Abstract
Archaeal genes for ammonia oxidation are widespread in the marine environment, but direct physiological evidence for ammonia oxidation by marine archaea is limited. We report the enrichment and characterization of three strains of pelagic ammonia-oxidizing archaea (AOA) from the North Pacific Ocean that have been maintained in laboratory culture for over 3 years. Phylogenetic analyses indicate the three strains belong to a previously identified clade of water column-associated AOA and possess 16S ribosomal RNA genes and ammonia monooxygenase subunit a (amoA) genes highly similar (98–99% identity) to those recovered in DNA and complementary DNA clone libraries from the open ocean. The strains grow in natural seawater-based liquid medium while stoichiometrically converting ammonia (NH3) to nitrite (NO2−). Ammonia oxidation by the enrichments is only partially inhibited by allylthiourea at concentrations known to completely inhibit cultivated ammonia-oxidizing bacteria. The three strains were used to determine the nitrogen stable isotope effect (15ɛNH3) during archaeal ammonia oxidation, an important parameter for interpreting stable isotope ratios in the environment. Archaeal 15ɛNH3 ranged from 13‰ to 41‰, within the range of that previously reported for ammonia-oxidizing bacteria. Despite low amino acid identity between the archaeal and bacterial Amo proteins, their functional diversity as captured by 15ɛNH3 is similar.
Keywords: thaumarchaea, nitrification, Pacific ocean, isotope effects
Introduction
Mesophilic archaea are ubiquitous and abundant members of diverse marine environments including coastal waters (Mincer et al., 2007; Beman et al., 2010), marine sediments, estuaries (Mosier and Francis, 2008; Bernhard et al., 2010; Urakawa et al., 2010), stratified basins (Coolen et al., 2007; Lam et al., 2007) and open ocean water columns (Beman et al., 2008; Church et al., 2010; Santoro et al., 2010). The recent cultivation of the first mesophilic marine archaeon, Nitrosopumilus maritimus, (Konneke et al., 2005; Martens-Habbena et al., 2009), two thermophilic archaea, Nitrosocaldus yellowstonii and Nitrosophaera gargensis (Hatzenpichler et al., 2008; de la Torre et al., 2008) and a freshwater archaeon, Nitrosoarchaeum limnia (Blainey et al., 2011) established that at least some of these organisms are chemolithoautotrophic ammonia oxidizers. Several studies coupling gene- and cell-based quantification of putative ammonia-oxidizing archaea (AOA) to rate measurements (Wuchter et al., 2006; Beman et al., 2008; Santoro et al., 2010) suggest that nitrification by AOA in the ocean is significant. However, direct demonstration of ammonia-oxidizing activity by the AOA genotypes encountered in the open ocean has not yet been demonstrated.
Ammonia oxidation is the first step of nitrification, a key remineralization reaction in the sea. Work by Olson (1981) and Ward et al. (1982) using 15N tracers first suggested that nitrification in the upper ocean could be a significant source of regenerated nutrients for primary production. The quantitative role, however, of nitrification in the euphotic zone is still uncertain. Recent syntheses of nitrification rates made using isotope tracers (Yool et al., 2007; Clark et al., 2008) suggest that globally, nitrification may be a significant source of regenerated N for primary production, supplying as much as half of the necessary nitrate. This has important implications for estimates of carbon export based on new production (Eppley and Peterson, 1979); an underestimation of nutrient regeneration would lead to an overestimation of carbon export from the surface ocean (Ward, 2002).
Although instantaneous rate measurements with 15N-labeled compounds provide important insights into the environmental controls on nitrification, they capture only a ‘snapshot' view of this undoubtedly time-varying process. An alternative way of quantifying the importance of nitrification is using natural abundance stable isotope ratios to infer the relative importance of different nitrogen cycling processes. In particular, nitrogen and oxygen stable isotope ratios (δ15N and δ18O) in marine nitrate (NO3−) can integrate information about the relative sources (nitrification, upward diffusion, atmospheric deposition) and sinks (phytoplankton uptake) of NO3− in the euphotic zone (Sigman et al., 2005; Wankel et al., 2007; Casciotti et al., 2008; DiFiore et al., 2009) and provide an independent constraint on the importance of nitrification to new production. In order to interpret these measurements, however, the isotope effects for the various sources and sinks of NO3− must be known. The isotope effects for NO3− uptake by phytoplankton (Needoba et al., 2003; Granger et al., 2004) and NO2− production by nitrifying bacteria (Mariotti et al., 1981; Casciotti et al., 2003, 2010) are relatively well studied, but there are no data on the isotope effects for ammonia oxidation by AOA.
