Abstract
We investigated the genomic diversity of a local population of the symbiotic bacterium Sinorhizobium medicae, isolated from the roots of wild Medicago lupulina plants, in order to assess genomic diversity, to identify genomic regions influenced by duplication, deletion or strong selection, and to explore the composition of the pan-genome. Partial genome sequences of 12 isolates were obtained by Roche 454 shotgun sequencing (average 5.3 Mb per isolate) and compared with the published sequence of S. medicae WSM 419. Homologous recombination appears to have less impact on the polymorphism patterns of the chromosome than on the chromid pSMED01 and megaplasmid pSMED02. Moreover, pSMED02 is a hot spot of insertions and deletions. The whole chromosome is characterized by low sequence polymorphism, consistent with the high density of housekeeping genes. Similarly, the level of polymorphism of symbiosis genes (low) and of genes involved in polysaccharide synthesis (high) may reflect different selection. Finally, some isolates carry genes that may confer adaptations that S. medicae WSM 419 lacks, including homologues of genes encoding rhizobitoxine synthesis, iron uptake, response to autoinducer-2, and synthesis of distinct polysaccharides. The presence or absence of these genes was confirmed by PCR in each of these 12 isolates and a further 27 isolates from the same population. All isolates had rhizobitoxine genes, while the other genes were co-distributed, suggesting that they may be on the same mobile element. These results are discussed in relation to the ecology of Medicago symbionts and in the perspective of population genomics studies.
Keywords: population genomics, genome evolution, horizontal gene pool, population structure, Sinorhizobium medicae
Introduction
There have been many studies of genetic diversity within a bacterial species, and the majority have adopted one of three approaches. Innumerable ‘classical' studies have characterized isolates by their genotypes at a small number of marker loci. Recent advances in sequencing technology have facilitated two additional approaches: sequencing the entire genomes of multiple isolates or sequencing randomly from an entire bacterial community (metagenomics). While there are now many species for which several isolates have been sequenced, these isolates have generally been chosen from the global diversity of the species, and do not represent a sample of a defined local population. Comparison of such genomes has revealed extensive polymorphism affecting both genome content and gene sequences (Tettelin et al., 2005; Lefebure and Stanhope, 2007). Based on such observations, the pan-genome of a species has been defined as the total gene repertoire of the species, including both core genes, which are present in the genome of (almost) all individuals within the species, and accessory genes, which have a sparser distribution. Theoretical studies suggest that the frequency of a gene in a bacterial population would depend on parameters related to its inheritance mechanism and its fitness effect on the recipient genome (Berg and Kurland, 2002; Novozhilov et al., 2005), as would allele frequency at a given locus (Mes, 2008). Population genomics approaches using full genomes are giving insights into the parameters that shape the diversity of bacterial species (Didelot et al., 2007), but estimates of diversity and recombination are not easily interpreted when the isolates are sparsely sampled from a global distribution. By contrast, metagenomic studies do provide samples of a local population, and gene-based estimates of diversity, but the partitioning of these sequences into discrete genomes is not known. This hampers studies on recombination and selection, although some interesting analysis is still possible (Allen et al., 2007; Eppley et al., 2007). These considerations raise questions about the sampling strategies employed to study the genomic diversity of bacteria (Rocha, 2008). Furthermore, it has been proposed that gene and allele frequencies obtained from genomic data might be used in top-down approaches (similar to association mapping) to identify, without a priori knowledge, genes that could be involved in bacterial adaptations (Falush and Bowden, 2006).
In this context, we have acquired genomic data for rhizobia, which are ecologically important bacteria that have been extensively studied by both functional and evolutionary approaches, to assess the results that can be obtained from a population genomic survey. Rhizobia constitute a functional group that includes both Alphaproteobacteria and Betaproteobacteria able to form a nitrogen-fixing endosymbiosis with a legume host plant. To establish this interaction, bacteria trigger the organogenesis of nodules, which are usually located on the roots of the host. The association between plants belonging to the genus Medicago, such as alfalfa (Medicago sativa) or barrel medic (Medicago truncatula), and their specific symbionts, Sinorhizobium meliloti and Sinorhizobium medicae, is one of the best-studied systems. Bacterial genetic and post-genomic approaches have demonstrated the critical role played by genes involved in the synthesis of nodulation factors (that is, nod genes) and surface polysaccharides (for example, exo or rkp genes) in providing signals for the recognition of S. meliloti and S. medicae by their host (Jones et al., 2007). While most functional studies have used S. meliloti as a model bacterium because the genome of the strain 1021 was available (Galibert et al., 2001), more studies in future might be performed on S. medicae now that the genome sequence of the strain WSM 419 has been released (Reeve et al., 2010), because this bacterium, isolated in Sardinia, is a better symbiont than S. meliloti 1021 for the model legume M. truncatula (Terpolilli et al., 2008).
S. meliloti and S. medicae are closely related species that form a tight phylogenetic clade together with Sinorhizobium arboris (Martens et al., 2007). The genomes of S. meliloti 1021 and S. medicae WSM 419 share similar architectures. Both include three main replication units: (i) a chromosome which harbours most of the housekeeping genes; (ii) a chromid (Harrison et al., 2010), where many genes involved in polysaccharide synthesis are clustered (pSMED01 for S. medicae WSM 419 and pSymB for S. meliloti 1021); and (iii) a megaplasmid, where nod genes and genes involved in nitrogen fixation are located (pSMED02 for WSM 419 and pSymA for 1021). The genome of S. medicae WSM 419 also includes a 219-kb plasmid called pSMED03. Although S. meliloti 1021 does not have any comparable plasmid, additional plasmids have also been observed in some other strains of this species (Mercado-Blanco and Olivares, 1993, 1994; Stiens et al., 2006, 2007; Kuhn et al., 2008). Based on sequencing of several loci, S. medicae is less diverse than S. meliloti, especially at chromosomal loci (Bailly et al., 2006; van Berkum et al., 2006). Indications of directional and balancing selection have been identified for the nod gene region on pSMED02/pSymA and the region involved in polysaccharide synthesis on pSMED01/pSymB, respectively, in sympatric populations of S. meliloti and S. medicae (Bailly et al., 2006). Such diversity patterns suggest that recombination has an important role in shaping the diversity of both species. Within each species, linkage disequilibrium analyses suggest that homologous recombination occurs preferentially at loci located on the chromid and megaplasmid, rather than the chromosome (Bailly et al., 2006).
The diversity of the accessory genome of S. meliloti has been studied using various approaches. Novel genomic sequences have been described both from the cryptic plasmids of the strain SM11 (Stiens et al., 2006, 2007) and also from a representational difference analysis based on the strain ATCC 9930 (Guo et al., 2005). These new sequences occur with a wide range of frequencies in natural populations of S. meliloti (Guo et al., 2005; Kuhn et al., 2008). Some of this genetic material might have an important role in the adaptation of Medicago symbionts. Finally, the analysis of the genome content of four natural isolates of S. meliloti using comparative genomic hybridization revealed that at least 12% of the genes of the model strain S. meliloti 1021 have been recently duplicated, gained or lost during the diversification of the species (Giuntini et al., 2005). Altogether, these data suggest that high rates of gain and loss of genetic material influence the evolution of Medicago symbionts.
