences are highly conserved among the species and different only at the position of 161 (indicated by gray box).
Figure 2. Expression of GST-USP46 fusion protein.
GST-USP46 fusion protein was expressed in Escherichia coli strain DH5α. Bacteria harboring recombinant plasmid pGEX-Usp46 were grown up and the expression of GST or GST-USP46 was induced by IPTG (lanes 2 and 4). Attempts to purify the fusion protein were failed, because the protein remained insoluble even after trying a lot of detergents including Triton-100, Urea, and N-lauroyl sarcosinate (data not shown).
Deubiquitinating enzyme activity of USP46
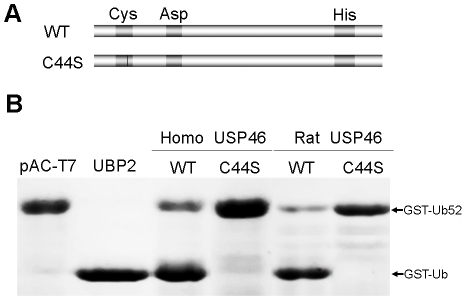
To investigate whether Usp46 has deubiquitinating enzyme activity, we performed USP cleavage assay using GST-Ub52 as a model substrate. After induced by IPTG, UBP2 cleaved GST-Ub52 efficiently to yield a product of the expected size of 36 KDa (Fig. 3 B; lane 2). Both human and rat USP46 resulted in the same cleavage product with UBP2, albeit at a reduced efficiency (Fig. 3 B; lane 3, 5). As expected, when Cys 44 was replaced by serine, the activity of USP46 disappeared (Fig. 3 B; lane 4, 6). Thus, USP46 has deubiquitinating enzyme activity.
Figure 3. Both human and rat USP46s have deubiquitinating activity toward the model substrate of GST-Ub52.
(A) Constructs of wild-type (WT) and mutant USP46 (C44S) for the USP cleavage assay. (B) Cleavage of model substrate GST-Ub52 by USP46. The GST-Ub52 was coexpressed with indicated proteins induced by IPTG in E coli BL21. The arrows indicate the GST-Ub52 (about 42 kDa) and its cleavage products GST-Ub (about 36 kDa), which were purified by glutathione-agarose beads and detected by Coomassie Brilliant Blue. UBP2 (a yeast deubiquitinase) and pAC-T7 (an empty plasmid vector) are used as positive and negative controls, respectively.
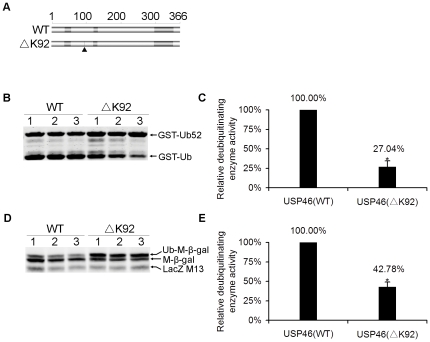
To determine whether Lysine 92 residue affects the deubiquitinating enzyme activity of USP46, we constructed the Lys 92 deletion mutant by site-directed mutagenesis. We employed the sensitive and quantitative Odyssey Infrared Imaging System for Western blot assay based on the intensity of fluorescence of protein band, which can reflect the deubiquitinating enzyme activity. We found that the deubiquitinating enzyme activity of USP46 (ΔK92) was decreased significantly compared with that of wild-type (Fig. 4), detected by USP cleavage assay using both GST-Ub52 and Ub-Met-β-gal as substrates (see Fig. 4 B, D). Results from three independent experiments showed that the relative deubiquitinating enzyme activity decreased to 27.04%±7.50% (mean±SEM) in USP46 (ΔK92) mutant compared with wild-type (P = 0.010) using GST-Ub52 as substrate (see Fig. 4 C), while 42.78%±6.47% (mean±SEM; P = 0.013) using Ub-Met-β-gal as substrate (see Fig. 4 E). The expression levels of USP46 were no significant differences between wild-type and ΔK92 mutant (see Fig. S1). This result suggests that the Lys 92 deletion influences USP46 deubiquitinating enzyme activity significantly.
Figure 4. Deubiquitinating enzyme activity of USP46 (ΔK92) declined significantly.