In this study, we describe the enrichment and characterization of three strains of AOA from the water column of the North Eastern Pacific Ocean that oxidize NH3 to NO2−. We used the three strains to estimate the nitrogen kinetic isotope effect (15ɛNH3) during ammonia oxidation by AOA. Our results extend the ability for chemoautotrophic ammonia oxidation within the Archaea and expand the number of marine archaea for which we have both phylogeny and confirmed metabolic function. We provide important constraints for the interpretation of natural abundance stable isotope ratios for compounds affected by these ubiquitous organisms in the marine nitrogen cycle.
Materials and methods
Enrichment and cultivation
Enrichment cultures were initiated with water collected from the North Eastern Pacific approximately 300 km from shore (California Cooperative Oceanic Fisheries Investigations station 67–90; 35.46°N, 124.91°W) aboard the R/V Western Flyer during cruise CN107 in July 2007 (Table 1). Seawater was collected from 25, 75, 150 and 500 m depths using a standard 10 l Niskin rosette sampler equipped with a conductivity-temperature-depth sensor package. Seawater was stored at 22 °C (25 and 75 m waters) and 4 °C (150 and 500 m waters) in 500 ml acid-cleaned polycarbonate bottles. After 20 months of incubation, filter sterile ammonium chloride (NH4Cl) was added to a final concentration of 10 μmol l−1 and the enrichments were monitored for production of NO2− using standard colorimetric methods (Strickland and Parsons, 1968). At that time, the 150 m enrichment was moved to 13 °C and maintained at that temperature. Hereafter, the successful enrichments are referred to as CN25, CN75 and CN150 referring to the cruise name and depth of the enrichment inoculums.
Table 1. In situ conditions for the starting material for North Pacific Ocean seawater enrichments used for archaeal kinetic isotope fractionation (15ɛNH3) determination.
| Enrichment | Depth (m) | Temp (°C) | [NH4+] (nmol l−1) | [NO3−] (μmol l−1) |
|---|---|---|---|---|
| CN25 | 25 | 14 | 40 | 0.5 |
| CN75 | 75 | 12 | <15 | 2.2 |
| CN150 | 150 | 10 | <15 | 15.1 |
Cultures were maintained with transfers of 10–20% (v/v) late exponential phase culture into oligotrophic North Pacific (ONP) medium consisting of: 0.2 μm-filtered North Pacific surface seawater amended with 10–100 μmol l−1 NH4Cl, 1 ml l−1 chelated trace elements solution (Balch et al., 1979), 15 μmol l−1 KH2PO4 and 100 μg ml−1 each streptomycin and ampicillin. Maintenance cultures were grown in 200 ml volumes in 250 ml acid-cleaned polycarbonate bottles. Surface seawater for ONP medium was obtained either from the initial collection site or from the Pacific hydrographic station SAFe (Johnson et al., 2007).
Abundance of archaeal and bacterial cells in the enrichments was periodically monitored in 1 ml formaldehyde-fixed culture volumes filtered onto 25 mm diameter, 0.2 μm pore size polycarbonate filters (Millipore GTTP, Billerica, MA, USA) with catalyzed auto reporter deposition-fluorescent in situ hybridization (CARD-FISH) and epifluorescence microscopy using probes Cren537–554 and EUB338 I-III (Teira et al., 2004).