In this study, we investigate the genomic diversity of a local population of S. medicae in symbiosis with M. lupulina, the only native Medicago species that is widespread in the United Kingdom. We compare partial genome sequences of 12 isolates with the reference strain WSM 419, and examine the distribution of accessory genes in a larger sample, in order to (i) gain insights into the evolutionary dynamics of the different replication units, (ii) look for evidence of past selection, and (iii) explore the properties of the pan-genome of S. medicae. We compare our results with current hypotheses on the ecology and the evolution of Medicago symbionts and discuss them in the perspective of population genomic approaches.
Materials and methods
Bacterial collection DNA preparation and sequencing
The nodules of six M. lupulina plants, growing on a one metre squared area of roadside vegetation (grasses and herbs) located between Wentworth College and Walmgate Stray at the University of York, UK (53° 56′ 44″ N, 1° 03′ 35″ W), were harvested on 22 March 2008. Each nodule was sterilized in a 5% (w/v) calcium hypochlorite solution for 5 min and subsequently rinsed three times in sterile water. Independent nodules were crushed in 100 μl sterile water using autoclaved plastic grinders and plated on TY agar medium. Single colonies were plated three consecutive times in order to ensure that a single bacterial isolate was obtained from each nodule.
Each bacterial strain was grown in 15 ml of liquid TY (Beringer, 1974) to perform DNA extractions. The liquid culture was centrifuged for 10 min at 4000 r.p.m. After resuspending the culture, DNA was extracted with the FastDNA Kit (Qbiogene, Carlsbad, CA, USA) according to the manufacturer's instructions. The quality and quantity of DNA were checked by spectrophotometer at 260 and 280 nm (Nanodrop, Thermo Scientific, Wilmington, DE, USA). Partial sequences of the 16S rRNA genes were obtained for all strains as described by Weisburg et al. (1991).
From a total of 39 isolates, all of which were putatively S. medicae because their 16S rRNA sequences were identical to that of the type strain, we selected a random sample of 12 strains to be sequenced using a GS FLX genome sequencer (Roche 454 Life Sciences, Branford, CT, USA). DNA samples from the different strains were tagged using the standard multiplex identifiers (MID, Roche) to obtain a single library including the DNA of the different strains. This library was sequenced using the LR70 kit (Roche) on half a plate of the GS FLX genome sequencer. Data are available from the Sequence Read Archive of the International Nucleotide Sequence Database Collaboration as study accession ERP000630.
Presence/absence of genome regions shared with WSM 419
After filtering identifiers from the GS FLX sequence data set, sequence reads were mapped against the genome of S. medicae WSM 419 (accession numbers NC_009636, NC_009620 to NC_009622) using the gsMapper software (Roche) with default parameters except that identity thresholds were fixed at 70 base pairs (bp) and 80%. For each strain, we identified major duplications and indels (insertions in WSM 419 or deletions in our strains since the most recent common ancestor). We computed the number of read starts (that is, 5′ ends) matching within 10 kilobase (kb) windows located every 10 kb along the genome of WSM 419. This window size was chosen to provide, for each strain, enough read starts in each window to be able to perform a statistical test to detect major indels and duplication events with high confidence. Inevitably, the large windows and low coverage mean that small deletions, insertions or duplications will be underestimated. Read numbers were weighted by 1/X, X being the number of locations where a repeated sequence was mapped. The sum of weighted read numbers was computed for the different windows and rounded. As most of the genes on the chromosome of S. medicae WSM 419 (and of S. meliloti 1021) are single copy, a Poisson distribution for each strain was fitted, using a maximum likelihood approach (Wessa, 2008), to the computed density of read starts in windows of the WSM 419 chromosome. Assuming this distribution, windows presenting a significant lack or increase of the number of read starts (that is, windows affected by major duplication or indel events) were identified on each of the WSM 419 replication units using a 0.1% threshold modified to take into account multiple tests. 95% confidence intervals of the fraction of windows for which at least one indel or duplication event would be observed among the 12 strains on a given replication unit were obtained using a Monte–Carlo re-sampling procedure based on the observed frequency of duplication and deletion events.
Genetic divergence for genome regions shared with WSM 419
Based on mapping data, a strict consensus sequence was obtained for the genome of each of the 12 strains to study sequence divergence. At this step, 10 kb windows presenting major indel or duplication events, as defined in the preceding section, were removed from the analysis. For each of the remaining windows, the pairwise divergence between wild strains, or between each wild strain and the reference strain WSM 419, was obtained by dividing the number of differences by the sequence length which was aligned. The differentiation between the reference strain WSM 419 and the population we sampled was measured using the KST statistic (Hudson et al., 1992). Mann–Whitney U-tests were used to compare the distribution of both the average divergence among the sampled strains and the average KST value among windows located on different replication units. Two different approaches were used to observe whether the phylogenetic information contained in pairwise divergence matrices was congruent among 10 kb windows and among replicons. First, a principal component analysis (PCA) was performed on pairwise divergence matrices. To this end, each distance matrix was normalized by its highest divergence to avoid overweighting of either rapidly evolving or recombination-prone windows. For the PCA, each window was treated as an independent observation of the variables, which were the set of normalized pairwise divergences. For each replicon, we investigated the heterogeneity of distance matrices between windows by computing the Euclidean distances between the projections of windows. The variances of these distances were compared among replication units using Levene's test, which does not assume distributions are normal. In addition, a Neighbour-Net network (Bryant and Moulton, 2004) was obtained, based on the average of the raw window-based pairwise divergence matrices, to illustrate the genetic relationships between the sampled strains and the reference strain WSM 419 for each replication unit.
Population-specific genes
To find accessory genes that were specific to the population, we assembled the reads that were not mapped, or only partially mapped, to the WSM 419 reference genome. Contigs larger than 500 bp were obtained using the gsAssembler tool (Roche) based on default parameters, except that the identity threshold was fixed at 70 bp and 80%. Contigs were annotated based on BlastX searches of the NCBI nr protein database. We selected eight contigs that included homologues of potentially adaptive genes annotated in other species of rhizobia (Table 1) and designed 20 pairs of PCR primers (Table 2) to amplify coding and intergenic regions. We used PCR to screen all the 39 strains that we initially sampled. All reactions were in 35 μl Green GoTaq Flexi buffer (Promega, Madison, WI, USA) with 0.4 μ each primer (Eurofins, Ebersberg, Germany), 250 μ each dNTP, 1.5 m Mg2+, 0.875 units GoTaq polymerase (Promega), 25 ng DNA template; initial denaturation for 2 min at 95 °C, 35 cycles of 1 min at 94 °C, annealing for 1 min at 55 °C and extension at 72 °C for 1 min, with a final 5 min extension at 72 °C. Results were assessed by agarose gel electrophoresis of PCR products. Strains that yielded PCR amplicons of the expected length were considered positive for that primer pair.