(A) Constructs of wild-type (WT) and mutant USP46 (ΔK92) for the USP cleavage assay. The deleted position was indicated by filled triangle. (B) A comparison of the deubiquitinating enzyme activity of wild-type and mutant USP46 (ΔK92) detected by USP cleavage assay using GST-Ub52 as a model substrate (See figure 3 for details). The arrows indicate the GST-Ub52 fusion protein and the cleaved GST-Ub products, respectively. The lane 1, 2, and 3 indicate three independent experiments of the assay. (C) Quantification of enzymatic activity in panel (B). The activity was expressed as the ratio of the intensity of GST-Ub and GST-Ub52 bands. The deubiquitinating enzyme activity of wild-type was defined as 100%. The relative deubiquitinating enzyme activity of ΔK92 mutant was measured compared to the activity of wild-type. Data was shown as mean±SEM of three independent experiments of lane 1, 2, and 3 derived from (B). Values that differ significantly from wild-type (Student's t test) are indicated as asterisk (P<0.05). (D) A comparison of the deubiquitinating enzyme activity of wild-type and mutant USP46 (ΔK92) detected by USP cleavage assay using Ub-Met-β-gal as a model substrate. The arrows indicate the ubiquitin-β-galactosidase fusion protein (Ub-Met-β-gal), the cleaved β-galactosidase moiety (Met-β-gal) and endogenous β-galactosidase (LacZ-M13) fragments. The lanes 1, 2, and 3 indicate three independent experiments of the assay. (E) Quantification of enzymatic activity in panel (D). The activity was expressed as the ratio of the fluorescence intensity of Met-β-gal and Ub-Met-β-gal bands. The deubiquitinating enzyme activity of wild-type was defined as 100%. The relative deubiquitinating enzyme activity of ΔK92 mutant was measured compared to that of wild-type. Data was shown as mean±SEM of three independent experiments of lane 1, 2, and 3 derived from (D). Values that differ significantly from wild-type (Student's t test) are indicated as asterisk (P<0.05).
Multi-tissues expression of Usp46
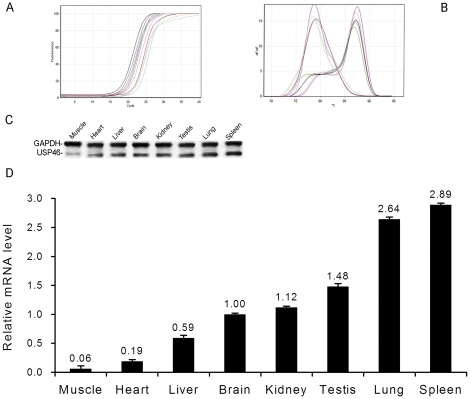
In order to investigate the expression level of Usp46 in a variety of tissues in rat, real-time RT-PCR analysis using 2−ΔΔCt method was carried out. Approximately, the mRNA of Usp46 mainly expressed for brain. So we use the expression level of brain as standard, and compared other tissues with brain. The result showed that Usp46 weakly expressed in skeletal muscle, heart, and liver, which exhibited 0.06, 0.19, and 0.59-fold lower level than brain. While Usp46 expression was observed to be moderate expressed in various organs including kidney and testis (1.12 and 1.48-fold), Usp46 expression was observed to be marginally higher in lung and spleen, which is 2.64 and 2.89-fold higher than brain (Fig. 5). Real-time RT-PCR analysis revealed that the expression of Usp46 gene is widely distributed in rat tissues, indicating that this enzyme may play a role in these tissues.
Figure 5. Multi-tissue expression of Usp46 mRNA.
Total RNA was prepared from various tissues and the relative expression levels were analyzed by real- time PCR using GAPDH as an internal control. (A) Fluorescence signals of Usp46 and GAPDH collected by real-time RT-PCR. (B) Melting curves of Usp46 (left) and GAPDH (right) to show the reproducibility of the two genes. (C) Agarose gel analysis of final products of the real-time RT-PCR. The bands of GAPDH and Usp46 were indicated. (D) Bar graph showing the relative levels of Usp46 mRNA expression in various tissues analyzed by relative quantitative analysis of 2−ΔΔCt method. Note that expression levels were normalized based on the values obtained from the GAPDH gene, comparing with the value of Usp46 in brain, shown are the means and SEM of the data.
Discussion
In the present study, we report that USP46 has deubiquitinating enzyme activity detected by USP cleavage assay using GST-Ub52 as a model substrate. We find that, compared to wild type, the Lys 92 deletion mutant results in a decreased enzyme activity of 27.04%. This work indicates that behavioral despair associated mutant of the Lys 92 deletion of USP46 could influence enzyme activity and therefore might contribute to the understanding of the molecular mechanisms of the mental disorders associated with USP46. In addition, we also determined the relative expression levels of Usp46 in rat tissues using real-time RT-PCR. Usp46 mRNA is expressed in various tissues examined including brain, with the highest expression in spleen.