Phylogenetic analysis of 16S rRNA and amoA genes
DNA was extracted and purified from 25 ml of culture after vacuum filtration onto 25 mm diameter, 0.2 μm pore size Supor membrane filters (Pall, Port Washington, NY, USA) using DNeasy columns (Qiagen, Valencia, CA, USA) as previously described (Santoro et al., 2008). Genes encoding for the 16S ribosomal RNA (rRNA) and the α subunit of ammonia monooxygenase (amoA), believed to contain the catalytic site for ammonia oxidation (Hyman and Wood, 1985), were amplified using PCR. Target genes were amplified in 25 μl PCR reactions using the primers: 21F/1492R (archaeal 16S rRNA genes, DeLong, 1992), ArchamoAF/ArchamoAR (archaeal amoA, Francis et al., 2005), 27F/1492R (bacterial 16S rRNA) and amoAF*/amoAR (betaproteobacterial amoA, Stephen et al., 1999) using reaction and thermocycling conditions described in the original papers or as modified in Santoro et al., 2010. Gammaproteobacterial amoA genes were amplified using the amoA3F/amoB4R primer set (Purkhold et al., 2000) using the following thermocycling profile: 95 °C for 3 min followed by 35 cycles of: 95 °C for 30 s, 50 °C for 45 s and 72 °C for 1 min. Genomic DNA from Nitrosococcus oceani was used as a positive control. PCR products were pooled, purified (MinElute PCR Purification Kit, Qiagen), and cloned using the TOPO-TA cloning kit with pCR4 vector and MACH1 competent cells (Invitrogen, Carlsbad, CA, USA). Plasmids were purified (Mini Prep Spin Kit, Qiagen) and sequenced on an ABI 3730xl sequencer. Twelve archaeal 16S rRNA and amoA clones and 24 bacterial 16S rRNA clones were sequenced per enrichment.
16S rRNA gene sequences were imported into the ARB software program (Ludwig et al., 2004) and aligned to a subset of the SILVA SSU reference database, February 2009 release (Pruesse et al., 2007). Archaeal amoA sequences were manually aligned and imported into an ARB database maintained by our laboratory containing approximately 2000 environmental amoA sequences. Phylogenetic trees were constructed using 1268 nucleotide positions (16S rRNA) or 489 nucleotide positions (amoA) using maximum likelihood (RAxML) analyses implemented using the Cyberinfrastructure for Phylogenetic Research (CIPRES) Portal v3.0 (www.phylo.org) and visualized using FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/). Archaeal 16S rRNA gene and amoA sequences from this study were deposited in GenBank under the accession numbers HQ338108–HQ338109 and JF521547–JF521549.
Growth curves
Growth curves were determined for CN25 and CN75 at 22 °C in replicate 200 ml volumes of ONP medium containing 50 μmol l−1 NH4+. At each time point, 15 ml of culture was removed. Ten milliliters of culture were frozen for later [NO2−] and [NH4+] determination. The remaining 5 ml of culture were immediately fixed with formaldehyde (2% final concentration) for four hours and replicate volumes (0.5–2 ml) of fixed culture were vacuum filtered onto 25 mm, 0.2 μm pore size polycarbonate membrane filters (Millipore) and frozen for later CARD-FISH analysis as described above. A minimum of 10 fields of view were counted from each filter using the 100 × objective on a Zeiss Axio Scope (Carl Zeiss, Jena, Germany) epifluorescence microscope. [NH4+] and [NO2−] determinations were made in duplicate using phthaldialdehyde fluorescence (Holmes et al., 1999) and azo dye colorimetry (Strickland and Parsons, 1968), respectively.
Allylthiourea inhibition experiments
To assess the effect of the metal chelator allylthiourea (ATU) on archaeal ammonia oxidation, strain CN25 was grown in triplicate 50 ml volumes in 60 ml polycarbonate screw cap bottles for each of three treatments: no ATU addition (control), 10 mg l−1 ATU (86 μmol l−1), and 100 mg l−1 (860 μmol l−1). ATU additions were made from a 10 mg ml−1 stock solution of ATU (Sigma) dissolved in water. Ammonia oxidation was monitored by the production of NO2−, determined using azo dye colorimetry in 1 ml volumes (Strickland and Parsons, 1968). ATU was added to the treatment bottles after initial NO2− production was detected in each flask (day 4). Inhibition for the ATU treatments was calculated as the percent decrease in slope of a linear regression fit to the ln[NO2−] over time compared with the control bottles.
Nitrogen isotope effect estimates
For each experiment, 25 ml of enrichment culture was inoculated into 175 ml of ONP medium containing 10–75 μmol l−1 NH4+ in a 250 ml acid-washed, screw-cap polycarbonate bottle. Growth was not observed when experiments were initiated with washed cell suspensions collected by filtration (data not shown), thus all experiments started with an initial [NO2−] between 2 and 10 μmol l−1 as well as residual NH4+ from maintenance cultures, in some cases. Temperature was maintained at 22 °C for CN25 and CN75 and 13 °C for CN150. NO2− production was monitored and 15 ml of enrichment culture was removed at each time point and frozen at −20 °C.