Table 1. Genomic regions screened by PCR in isolates of Sinorhizobium medicae, with information on genes in the best Blast hit in the NCBI nr nucleotide database.
| Region | Best hit genome | Gene ID | Gene | Function |
|---|---|---|---|---|
| RTXa | Bradyrhizobium japonicum USDA110 | blr2077 | rtxA | Dihydroxyacetonephosphate aminotransferase |
| RTXb | Burkholderia phymatum STM 815 | Bphy_7779 | rtxB | Dihydroxyrhizobitoxin synthase |
| RTXc | B. phymatum STM 815 | Bphy_7780 | rtxC | Dihydroxyrhizobitoxin desaturase |
| REPa | Sinorhizobium meliloti GR4 (pRmeGR4a) | ABA55661 | repC | Plasmid replication initiation |
| REPb | S. meliloti SM11 (pSmeSM11a) | ABA56127 | exoI | Exonuclease |
| SIDa | S. fredii NGR234 (pNGR234b) | NGR_b03560-70 | Putative siderophore biosynthesis and transport proteins | |
| SIDb | S. fredii NGR234 (pNGR234b) | NGR_b03580 | Putative iron ABC transporter solute-binding protein | |
| SIDc | S. fredii NGR234 (pNGR234b) | NGR_b03590 | Putative TonB-dependent ligand-gated channel | |
| AITa | S. meliloti 1021 (pSymB) | SM_b21022 | aitK | Autoinducer-2 (AI-2) kinase (C-terminal part) |
| AITb | S. meliloti 1021 (pSymB) | SM_b21022-3 | Intergenic | |
| AITc | S. meliloti 1021 (pSymB) | SM_b21023 | aitG | Autoinducer processing (lsrG homologue) |
| AITd | S. meliloti 1021 (pSymB) | SM_b21022 | aitK | Autoinducer-2 (AI-2) kinase (N-terminal part) |
| RKPa | S. meliloti 1021 (pSymB) | SM_b20823 | rkpZ2 | Lipopolysaccharide processing protein |
| RKPb | S. meliloti 1021 (pSymB) | SM_b20823-4 | Intergenic | |
| RKPc | S. meliloti 1021 (pSymB) | SM_b20824 | Putative glycosyltransferase for capsule biosynthesis | |
| RKPd | S. meliloti 1021 (pSymB) | SM_b20824-5 | Intergenic | |
| RKPe | S. meliloti 1021 (pSymB) | SM_b20825 | Putative acetyltransferase | |
| RKPf | S. meliloti 1021 (pSymB) | SM_b20828, 21663 | Unknown functions, putatively related to secretion | |
| RKPg | S. meliloti 1021 (pSymB) | SM_b20828-9, 21663-5 | Unknown functions, putatively related to secretion | |
| RKPh | S. meliloti 1021 (pSymB) | SM_b20829 | Putative secreted calcium-binding protein |
Table 2. PCR primers used to screen isolates of Sinorhizobium medicae.
| F primer sequence | R primer sequence | Contig | F location | R location | Length of product | |
|---|---|---|---|---|---|---|
| RTXa | AGTCTGTCCTCTACCGTAAG | GTATAAGTGTAGCCGTCCTC | 30 | 3572–3591 | 4850–4869 | 1297 |
| RTXb | CTGCCGCAAGCCAATTCAGG | CAATGCCGCGAGCGATCTCT | 30 | 4647–4666 | 6109–6128 | 1481 |
| RTXc | GTCCGCCATACATTGATCTC | TCTGCTGTTCGGCTTCATAC | 30 | 5995–6014 | 7055–7074 | 1079 |
| REPa | CACACGAACGGCGAAGAATA | ATTCGCTGATTCCGGATGCT | 165 | 855–874 | 2483–2502 | 1647 |
| REPb | CATGGAGGTCAGACATTGAG | CCACTACGCCATATTGGCTA | 165 | 2357–2376 | 2891–2910 | 553 |
| SIDa | GTGCGCGATGTCAACAACAG | CAGCGGATTCGGCAAGTGAT | 211 | 716–735 | 2196–2215 | 1499 |
| SIDb | TCGGCGTTCCAAGCCGTGAT | TACCGCCGAACGGAGATAGC | 211 | 3465–3484 | 4667–4686 | 1221 |
| SIDc | GACGCCGGACCAGTAGTCAA | GCGTGGCCTTAACCGTTAGC | 211 | 5157–5176 | 5613–5632 | 475 |
| AITa | TCTATGAGCGCATCCACAGG | CGCAAGGATCTGTGACCATT | 228 | 8–27 | 824–843 | 724 |
| AITb | GCAATCGGTGTCGTGAATGA | ATCGGCCATCTCGACGTTAT | 228 | 304–323 | 1427–1446 | 1142 |
| AITc | AGGAGAACGCGGCGATTGTC | GCCGTCTTCCAAGCCTGCTA | 228 | 716–735 | 1468–1487 | 771 |
| AITd | GCTCGATGGACTTCGATGTG | CTCGGTGACGCTGCCGATAA | 62, 228 | 182–201 | 206–225 | 542 |
| RKPa | GCGTTGATTCGGCTGCCGAT | AGACCTTCATACGCGACTGG | 59 | 94–113 | 914–933 | 839 |
| RKPb | GTATTGCCACGCGAGGACAC | GTTCAGTCGCCGCAGCTATG | 59 | 797–816 | 1794–1813 | 1016 |
| RKPc | TCTATCGCCTCGATCAGTCT | TGCGTGTCTTCCATCTCTAC | 59 | 1559–1578 | 3004–3023 | 1464 |
| RKPd | ATCGCTCGTGGTAGAGATGG | CGCAAGAACCGTCCTAACAG | 59, 179 | 2994–3013 | 147–166 | 989 |
| RKPe | GGAAGTAGTTGCGGCGCTTG | TCCTTCTTGGCGTGTATCGG | 179 | 289–308 | 994–1013 | 724 |
| RKPf | GCGGCATTAAGGCGGTGTCT | TCCGGTTAAGCGCATGGTCC | 47 | 593–612 | 948–967 | 374 |
| RKPg | TTGACTTCGGCGTGAACTCC | TACGACAGCGATGGCAATGG | 47 | 725–744 | 2191–2210 | 1485 |
| RKPh | AATCGGCTGCGGTGATGCTA | GCGGCATTCGGCAATCTTGA | 47 | 2129–2148 | 3368–3387 | 1258 |
Results
We isolated 39 bacterial strains from root nodules of M. lupulina. All these strains putatively belong to the species S. medicae according to the 100% identity observed between 1400 bp of their 16S rDNA sequences and the 16S rDNA of the type strain A321 (GenBank accession L39882, 1423 bp). There are three nucleotide differences between the 16S rDNA sequences of the type strains of S. medicae and its closest relative S. meliloti (Rome et al., 1996); our isolates all had the S. medicae variants. Twelve of these strains were randomly chosen for partial genome sequencing. We obtained from 13 351 (strain MLX_12) to 36 676 (strain MLX_11) sequence reads per strain (Table 3). The average length of these sequences was 223 bp. If we assume that the total genome size of each isolate is similar to that of S. medicae WSM 419 (6.8 Mb), the sequence coverage we obtained for each genome would vary from 0.44- to 1.20-fold.
Table 3. Shotgun sequencing of 12 isolates of Sinorhizobium medicae using the Roche 454 FLX platform.
| Isolate | Total sequence (bp) | No. of reads | Mapped readsa |
|---|---|---|---|
| MLX_01 | 7 110 888 | 31 539 | 27 239 |
| MLX_02 | 4 558 396 | 20 323 | 18 226 |
| MLX_03 | 6 028 954 | 27 017 | 23 094 |
| MLX_04 | 6 566 082 | 29 540 | 26 529 |
| MLX_05 | 3 224 492 | 14 471 | 12 109 |
| MLX_06 | 3 028 454 | 13 671 | 11 903 |
| MLX_07 | 4 597 694 | 20 496 | 16 511 |
| MLX_08 | 5 534 095 | 24 714 | 21 059 |
| MLX_09 | 7 221 463 | 32 500 | 27 587 |
| MLX_10 | 4 812 898 | 21 684 | 17 455 |
| MLX_11 | 8 200 909 | 36 676 | 31 419 |
| MLX_12 | 2 970 560 | 13 351 | 11 365 |
| Total | 63 854 885 | 2 85 982 | 2 44 496 |
Number of reads mapping to the genome of S. medicae WSM 419.