Our data reveal that USP46 has deubiquitinating enzyme activity detected by USP cleavage assay using GST-Ub52 as a model substrate. Our findings are in good agreement with previous study [20] in the enzyme activity of USP46 detected by the cleavage assay using Ub-Met-β-gal as a substrate using western bolt method. The both methods co-expressed USP and model ubiquitin fusion protein substrate in a bacteria based system, though different model substrate (GST-Ub52 and Ub-Met-β-gal) were used, suggesting that USP46 has deubiquitinating enzyme activity by itself toward the model substrate in bacterial cells.
On the other hand, Cohn MA et al [21] reported that both USP46 and USP12 deubiquitinating enzymes have weak activity by themselves, and that their activities are stimulated by UAF1 strongly. They conclude that the enzyme activity of both USP46 and USP12 are regulated by UAF1. In the study, they assessed the deubiquitinating activity by an in vitro deubiquitinating enzyme assay using ubiquitin-AMC [22] as a substrate. The differences in the detection systems might account for the difference in enzyme activity of USP46 between those studies. Cohn et al hired an in vitro system using ubiquitin-AMC as a model substrate. The system is relative new, and needs purified enzyme from Sf9 insect cells, since the most USPs is insoluble when expressed in E. coli. This is also the case in USP46 (Fig. 2). In contrast, both the current study and Quesada et al [20] used a bacterial expression system using either Ub-Met-β-gal or GST-Ub52 as model substrates. In those systems, the enzyme substrate interaction happens in bacterial cells and no protein purification is needed. In addition, like rat USP46, both human and mouse USP46 are active toward to the model substrate in our detection system (Fig. 3), indicating the USP cleavage assay is a stable method to detect deubiquitinating enzyme activity of USP46.
Usp46 with a Lys 92 deletion is closely associated with loss of ‘behavioral despair’ under inescapable stresses in addition to abnormalities in circadian behavioral rhythms and the GABAergic system [17]. However, the effects of Lys 92 deletion on the deubiquitinating enzyme activity of USP46 were not previously defined. Our results indicate that after deletion of Lys 92, the deubiquitinating enzyme activity of USP46 decreased to 27.04% compared with wild-type by GST-Ub52 assay, while 42.78% by Ub-Met-β-gal assay (Fig. 4). The differences in the detection systems might account for the difference in enzyme activity of USP46 between GST-Ub52 assay and Ub-Met-β-gal assay, but these two assays both revealed that after deletion of Lys 92, the deubiquitinating enzyme activity of USP46 declined significantly, supporting the notion of USP46 as a candidate gene regulating behavioral despairs.
More recently, Itaru Kushima, Branko Aleksic et al. explored an association of USP46 with bipolar disorder and schizophrenia in a Japanese population [23]. They found nominal evidence for an association of rs12646800 and schizophrenia. This association was not significant after correction for multiple testing. No significant association was detected for bipolar disorder. In conclusion, their data argue against the presence of any strong genetic susceptibility factors for bipolar disorder or schizophrenia in the region USP46. However, our finding, that the Lys 92 deletion of USP46 influences enzyme activity, provides a molecular clue in the interpretation how the enzyme regulates the pathogenesis of mental illnesses. In any case, further investigation is clearly needed to determine how the USP46 mutation affects the GABAergic system and involves in mental illnesses.
In conclusion, our data indicate that USP46 in solitary conditions has deubiquitinating enzyme activity detected by USP cleavage assay using GST-Ub52 as a model substrate, which is a simple and stable method to testing the enzymatic activity of USP46. The Lys 92 deletion of USP46 could influence enzyme activity and might contribute to the understanding of the neural and genetic mechanisms that underlie the mental disorders associated with this gene, thereby provide a molecular clue how the enzyme regulating the pathogenesis of mental illnesses.