Initial and time-course measurements of [NH4+] and [NO2−] were made in duplicate using phthaldialdehyde fluorescence (Holmes et al., 1999) and azo dye colorimetry (Strickland and Parsons, 1968), respectively, on 2 ml sample volumes. Error in replicate [NH4+] analyses ranged from 2% to 7% error in replicate [NO2−] analyses was 0% to 8%. Isotopic measurements of NO2− were made by converting NO2− to nitrous oxide (N2O) using the ‘azide method' (McIlvin and Altabet, 2005) with 10–20 nmol N per sample. The resulting N2O was captured using a custom purge and cryogenic trapping system (Casciotti et al., 2002; McIlvin and Casciotti, 2010, 2011) and analyzed using a Finnigan DeltaPLUS XP isotope ratio mass spectrometer. Each sample was analyzed in duplicate against RSIL nitrite reference materials N23, N7373 and N10219 run in parallel (Casciotti et al., 2007). Results are reported using delta notation: δ15NNO2 (‰ vs AIR)=[(15RNO2/15RAIR)−1] × 1000, where 15R=15N:14N and AIR is standard atmospheric N2. Error in replicate measurements of δ15NNO2 was 0.3‰ or better for all experiments.
Calculation of 15ɛNH3 requires knowledge of the isotopic composition of the NO2− produced over time since an arbitrary starting point. If the initial [NO2−] is zero, then the measured δ15NNO2 (δ15NNO2total) is equal to that produced (δ15NNO2produced). If there is NO2− initially present, as in our experiments, the δ15NNO2produced at any time since the initial time point (i) must be calculated from δ15NNO2total using the following equations:
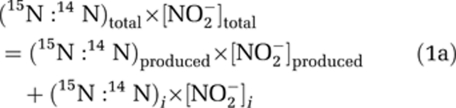 |
where (15N:14N)total=(15N:14N)AIR × (δ15NNO2total/1000+1) Rearranging:
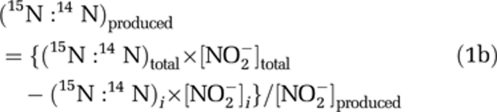 |
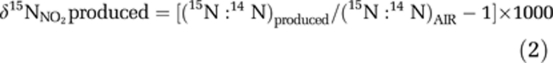 |
The isotope effect (15ɛNH3) was then calculated from the δ15NNO2produced data using the Rayleigh accumulated product equation (after Mariotti et al., 1981):
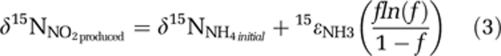 |
where f = [NH4+]/[NH4+]initial. By Eqn (3), the slope of a best fit line on a plot of δ15NNO2 vs f *ln(f)/(1-f) yields 15ɛNH3 and the y intercept corresponds to the starting δ15NNH4. As each experiment was initiated with a mixture of NH4+ carried over from the enrichment transfer and ‘new' NH4+ from freshly prepared medium (δ15N∼−3‰), minor variations in the δ15NNH4 (and the y intercept) are expected between experiments and therefore each experiment was fitted separately. Fitting and 95% confidence interval calculations were done using IGOR Pro software (v5, WaveMetrics, Inc., Lake Oswego, OR, USA).
Use of the Rayleigh model presumes that ammonia oxidation proceeds as a pseudo one-step reaction with no accumulation of an intermediate product between NH3 and NO2− and no back-reaction of the products (Casciotti et al., 2003). When estimated in this way, several isotope effects are incorporated into 15ɛNH3 including a ∼20–35‰ equilibrium isotope effect between NH4+ and NH3 (Casciotti et al., 2011), as well as any isotope effects for NH3 or NH4+ diffusion. Errors in 15ɛNH3 resulting from these assumptions, as well as errors resulting from uncertainties in the archaeal ammonia oxidation pathway, are discussed below.
Results
Phylogeny and physiology
Following 20 months of incubation in the dark with no amendments, archaeal amoA genes were detected by PCR in all four seawater ‘enrichment' bottles (25, 75, 150 and 500 m). On addition of 10 μmol l−1 NH4+, NO2− production was observed in the enrichments from 25, 75 and 150 m within 1 month (data not shown). No NO2− production was observed in the 500 m bottle. The NO2− producing enrichments are referred to as CN25, CN75 and CN150. All three CN enrichments contained a high proportion of cells hybridizing with the CARD-FISH probe suite CREN537–554 and PCR amplifiable archaeal amoA genes. No amoA genes from either gamma or betaproteobacteria were detected.