Overall, 85.5% of the reads could be mapped to the genome sequence of S. medicae WSM 419. The relative number of reads mapping to the chromosome, to pSMED01, and to pSMED02 was similar to the relative sizes of these replicons (55.5%, 23.0% and 18.3% of the WSM 419 genome, respectively). This implies that these three replicons (or the equivalents in our isolates) have a similar copy number per cell. The 213-kb pSMED03 makes up just 3.2% of the WSM 419 genome but attracted between 5% and 10% of the mapped reads from each strain, suggesting a 2- or 3-fold higher copy number.
Presence/absence of genome regions shared with WSM 419
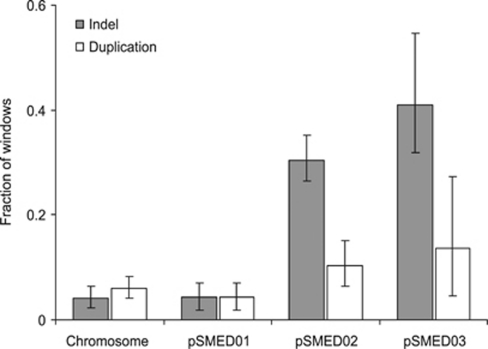
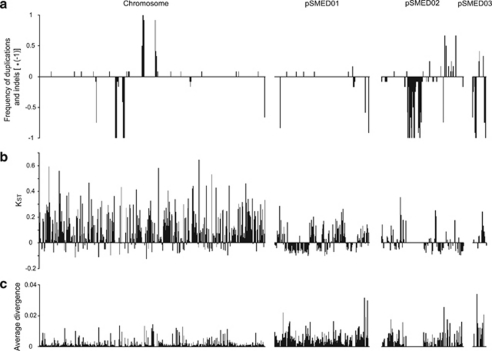
Reads from each strain were mapped to 10 kb windows along the genome of WSM 419. Indel and duplication events were identified by statistically significant deficiencies or excesses of reads. The proportion of windows for which an indel or duplication event was detected for at least one strain was computed for the different replication units of WSM 419, together with the associated 95% confidence intervals (Figure 1). Among the four replication units of the WSM 419 genome, there is no significant difference in the fraction of windows affected by at least one duplication event, but the fraction of windows for which at least one indel event was inferred is significantly lower for the chromosome and the chromid pSMED01 than for the plasmids pSMED02 and pSMED03. The frequency of indels and duplications among the 12 sampled strains is presented in Figure 2. While events seem to be clustered, especially on pSMED02, differences in the frequency of indels in the population indicate that several independent events are causing the diversity pattern (Figure 2).
Figure 1.
Fraction of 10 kb windows for which a deletion or duplication event has been inferred in at least one of the 12 sampled strains. Computations are based on the mapping of sequence reads against each replication unit of S. medicae WSM 419. 95% confidence intervals are based on a bootstrap procedure.
Figure 2.
Distribution of various diversity statistics along the genome in the observed population of S. medicae. (a) Positive values show the frequencies of strains harbouring duplication for a given window when compared with S. medicae WSM 419, while negative values illustrate the frequencies of strains harbouring a deletion. (b) KST values that illustrate the differentiation between the population we sampled and the reference strain S. medicae WSM 419 for the windows showing neither duplication nor deletion events. (c) The average divergence among the strains we sampled is illustrated by bars for the windows showing neither duplication nor deletion events. Statistics are computed for 10 kb windows on the chromosome, pSMED01, pSMED02 and pSMED03. Graphs are drawn to scale.
Genetic divergence for genome regions shared with WSM 419
Pairwise divergence among the sampled strains for a given 10 kb window was inferred from pairwise alignments that covered, on average, 1488 bp with a s.d. of 641 bp among windows for all the windows and 1526 bp with a s.d. of 496 bp among the windows which did not show evidence of major indel. The average divergence among strains is low (Table 4); sequences mapping to the chromosome showed least divergence, and those mapping to pSMED03 showed the most. The relative divergence of the reference strain WSM 419 from the population we sampled was greatest for the chromosome and least for pSMED1 and pSMED2, as estimated by the KST statistic (Table 4). The low genetic divergence on the chromosome between S. medicae WSM 419 and the population we sampled (0.0029 substitutions per bp), compared with the genetic divergence of around 0.1 substitutions per bp described between S. medicae and S. meliloti isolates based on multilocus sequence typing data (Bailly et al., 2006; van Berkum et al., 2006; Martens et al., 2007), confirms our initial assumption that our isolates were all S. medicae.
Table 4. Statistics based on reads that mapped to the four replication units of the S. medicae WSM 419 genome.
| WSM 419 replication unit | Average divergence among strains (mutations/site) | Differentiation between WSM 419 and the population (KST) | Phylogenetic diversity (mean distance between windows in PCA) |
|---|---|---|---|
| Chromosome | 0.00232a | 0.122d | 2.89g |
| pSMED01 | 0.00626b | 0.017e | 6.14h |
| pSMED02 | 0.00610b | 0.008e | 3.91i |
| pSMED03 | 0.01430c | 0.070d,f | 3.55i |
Abbreviation: PCA, principal component analysis.
Values that do not share same letter are significantly different (P<0.001 for a–e and g–h, P<0.05 for f and i).
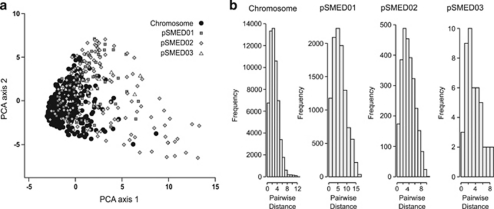
We investigated the degree to which the phylogenetic signal was consistent across the genome by means of a PCA. The PCA based on divergence matrices obtained from 10 kb windows reveals that matrices obtained from the chromosome are more homogeneous than matrices obtained from the megaplasmid pSMED02 and the plasmid pSMED03, while the chromid pSMED01 gave the most heterogeneous results (Figure 3a). Consistently, Levene's test revealed that the distribution of Euclidean distances between the projections of pairs of windows was different among replication units (P<0.05), with the exception of the comparison between pSMED02 and pSMED03 (P>0.75) (Figure 3b; Table 4).
Figure 3.
(a) PCA illustrating the distribution of genetic divergence between S. medicae isolates among 10 kb windows distributed along the genome of S. medicae WSM 419. (b) Distribution of Euclidean distances between projections of 10 kb windows and the centroid of the cloud observed for each replication unit.
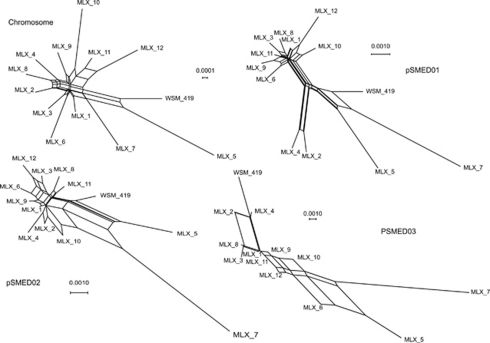
This suggests that the replication units support different phylogenetic relationships. In order to illustrate these differences, Neighbour-Net phylogenetic networks were obtained for the chromosome, pSMED01, pSMED02 and pSMED03 (Figure 4). As expected, these differ substantially. The network obtained from the chromosome suggests that most of our strains belong to the same broad cluster, the reference strain WSM 419 and MLX 5 forming a loose outgroup. The network obtained from pSMED02 still suggests that most of our strains belong to the same cluster, but the strains MLX 5 and MLX 7 are clustered together and are more divergent from the other MLX strains than the reference WSM 419. The network obtained from pSMED01 is similar to the network obtained from pSMED02 with the exception of MLX 2 and MLX 4 which form a tight cluster, far apart from either the group containing MLX 5, MLX 7 and the reference WSM 419 or the group including the other MLX strains. Finally, the network obtained from pSMED03 is based on less information and is poorly resolved, but groups together the strains WSM 419, MLX 2 and MLX 4 on one side and on the other MLX 6, MLX 5 and MLX 7, the other strains branching between the two groups.