Supporting Information
The expression levels were no significant differences between wild-type and ΔK92 mutant USP46. E. coli strain BL21 (DE3) cells harboring pGEX-Ub52 were co-transformed with either pAC-T7-Usp46 (wild-type) or pAC-T7-Usp46 (ΔK92). Protein expression was induced by IPTG and the cells were further incubated for 3 hours. Total protein extracts were subjected to SDS-PAGE and analyzed by Western blot using anti-T7 rabbit polyclonal antibody. The T7-USP46 was indicated.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by the National Natural Science Foundation of China (30840035 and 30970915). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.D'Andrea A, Pellman D. Deubiquitinating enzymes: a new class of biological regulators. Crit Rev Biochem Mol Biol. 1998;33:337–352. doi: 10.1080/10409239891204251. [DOI] [PubMed] [Google Scholar]
- 2.Reyes-Turcu FE, Wilkinson KD. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem Rev. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takayama Y, Toda T. Coupling histone homeostasis to centromere integrity via the ubiquitin-proteasome system. Cell Div. 2010;5:18. doi: 10.1186/1747-1028-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilroy GE, Zhang X, Floyd ZE. PPAR-gamma AF-2 domain functions as a component of a ubiquitin-dependent degradation signal. Obesity (Silver Spring) 2009;17:665–673. doi: 10.1038/oby.2008.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi T, Kimura J, Miki Y, Yoshida K. The deubiquitinating enzyme USP11 controls an IkappaB kinase alpha (IKKalpha)-p53 signaling pathway in response to tumor necrosis factor alpha (TNFalpha). J Biol Chem. 2007;282:33943–33948. doi: 10.1074/jbc.M706282200. [DOI] [PubMed] [Google Scholar]
- 6.Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, et al. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–731. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- 7.Bernardi R, Liebermann DA, Hoffman B. Cdc25A stability is controlled by the ubiquitin-proteasome pathway during cell cycle progression and terminal differentiation. Oncogene. 2000;19:2447–2454. doi: 10.1038/sj.onc.1203564. [DOI] [PubMed] [Google Scholar]
- 8.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Singhal S, Taylor MC, Baker RT. Deubiquitylating enzymes and disease. BMC Biochem. 2008;9(Suppl 1):S3. doi: 10.1186/1471-2091-9-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onno M, Nakamura T, Mariage-Samson R, Hillova J, Hill M. Human TRE17 oncogene is generated from a family of homologous polymorphic sequences by single-base changes. DNA Cell Biol. 1993;12:107–118. doi: 10.1089/dna.1993.12.107. [DOI] [PubMed] [Google Scholar]
- 12.Papa FR, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Schrodi S, Rowland C, Tacey K, Catanese J, et al. Genetic evidence for ubiquitin-specific proteases USP24 and USP40 as candidate genes for late-onset Parkinson disease. Hum Mutat. 2006;27:1017–1023. doi: 10.1002/humu.20382. [DOI] [PubMed] [Google Scholar]
- 14.Shanmugham A, Ovaa H. DUBs and disease: activity assays for inhibitor development. Curr Opin Drug Discov Devel. 2008;11:688–696. [PubMed] [Google Scholar]
- 15.Wu YR, Chen CM, Chen YC, Chao CY, Ro LS, et al. Ubiquitin specific proteases USP24 and USP40 and ubiquitin thiolesterase UCHL1 polymorphisms have synergic effect on the risk of Parkinson's disease among Taiwanese. Clin Chim Acta. 2010;411:955–958. doi: 10.1016/j.cca.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Cryan JF, Mombereau C. In search of a depressed mouse: utility of models for studying depression-related behavior in genetically modified mice. Mol Psychiatry. 2004;9:326–357. doi: 10.1038/sj.mp.4001457. [DOI] [PubMed] [Google Scholar]
- 17.Tomida S, Mamiya T, Sakamaki H, Miura M, Aosaki T, et al. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat Genet. 2009;41:688–695. doi: 10.1038/ng.344. [DOI] [PubMed] [Google Scholar]
- 18.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, et al. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian QB, Okano A, Nakayama K, Miyazawa S, Endo S, et al. A novel ubiquitin-specific protease, synUSP, is localized at the post-synaptic density and post-synaptic lipid raft. J Neurochem. 2003;87:665–675. doi: 10.1046/j.1471-4159.2003.02024.x. [DOI] [PubMed] [Google Scholar]
- 20.Quesada V, Diaz-Perales A, Gutierrez-Fernandez A, Garabaya C, Cal S, et al. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem Biophys Res Commun. 2004;314:54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 21.Cohn MA, Kee Y, Haas W, Gygi SP, D'Andrea AD. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem. 2009;284:5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang LC, Melandri FD, Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 23.Kushima I, Aleksic B, Ito Y, Nakamura Y, Nakamura K, et al. Association study of ubiquitin-specific peptidase 46 (USP46) with bipolar disorder and schizophrenia in a Japanese population. J Hum Genet. 2010;55:133–136. doi: 10.1038/jhg.2009.139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression levels were no significant differences between wild-type and ΔK92 mutant USP46. E. coli strain BL21 (DE3) cells harboring pGEX-Ub52 were co-transformed with either pAC-T7-Usp46 (wild-type) or pAC-T7-Usp46 (ΔK92). Protein expression was induced by IPTG and the cells were further incubated for 3 hours. Total protein extracts were subjected to SDS-PAGE and analyzed by Western blot using anti-T7 rabbit polyclonal antibody. The T7-USP46 was indicated.
(DOC)