After approximately 1 year of routine transfers into ONP medium (nearly 3 years after initial collection), the enrichments were highly enriched in archaeal cells (Figure 1) and the basic phylogeny and physiology of the enrichments were characterized. A consensus archaeal 16S rRNA gene sequence was obtained from 12 clones from each enrichment. One sequence from CN25 and two sequences from CN75 contained single nucleotide changes (all at different positions) from this consensus sequence. Phylogenetic analyses place all three sequences within the Marine Group I archaea, now proposed as a new archaeal kingdom—the Thaumarchaeota (Brochier-Armanet et al., 2008; Spang et al., 2010; Walker et al., 2010) (Figure 2a). The 16S rRNA gene sequences from CN25 and CN75 are 100% identical to each other, and 92% identical to N. maritimus. Closest GenBank BLAST matches to the CN25 16S rRNA gene sequence are 98% identical and include sequences from environmental clone libraries from 200 m depth off the Oregon coast (fosmid 4B7, U40238, (Stein et al., 1996)) and an unpublished deep sea hydrothermal vent clone from Suiyo Seamount (AB194001). The CN150 16S rRNA gene is 98% identical to N. maritimus and 99% identical to several sequences from the Sargasso Sea metagenome (AACY020033564, (Venter et al., 2004)).
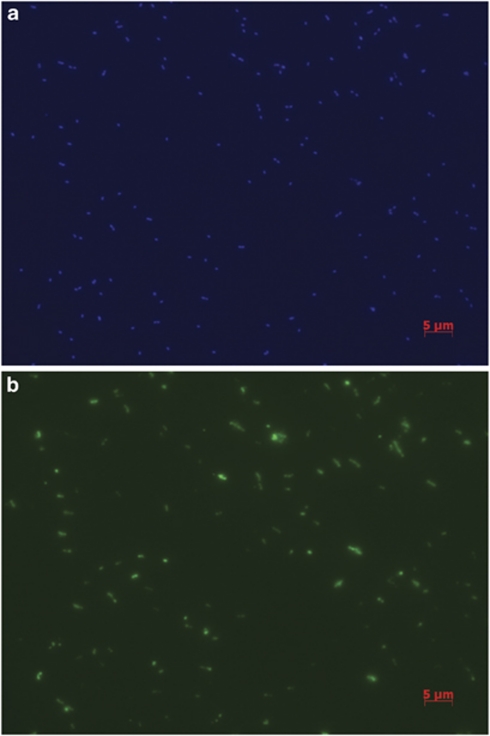
Figure 1.
Fluorescence microscopy images of enrichment CN25 stained with (a) 4,6-diamidino-2-phenylindole (DAPI) and (b) CARD-FISH using the Cren537–554 probe suite. Both images are from the same field of view; the scale bar represents 5 μm.
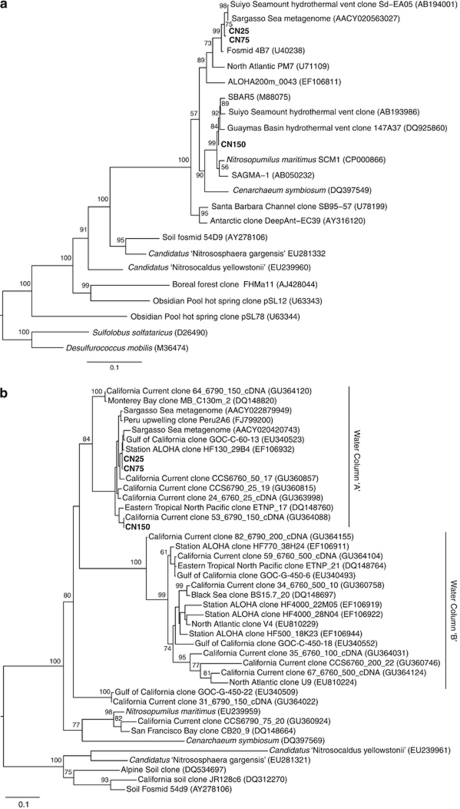
Figure 2.
Phylogenetic placement of the CN enrichments based on (a) 16S rRNA gene sequences (1268 nucleotide positions) and (b) amoA gene sequences (489 nucleic acid positions). The CN enrichment sequences are shown in bold. Trees were constructed using maximum likelihood methods (RAxML) using Sulfolobus solfataricus as the outgroup for the 16S rRNA tree and the soil and hot spring sequences as the outgroup for the amoA tree. Bootstrap support values (>70%) are shown at nodes; some values have been removed from minor nodes for clarity. An expanded amoA tree with additional sequences from the study environment is included as Supplementary Figure 1.