Figure 4.
Neighbour-Net network obtained from the average matrix of DNA sequence divergence among S. medicae strains for each of the replication units.
Population-specific genes
A total of 41 485 reads were not mapped, or only partially mapped, against the genome of S. medicae WSM 419. They were assembled into contigs, of which 257 were at least 1 kb in length. We selected eight contigs that spanned five potentially adaptive gene clusters with homologues in some other rhizobia (Table 1) and used 20 sets of PCR primers (Table 2) to assess their distribution across strains (Table 5). For all sequenced strains, there was complete concordance between the presence of a PCR product of the expected size and of reads that mapped to the corresponding contig, with the sole exception of MLX 4 and MLX 7, which had some reads in the REP region but no PCR products. The reads did not cover the PCR primer sites: we presume that these sites were absent or diverged in these two isolates.
Table 5. Distribution among the S. medicae isolates of five gene regions that are absent from S. medicae WSM 419 but have homologues in other species.
|
Classa |
No. of isolates |
RTX |
|
|
|
REP |
|
|
SID |
|
|
|
AIT |
|
|
|
|
RKP |
|
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | a | b | c | S | a | b | S | a | b | C | S | a | B | c | d | S | a | b | c | d | e | f | g | h | ||
| I | 9 | Y | + | + | + | Y | + | + | Y | + | + | + | Y | + | + | + | + | Y | + | + | + | + | + | + | + | + |
| I | 21 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| II | 1 | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | |||||
| III | 3 | Y | + | + | + | ? | − | − | N | − | − | − | N | − | − | − | − | N | − | − | − | − | − | − | − | − |
| III | 6 | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
For all 39 isolates, the presence of sequences was tested by PCR using 20 different primer pairs (+, PCR product; −, no product). For the 12 sequenced isolates, the presence of matching shotgun sequencing reads in each of the five regions is also indicated (Y if at least one read).
Isolates fell into three classes with respect to their PCR results, as shown in the table.
Class I: MLX 1, 3, 5, 6 and 8−12 (sequenced); MLX 13, 15−18, 21, 23−25, 26−29, 31−37 and 39 (not sequenced).
Class II: MLX 20 (not sequenced).
Class III: MLX 2 (sequenced but no reads in REP region), 4 and 7 (sequenced, with reads in REP); MLX 14, 19, 22, 26, 30 and 38 (not sequenced).
All 12 sequenced strains contributed to an 8-kb contig (RTX) that includes genes homologous to the rtxA, rtxC and rtxD genes of Bradyrhizobium elkanii USDA 94 and B. japonicum USDA 110 with amino-acid identities between 50% and 65%. There is no evidence of any sequence polymorphism in these genes among the S. medicae isolates. PCR based on the rtxA and rtxC homologues (primers RTXabc) demonstrated that these sequences are present in all 39 isolates. The B. elkanii rtxA gene encodes a bifunctional protein, the N-terminus being dihydroxyacetonephosphate aminotransferase and the C-terminus dihydroxyrhizobitoxin synthase. In the S. medicae contig, the corresponding sequences are encoded in two separate genes, which we call rtxA and rtxB.
The other regions show much greater sequence similarity to the homologous sequences in other Sinorhizobium species, but none of them are present in all the S. medicae isolates (Table 5). A cluster of genes involved in polysaccharide synthesis on pSymB of S. meliloti 1021 is quite different from the genes in the corresponding region of S. medicae WSM 419, but the majority of our S. medicae isolates have a set of genes (RKP) very similar to those of 1021 (contigs 59 and 179 with 99.6% DNA identity, contig 47 with >98.5% and some polymorphism among strains).
Another region of S. meliloti 1021 pSymB has genes for synthesis of autoinducer-2 (Pereira et al., 2008), and the aitK and aitG genes (SMb21022 and SMb21023 on pSymB in S. meliloti 1021) are also found in our S. medicae population on contig 228 (AIT, 96% DNA identity). The AI-2 cluster in S. meliloti 1021 has seven genes, and we subsequently found another contig (62, 96% identity) that carries the 5′ part of aitK plus aitR, aitA and aitC in the same configuration as in 1021. The contig does not extend to aitD or aitB, but all the relevant strains have individual reads matching one or both of these genes, while the AIT-negative strains have none. On the other hand, matches to S. meliloti SM_b20498 are weaker (88–93% identity) and found in AIT-negative as well as AIT-positive strains. Pereira et al. (2008) called this gene aitF and hypothesized that it was the orthologue of the Salmonella gene lsrF, implicated in AI-2 processing, but our result suggests that it is not specific to the AI-2 system in Sinorhizobium.
The REP contig carries a gene that is most similar (92% DNA identity) to the repC plasmid replication gene of pRmeGR4a, a plasmid found in S. meliloti GR4 (Izquierdo et al., 2005), but the gene next to it resembles (93% DNA identity) the exonuclease gene exoI that is next to repC in pSmeSM11a, a plasmid in another strain of S. meliloti (Stiens et al., 2007).
Finally, contig 211 (SID) has four genes that are similar (88% DNA identity) to genes on plasmid b of Sinorhizobium sp. NGR234 putatively involved in iron uptake.
All five of these gene clusters show a very similar distribution across our S. medicae strains: 29 strains have all of them, 9 have none, while 1 strain, MLX20, has REP and SID but not AIT or RKP (Table 5).
Discussion
Sampling strategies for bacterial population genomics
As we discussed in the Introduction, genomes from a global collection of species can be valuable, for example, for assessing the pan-genome (Tettelin et al., 2005) or to design molecular markers (Pearson et al., 2009), whereas a metagenomic approach can provide information about diversity in a local context (Allen et al., 2007; Eppley et al., 2007). Both these strategies have limitations, however, and do not directly address the manner in which genetic information is distributed among members of a bacterial species coexisting and potentially encountering each other in a local environment. The approach we adopted, therefore, was to first obtain multiple isolates from a single site, and then to sample the genome sequence of each. Even though the coverage of each genome was low (averaging around 0.8 × ), this was sufficient to estimate nucleotide divergence and detect insertion and deletion across the whole genome down to a scale of 10 kb. It was also sufficient to detect the presence or absence of genes in individual strains with reasonable confidence, as confirmed by the concordance between read coverage and PCR product for each of the accessory gene regions we assessed (Table 5). The analysis was aided by the relatively low genomic diversity in S. medicae compared with some other bacterial species. A sample size of 12 is small by population genetics standards, of course, but it is sufficient to demonstrate that a genome-wide approach to population diversity is now feasible. Larger samples and higher coverage are, of course, becoming increasingly practicable as sequencing technologies continue to advance.
Spatial scale is, of course, important. The choice of one square metre was dictated, in part, by the need to obtain enough nodules for analysis, but previous studies suggest that the diversity of a rhizobium population is not greatly influenced by scale within the range of centimetres to metres. Young et al. (1987) demonstrated that, for Rhizobium leguminosarum in a pea field, each nodule was essentially an independent sample and the diversity among isolates from a single plant was almost the same as between plants across a metre, while samples 20 m apart were not significantly different. Genotype frequencies do, however, vary significantly when sites are many kilometres apart, reflecting environmental differences (Harrison et al., 1989).
Impact of gene transfer on S. medicae evolution
The mapping of sequence reads against the reference genome of S. medicae WSM 419 suggests that most indels, that is, gene gain by horizontal transfer and gene loss, have affected pSMED02 and pSMED03. This is concordant with the observation that most indel events affecting the S. meliloti genome occur on pSymA (Giuntini et al., 2005), and with the low proportion of homologous genes observed between pSMED02 of S. medicae and pSymA of S. meliloti when compared with the strong synteny between pSMED01 and pSymB and between the chromosomes of S. meliloti 1021 and S. medicae WSM 419 (data not shown).
Sequence divergence is quite low all along the chromosome of the S. medicae strains we sampled, but more than two orders of magnitude above the expected error rate of the sequencing method for nucleotide substitutions (Droege and Hill, 2008). Conversely, it is higher on the chromid pSMED01 and megaplasmid pSMED02. Furthermore, PCA demonstrates that divergence patterns are more heterogeneous on these replicons than on the chromosome. This is in agreement with previous observations obtained from French populations of either S. medicae or S. meliloti (Bailly et al., 2006), and suggests that recombination events are more important in shaping diversity patterns within or between chromids and plasmids than within the chromosome.
The concept of a ‘clonal complex' has been widely used in pathogen epidemiology to describe strains that are identical, or near identical, at a number of chromosomal loci, as determined by multilocus sequence typing. Strains that have identical sequences for at least six out of a standard set of seven housekeeping genes are considered to belong to the same clonal complex (Feil et al., 2004). Given that the average divergence between our S. medicae strains is only 0.00232 mutations/site for chromosomal sequences, it is likely that some belong to the same clonal complex, as has been observed for S. medicae isolates of diverse origins (van Berkum et al., 2006). Even this low level of divergence is not compatible with a single ancestor for all strains on a time scale of a few years, as in a true epidemic. Furthermore, our data show that strains that have all but identical chromosomes may differ substantially in their extrachromosomal sequences, so multilocus sequence typing does not paint a complete picture of population structure in bacteria that have a substantial accessory genome. Comparable evidence for diversity and transfer of extrachromosomal sequences in S. meliloti and S. medicae was provided by a recent study that extended the multilocus sequence typing approach beyond housekeeping genes (van Berkum et al., 2010).
Interestingly, a genomic architecture involving three large replication units (that is, chromosome, chromid(s) and/or megaplasmid) has evolved several times during the diversification of bacterial lineages (Harrison et al., 2010). In Burkholderia, Chain et al. (2006) also showed that the different replication units are evolving under independent evolutionary dynamics. Even though no significant fitness effect has been detected in S. meliloti strains characterized by atypical genome architectures (Guo et al., 2003), the organisation and the polymorphism of the S. medicae genome raise questions about the mechanisms leading to the differing gene content of its replicons and the possible adaptive nature of this organisation.
Selection and the genetic structures of S. medicae
The chromosomal phylogenetic network shows relatively long terminal branches as a result of strain-specific mutations, resulting in a star-like pattern. There is considerable reticulation, reflecting phylogenetic incongruence that is probably due to limited information, or possibly recombination, rather than saturation, given the low levels of divergence. Sequence divergence is lower on the chromosome than on the other replication units (note the differing scales in Figure 4). As the chromosome harbours most of the housekeeping genes in both S. meliloti and S. medicae, the low diversity on this replication unit would be consistent with the directional selective pressures (that is, purifying and/or transient-positive selection) that are expected for such gene content.
The KST values, which indicate the relative divergence of the Sardinian strain WSM 419 from our population, are lower for pSMED01 and pSMED02 than for the chromosome. A high KST indicates that the Sardinian strain is an outlier relative to the variation within our population, which would be an expected consequence of geographic isolation, although more strains would be needed to investigate this aspect of population structure convincingly. The lower KST values for pSMED01 and pSMED02 reflect the much higher diversity of these replicons within our population in comparison to the chromosome. Compared with this diversity, the divergence of WSM 419 is less marked. Indeed, for many 10 kb windows, KST is actually negative, indicating that the majority of MLX strains are more similar to WSM 419 than to some highly diverged MLX strains. This is also reflected in the phylogenetic networks for pSMED01 and pSMED02 (Figure 4), which place WSM 419 on a short branch near the centre of the network. In other words, strains within our sample of the population in one square metre of soil can be more diverged from each other in these replicons than from a strain originating >1600 km away in very different climatic conditions. It is usual that genes of accessory or unknown function, which are frequent on extrachromosomal replicons, are more polymorphic than core genes that may be under strong purifying selection (for example, Cooper and Feil (2006)), but it is interesting to observe that this polymorphism extends to local populations and not just wider strain collections. Of course, we are looking here at nucleotide divergence of genes that are shared by all strains, rather than at the presence or absence of potentially adaptive accessory genes, which are discussed in the next section.
The non-chromosomal window showing the lowest average divergence among the 12 sampled strains is located on pSMED02. This window includes, among others, some of the most important genes involved in symbiotic association: nodA, nodB and nodC (Perret et al., 2000). This diversity pattern is coherent with previous evidence of directional selection acting on nod genes of both S. meliloti and S. medicae (Bailly et al., 2006). Furthermore, the occurrence of indels and the heterogeneity of divergence patterns around this genomic area suggest that nod genes of S. medicae could be transferred as a genomic island, as described for Mesorhizobium spp. (Sullivan and Ronson, 1998).
A window showing one of the highest average levels of divergence among the 12 sampled strains is located on pSMED01. This window is located next to an indel event identified in several strains. This genomic area includes a number of genes involved in the synthesis of polysaccharides, among others an rkpZ gene found in 28 of the 39 S. medicae strains we sampled. The occurrence of such genes in the S. meliloti genome has been shown to influence several phenotypes including the host range of the bacteria or its phage tolerance (Williams et al., 1990; Brzoska and Signer, 1991; Reuhs et al., 1995; Sharypova et al., 2006). The pleiotropy of genes involved in the synthesis of polysaccharides and/or physical linkage could be involved in the different traces of balancing selection which have already been described in this part of pSMED01/pSymB (Bailly et al., 2006; Sun et al., 2006).
These two different examples indicate that selection can have an important role in shaping the diversity pattern of the chromid and plasmids of S. medicae. From a practical perspective, they strengthen the idea that genomic screening of diversity patterns could be used to identify genes that have a critical role in shaping bacterial ecological niches (Falush and Bowden, 2006). Higher coverage or a more complex sampling scheme would facilitate more advanced population genomic analyses.
New genes belonging to the S. medicae pan-genome
The deletion of an rkpZ copy in some of the strains is also linked to polymorphisms in the presence/absence of a number of other genes, some of which might be involved in symbiosis. The assembly of reads that were not mapped against the reference genome allowed the description of new genes belonging to the pan-genome of S. medicae. Most of these genes shared high sequence similarity (>90%) with the homologous genes identified in GenBank and their distribution in our S. medicae population is almost the same as that of the rkpZ region (Table 5). Among the potential symbiosis-related genes, we identified is a homologue of SM_b20825, which encodes a putative acetyltransferase belonging to the CysE/LacA/LpxA/NodL family that is essential for the O-antigen synthesis in Rhizobium etli CE3. Bacteria lacking the O-antigen region of their lipopolysaccharide are seriously impaired in their ability to invade developing root nodules, producing root nodules devoid of bacteria (Lerouge et al., 2003). The facts that the distribution of these genes is so strongly correlated, that they occur in strains that are genetically diverse in other respects, and that they include a plasmid replication gene, suggest that they may all be carried together on a transmissible plasmid, although this has not been confirmed directly.