The amoA sequences from CN25 and CN75 fall within a cluster of environmental sequences previously termed water column cluster ‘A'. They are also 100% identical at the amino acid level to several sequences from the Sargasso Sea metagenome (Venter et al., 2004) (Figure 2b, Supplementary Figure 1) and 84% and 95% identical at the nucleotide and amino acid levels, respectively, to the amoA sequence from N. maritimus. The CN150 amoA gene is also 84% identical to N. maritimus at the nucleic acid level. As CN25 and CN75 are identical at the 16S rRNA and amoA level, full genome sequencing may be necessary to resolve any strain-level differences between the two enrichments.
Bacterial 16S rRNA gene clone libraries from the CN enrichments did not contain 16S rRNA genes from any genera of known bacterial nitrifiers and contained sequences associated with the genera Erythrobacter and Gracilimonas. Bacterial amoA genes could not be detected using PCR amplification.
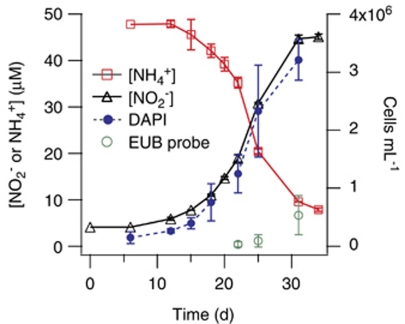
Stoichiometric conversion of NH4+ to NO2− was observed in all three enrichments coincident with an exponential increase in the number of archaeal cells, evidence that the CN strains couple ammonia oxidation to growth (Figure 3). All three CN strains were able to utilize NH4+ from the medium below our detection limit of ∼200 n. Bacterial cells, as quantified by CARD-FISH, were not detectible until the archaeal population entered late exponential-early stationary phase. CN25 and CN75, growing at 22 °C, had lag phases of 4–10 days on transferring 10–25% of the original culture volume into new medium, even when cultures were transferred during exponential phase growth. Transfer volumes of <10% failed to grow (data not shown). CN150, growing at 13 °C, had lag phases of up to 25 days.
Figure 3.
Growth of enrichment CN75 in ONP medium. Error bars denote 1 s.d. and in some cases are smaller than the point. EUB is the CARD-FISH probe suite EUB338 targeting all bacteria.
CN25 and CN75 were chosen for more detailed growth studies because of faster NH4+ oxidation rates, shorter lag phases, and more consistent growth. The CN25 growth rate at 22 °C was 0.15 day−1; the CN75 growth rate was 0.17 day−1, corresponding to doubling times of about 4–4.6 days. Stationary phase cell densities in CN25 and CN75 were 2.7–3.2 × 106 cells ml−1 with 86–97% of cells hybridizing with archaeal CARD-FISH probes depending on the growth stage of the enrichment.
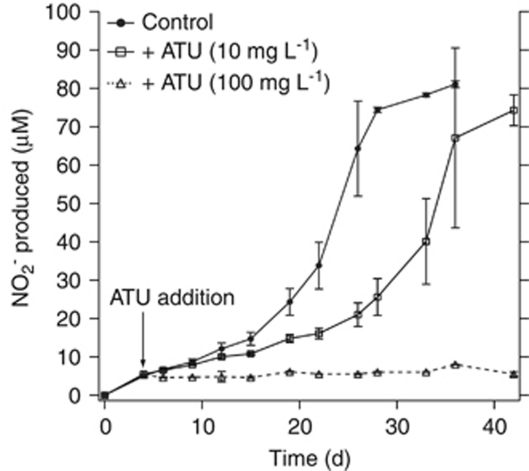
ATU is frequently used to inhibit nitrifying activity in environmental studies. We tested the effect of ATU on the ammonia oxidation rate for CN25. An ATU concentration of 10 mg l−1 (86 μmol l−1) inhibited the ammonia oxidation rate by 58% an ATU concentration of 100 mg l−1 (860 μmol l−1) completely inhibited ammonia oxidation (Figure 4).
Figure 4.
Ammonia oxidation by CN25 in the presence of 10 and 100 mg l−1 ATU. Each point represents the mean of three replicate bottles; error bars denote 1 s.d. from the mean.