Moreover, we also found rtxA, rtxB, rtxC and rtxD homologues in all 12 sequenced genomes. These genes are involved in the synthesis of rhizobitoxine in Bradyrhizobium elkanii (Yasuta et al., 2001). PCR results indicate that this gene cluster is present in all 39 sampled S. medicae strains. In B. elkanii, rhizobitoxine has been shown to enhance nodulation capabilities of the bacteria by inhibiting ethylene synthesis in plant tissues (Yuhashi et al., 2000). If the rtx homologues of S. medicae encode the same function, the strains we collected might have an increased capability to induce nodules on the roots of their host when compared with S. medicae WSM 419. Surprisingly, the RtxA sequence of our S. medicae strains is more diverged from that of Bradyrhizobium than are those found in Burkholderia phymatum, Pseudomonas savastanoi pv. savastanoi and various species of Xanthomonas (unpublished analysis of public genome sequences), indicating that rhizobitoxine production is an accessory function of considerable antiquity that has spread widely by horizontal gene transfer among plant-interacting bacteria, both symbionts and pathogens. Rhizobia also have another strategy for interfering with plant ethylene signalling. This involves the acdS gene, which encodes an enzyme that increases nodulation by metabolising a precursor of ethylene (Ma et al., 2004), and has been described from the plasmid pSmeSM11a. This gene was present in 89% of S. meliloti isolates sampled in Germany (Kuhn et al., 2008). Surprisingly, not one of the S. medicae strains that we sampled had this gene. This raises questions about the ecological factors that could limit the spread of such functions among rhizobia, as a potential trade-off between the fitness of symbiotic partners (Ratcliff and Denison, 2009).
No gene involved in increasing nodulation by influencing the ethylene pathways of the host was found in the reference genomes of the Medicago symbionts S. meliloti 1021 and S. medicae WSM 419. The high frequency of such genes in natural populations illustrates the outcome of a conflict of interest between rhizobia and their host regarding the investment of legumes in symbiotic functions. The existence of two alternative bacterial systems that can influence the concentration of ethylene in the roots of host plants reminds us that defining ecologically important functions is not straightforward, as different pathways can be involved in similar phenotypes. More generally, studies of accessory genome composition are important in defining the selective pressures that describe the ecological niche and drive the evolution of bacterial species. Indeed, one could argue that, while the core genome defines the taxonomy of bacteria, the accessory genome has an equal or greater importance in defining their ecological niche.
Acknowledgments
This study was funded by grant NE/D011485/1 from the Natural Environment Research Council. PWH was funded by a BBSRC research studentship. We thank the University of Liverpool Advanced Genomics Facility for carrying out the sequencing, and particularly Professor Neil Hall and Dr Margaret Hughes for their technical expertise.
References
- Allen EE, Tyson GW, Whitaker RJ, Detter JC, Richardson PM, Banfield JF. Genome dynamics in a natural archaeal population. Proc Natl Acad Sci USA. 2007;104:1883–1888. doi: 10.1073/pnas.0604851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly X, Olivieri I, De Mita S, Cleyet-Marel JC, Béna G. Recombination and selection shape the molecular diversity pattern of nitrogen-fixing Sinorhizobium sp associated to Medicago. Mol Ecol. 2006;15:2719–2734. doi: 10.1111/j.1365-294X.2006.02969.x. [DOI] [PubMed] [Google Scholar]
- Berg OG, Kurland CG. Evolution of microbial genomes: sequence acquisition and loss. Mol Biol Evol. 2002;19:2265–2276. doi: 10.1093/oxfordjournals.molbev.a004050. [DOI] [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Brzoska PM, Signer ER. lpsZ, a lipopolysaccharide gene involved in symbiosis of Rhizobium meliloti. J Bacteriol. 1991;173:3235–3237. doi: 10.1128/jb.173.10.3235-3237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PSG, Denef VJ, Konstantinidis KT, Vergez LM, Agullo L, Reyes VL, et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci USA. 2006;103:15280–15287. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JE, Feil EJ. The phylogeny of Staphylococcus aureus—which genes make the best intra-species markers. Microbiology. 2006;152:1297–1305. doi: 10.1099/mic.0.28620-0. [DOI] [PubMed] [Google Scholar]
- Didelot X, Achtman M, Parkhill J, Thomson NR, Falush D. A bimodal pattern of relatedness between the Salmonella Paratyphi A and Typhi genomes: convergence or divergence by homologous recombination. Genome Res. 2007;17:61–68. doi: 10.1101/gr.5512906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droege M, Hill B. The Genome Sequencer FLX System--longer reads, more applications, straight forward bioinformatics and more complete data sets. J Biotechnol. 2008;136:3–10. doi: 10.1016/j.jbiotec.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Eppley JM, Tyson GW, Getz WM, Banfield JF. Genetic exchange across a species boundary in the archaeal genus Ferroplasma. Genetics. 2007;177:407–416. doi: 10.1534/genetics.107.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Bowden R. Genome-wide association mapping in bacteria. Trends Microbiol. 2006;14:353. doi: 10.1016/j.tim.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F, Finan TM, Long SR, Pühler A, Abola P, Ampe F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- Giuntini E, Mengoni A, De Filippo C, Cavalieri D, Aubin-Horth N, Landry CR, et al. Large-scale genetic variation of the symbiosis-required megaplasmid pSymA revealed by comparative genomic analysis of Sinorhizobium meliloti natural strains. BMC Genomics. 2005;6:158. doi: 10.1186/1471-2164-6-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Sun S, Finan TM, Xu J. Novel DNA sequences from natural strains of the nitrogen-fixing symbiotic bacterium Sinorhizobium meliloti. Appl Environ Microbiol. 2005;71:7130–7138. doi: 10.1128/AEM.71.11.7130-7138.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Flores M, Mavingui P, Fuentes SI, Hernandez G, Davila G, et al. Natural genomic design in Sinorhizobium meliloti: novel genomic architectures. Genome Res. 2003;13:1810–1817. doi: 10.1101/gr.1260903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Lower RPJ, Kim NKD, Young JPW. Introducing the bacterial chromid: not a chromosome, not a plasmid. Trends Microbiol. 2010;18:141–148. doi: 10.1016/j.tim.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Harrison SP, Jones DG, Young JPW. Rhizobium population genetics: genetic variation within and between populations from diverse locations. J Gen Microbiol. 1989;135:1061–1069. [Google Scholar]
- Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- Izquierdo J, Venkova-Canova T, Ramírez-Romero MA, Téllez-Sosa J, Hernández-Lucas I, Sanjuan J, et al. An antisense RNA plays a central role in the replication control of a repC plasmid. Plasmid. 2005;54:259–277. doi: 10.1016/j.plasmid.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn S, Stiens M, Pühler A, Schlüter A. Prevalence of pSmeSM11a-like plasmids in indigenous Sinorhizobium meliloti strains isolated in the course of a field release experiment with genetically modified S. meliloti strains. FEMS Microbiol Ecol. 2008;63:118–131. doi: 10.1111/j.1574-6941.2007.00399.x. [DOI] [PubMed] [Google Scholar]
- Lefebure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8:R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge I, Verreth C, Michiels J, Carlson RW, Datta A, Gao MY, et al. Three genes encoding for putative methyl- and acetyltransferases map adjacent to the wzm and wzt genes and are essential for O-antigen biosynthesis in Rhizobium etli CE3. Mol Plant Microbe Interact. 2003;16:1085–1093. doi: 10.1094/MPMI.2003.16.12.1085. [DOI] [PubMed] [Google Scholar]
- Ma W, Charles TC, Glick BR. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl Environ Microbiol. 2004;70:5891–5897. doi: 10.1128/AEM.70.10.5891-5897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens M, Delaere M, Coopman R, De Vos P, Gillis M, Willems A. Multilocus sequence analysis of Ensifer and related taxa. Int J Syst Evol Microbiol. 2007;57:489–503. doi: 10.1099/ijs.0.64344-0. [DOI] [PubMed] [Google Scholar]
- Mercado-Blanco J, Olivares J. Stability and transmissibility of the cryptic plasmids of Rhizobium meliloti GR4. Arch Microbiol. 1993;160:477–485. [Google Scholar]
- Mercado-Blanco J, Olivares J. The large nonsymbiotic plasmid pRmeGR4a of Rhizobium meliloti GR4 encodes a protein involved in replication that has homology with the RepC protein of Agrobacterium plasmids. Plasmid. 1994;32:75–79. doi: 10.1006/plas.1994.1046. [DOI] [PubMed] [Google Scholar]
- Mes THM. Microbial diversity—insights from population genetics. Environ Microbiol. 2008;10:251–264. doi: 10.1111/j.1462-2920.2007.01449.x. [DOI] [PubMed] [Google Scholar]
- Novozhilov AS, Karev GP, Koonin EV. Mathematical modeling of evolution of horizontally transferred genes. Mol Biol Evol. 2005;22:1721–1732. doi: 10.1093/molbev/msi167. [DOI] [PubMed] [Google Scholar]
- Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, et al. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 2009;7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, McAuley JR, Taga ME, Xavier KB, Miller ST. Sinorhizobium meliloti, a bacterium lacking the autoinducer-2 (AI-2) synthase, responds to AI-2 supplied by other bacteria. Mol Microbiol. 2008;70:1223–1235. doi: 10.1111/j.1365-2958.2008.06477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff WC, Denison RF. Rhizobitoxine producers gain more poly-3-hydroxybutyrate in symbiosis than do competing rhizobia, but reduce plant growth. ISME J. 2009;3:870–872. doi: 10.1038/ismej.2009.38. [DOI] [PubMed] [Google Scholar]
- Reeve WG, O'Hara G, Chain P, Ardley J, Bräu L, Nandesena K, et al. Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM1325, an effective microsymbiont of annual Mediterranean clovers. Stand Genomic Sci. 2010;2:347–356. doi: 10.4056/sigs.852027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuhs BL, Williams MN, Kim JS, Carlson RW, Cote F. Suppression of the Fix-phenotype of Rhizobium meliloti exoB mutants by lpsZ is correlated to a modified expression of the K polysaccharide. J Bacteriol. 1995;177:4289–4296. doi: 10.1128/jb.177.15.4289-4296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. The organization of the bacterial genome. Annu Rev Genet. 2008;42:211–233. doi: 10.1146/annurev.genet.42.110807.091653. [DOI] [PubMed] [Google Scholar]
- Rome S, Fernandez MP, Brunel B, Normand P, Cleyet-Marel JC. Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int J Syst Bacteriol. 1996;46:972–980. doi: 10.1099/00207713-46-4-972. [DOI] [PubMed] [Google Scholar]
- Sharypova LA, Chataigne G, Fraysse N, Becker A, Poinsot V. Overproduction and increased molecular weight account for the symbiotic activity of the rkpZ-modified K polysaccharide from Sinorhizobium meliloti Rm1021. Glycobiology. 2006;16:1181–1193. doi: 10.1093/glycob/cwl042. [DOI] [PubMed] [Google Scholar]
- Stiens M, Schneiker S, Keller M, Kuhn S, Pühler A, Schlüter A. Sequence analysis of the 144-kilobase accessory plasmid pSmeSM11a, isolated from a dominant Sinorhizobium meliloti strain identified during a long-term field release experiment. Appl Environ Microbiol. 2006;72:3662–3672. doi: 10.1128/AEM.72.5.3662-3672.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiens M, Schneiker S, Pühler A, Schlüter A. Sequence analysis of the 181-kb accessory plasmid pSmeSM11b, isolated from a dominant Sinorhizobium meliloti strain identified during a long-term field release experiment. FEMS Microbiol Lett. 2007;271:297–309. doi: 10.1111/j.1574-6968.2007.00731.x. [DOI] [PubMed] [Google Scholar]
- Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Guo H, Xu J. Multiple gene genealogical analyses reveal both common and distinct population genetic patterns among replicons in the nitrogen-fixing bacterium Sinorhizobium meliloti. Microbiology. 2006;152:3245–3259. doi: 10.1099/mic.0.29170-0. [DOI] [PubMed] [Google Scholar]
- Terpolilli JJ, O'Hara GW, Tiwari RP, Dilworth MJ, Howieson JG. The model legume Medicago truncatula A17 is poorly matched for N2 fixation with the sequenced microsymbiont Sinorhizobium meliloti 1021. New Phytol. 2008;179:62–66. doi: 10.1111/j.1469-8137.2008.02464.x. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome'. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkum P, Elia P, Eardly BD. Multilocus sequence typing as an approach for population analysis of Medicago-nodulating rhizobia. J Bacteriol. 2006;188:5570–5577. doi: 10.1128/JB.00335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkum P, Elia P, Eardly BD. Application of multilocus sequence typing to study the genetic structure of megaplasmids in Medicago-nodulating rhizobia. Appl Environ Microbiol. 2010;76:3967–3977. doi: 10.1128/AEM.00251-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessa P.2008Maximum-likelihood Poisson Distribution Fitting (v1.0.2) Free Statistics Software (v1.1.23-r3) http://www.wessa.net/rwasp_fitdistrpoisson.wasp/ : Office for Research Development and Education.
- Williams MN, Hollingsworth RI, Klein S, Signer ER. The symbiotic defect of Rhizobium meliloti exopolysaccharide mutants is suppressed by lpsZ+, a gene involved in lipopolysaccharide biosynthesis. J Bacteriol. 1990;172:2622–2632. doi: 10.1128/jb.172.5.2622-2632.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuta T, Okazaki S, Mitsui H, Yuhashi K, Ezura H, Minamisawa K. DNA sequence and mutational analysis of rhizobitoxine biosynthesis genes in Bradyrhizobium elkanii. Appl Environ Microbiol. 2001;67:4999–5009. doi: 10.1128/AEM.67.11.4999-5009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JPW, Demetriou L, Apte RG. Rhizobium population genetics: enzyme polymorphism in Rhizobium leguminosarum from plants and soil in a pea crop. Appl Environ Microbiol. 1987;53:397–402. doi: 10.1128/aem.53.2.397-402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuhashi K-I, Ichikawa N, Ezura H, Akao S, Minakawa Y, Nukui N, et al. Rhizobitoxine production by Bradyrhizobium elkanii enhances nodulation and competitiveness on Macroptilium atropurpureum. Appl Environ Microbiol. 2000;66:2658–2663. doi: 10.1128/aem.66.6.2658-2663.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]