Nitrogen isotopic fractionation
To further investigate the physiology of the enriched strains, we determined their N kinetic isotope effect during ammonia oxidation (15ɛNH3). As in growth curve experiments, we observed near stoichiometric conversion of NH4+ to NO2− over the course of the isotope fractionation experiments, with conservation in the dissolved inorganic nitrogen pool (NH4++NO2−) ranging from 74% to 107% with a mean of 97% (Table 2).
Table 2. Experimental results for kinetic isotope effect (15ɛNH3) determinations during of archaeal ammonia oxidation.
| Enrichment | Experiment # | 15ɛNH3 (‰) | CI 95% (‰) | Initial [NH4+] (μmol l−1) | [N+N]final: [N+N]initial | NH4+ oxidation rate (μmol l−1 day−1)a |
|---|---|---|---|---|---|---|
| CN25 | 1 | 25 | 1 | 15 | 1.05 | 3.0 |
| 28 | 28 | 15 | 1.05 | 2.5 | ||
| 30 | 83 | 15 | 1.05 | 2.1 | ||
| 2 | 25 | 52 | 9 | 0.99 | 0.4 | |
| 3 | 26 | 21 | 16 | 0.74 | 0.8 | |
| 14 | 4 | 16 | 0.98 | 0.9 | ||
| 15 | 3 | 14 | 1.07 | 0.4 | ||
| 4 | 20 | 7 | 76 | 1.03 | 4.5 | |
| 21 | 6 | 77 | 1.02 | 4.5 | ||
| 20 | 4 | 77 | 1.02 | 4.5 | ||
| 20 | 4 | 76 | 0.97 | 4.4 | ||
| Mean±s.d. | 22±5‰ | |||||
| CN75 | 1 | 37 | 58 | 10 | 0.81 | 0.4 |
| 20 | 5 | 10 | 1.05 | 0.4 | ||
| 14 | NaNb | 9 | 1.07 | 0.5 | ||
| 2 | 23 | 20 | 16 | 0.90 | 0.8 | |
| 19 | 11 | 17 | 0.90 | 0.9 | ||
| 10 | 8 | 14 | 1.06 | 0.2 | ||
| Mean±s.d. | 21±10‰ | |||||
| CN150 | 1 | 16 | 3 | 25 | 1.04 | 0.4 |
| 20 | 1 | 25 | 1.03 | 0.6 | ||
| 2 | 18 | 4 | 46 | 0.94 | 3.4 | |
| 28 | 17 | 48 | 0.88 | 1.7 | ||
| 3 | 23 | 2 | 49 | 0.90 | 4.1 | |
| 28 | 4 | 50 | 0.88 | 4.5 | ||
| 22 | 4 | 52 | 0.85 | 3.7 | ||
| Mean ± s.d. | 22±5‰ |
Abbreviations: CI 95%, 95% confidence interval; [N+N], sum of the measured [NH4+] and [NO2−].
Approximate NH4+ oxidation rates were calculated from the change in [NO2−] and the time elapsed between the first and final timepoints, and may underestimate the actual rate in experiments where the final time point was after all NH4+ had been consumed.
Calculating a confidence interval was not possible for this experiment because only two points were available to calculate the slope.
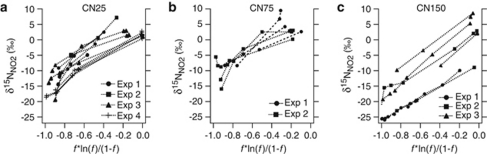
Results from replicate experiments are reported individually, as well as an unweighted average of experiments for a given enrichment (Table 2). 15ɛNH3 for CN25 ranged from 14 to 30‰ with an average of 22±5‰ (n=11), 15ɛNH3 for CN75 ranged from 10 to 37‰ with an average of 21±10‰ (n=6), and 15ɛNH3 for CN150 ranged from 16 to 28‰ with an average of 22±5‰ (n=7). Non-linearity was observed in most experiments with CN25 and CN75, with larger 15ɛNH3 (that is, greater slopes) at the beginning of the growth curve when a large fraction (f) of the initial NH4+ remained (Figures 5a and b). This leads to large uncertainties (expressed as 95% confidence intervals) in the slopes for many of the experiments with CN25 and CN75 (Table 2). We did not observe the same non-linearity of 15ɛNH3 in experiments with CN150 (Figure 5c).
Figure 5.
δ15NNO2 during ammonia oxidation in batch culture for the archaeal enrichments (a) CN25, (b) CN75 and (c) CN150. Kinetic isotope effects for ammonia oxidation (15ɛNH3) were calculated using linear regression of δ15NNO2 vs f * ln(f)/(1–f) as described in the text and are reported in Table 2. f is the fraction of the initial [NH4+] remaining in the culture.
Discussion
Phylogeny
We established three enrichments of marine archaea that stoichiometrically oxidize NH4+ to NO2−. Multiple lines of evidence suggest that the archaea are the active ammonia oxidizers in the enrichments. Archaeal cells increase exponentially concomitant with an exponential increase in NO2−, and amoA genes from the enrichments have a high identity to known archaeal ammonia oxidizers. The enrichments lack 16S rRNA and amoA genes associated with known γ- or β-proteobacterial ammonia oxidizers. Though no NO2− or NO3− production was observed in the enrichment from 500 m, this cannot be used to infer a lack of NH3-oxidizing ability in field populations at this depth, as active NH3 oxidation was measured in situ (Santoro et al., 2010).
Both 16S rRNA and amoA gene sequences from the CN strains suggest they are representative of organisms present and active in the open ocean. CN25 and CN75 are 97% identical to amoA sequences obtained in DNA-based clone libraries from the environment from which they were enriched (GU360825, (Santoro et al., 2010)) and CN150 is 100% identical to an amoA actively expressed in the environment (GU364088). The CN strains are also highly similar on the 16S and amoA level to abundant groups of open ocean archaea from a range of oceanic provinces including the Sargasso Sea, Fernandina Island and the coast of Africa as indicated by BLAST searches of the Global Ocean Survey dataset (Rusch et al., 2007). Previous amoA-based phylogenies of AOA have shown that water column-derived sequences fall into two clusters termed ‘A' and ‘B' (Francis et al., 2005) thought to represent a depth-dependent partitioning of AOA (Hallam et al., 2006; Mincer et al., 2007; Beman et al., 2008). The exact physiological basis for the partitioning of archaeal amoA genotypes is unknown, but could include adaptations to temperature, light or substrate availability. Based on amoA sequences, all three CN strains belong to the shallow water column clade ‘A'. Our data now associate a 16S rRNA genotype with at least some members of the clade ‘A' archaea that are distinct from N. maritimus and are likely to represent a new genus within the Thaumarchaea.
Physiology
The growth rate of CN25 and CN75 (0.17 day−1) in ONP medium is slower than growth rates reported for the cultivated AOA N. maritimus (0.65 day−1 at 30 °C, (Martens-Habbena et al., 2009) and N. yellowstonii (0.8 day−1 at 72 °C, (de la Torre et al., 2008) and cultivated marine ammonia-oxidizing bacteria (AOB) (0.34–0.77 day−1, (Prosser, 1990). These differences can be partially explained by the lower cultivation temperature of the CN enrichments (22 °C) relative to N. maritimus and N. yellowstonii. Further optimization of the culture medium may lead to increases in the growth rates reported here. The per-cell ammonia oxidation rate for the CN strains, ∼2 fmol NO2− cell−1 day−1, is lower than N. maritimus (13 fmol cell−1 day−1, (Martens-Habbena et al., 2009)), but similar to a mesocosm enrichment of archaea from North Atlantic (2–4 fmol cell−1 day−1)(Wuchter et al., 2006), and estimates of in situ per-cell activity rates in the California Current (0.2–15 fmol cell−1 day−1) (Santoro et al., 2010).
Multiple isolation strategies, including the addition of antibiotics and size fractionation through 0.45 μm filters, did not yield pure cultures of any of the CN strains. Dilution-to-extinction approaches were also not successful, as high dilutions of the CN strains did not grow (data not shown). N. yellowstonii and N. gargensis, the two cultivated thermophilic strains of AOA, also could not be established in pure culture (Hatzenpichler et al., 2008; de la Torre et al., 2008). This may be because there is a cooperative relationship between the bacteria and the archaea in these enrichment cultures, as has been reported in cultures of the marine chlorophyte Prochlorococcus (Morris et al., 2008), and low cell density dilutions that do not contain the associated heterotrophic bacteria are unable to grow. Interactions between the heterotrophic bacteria and autotrophic archaea in the enrichment, and in the ocean, will be an exciting area of future research.
We observed partial inhibition of archaeal ammonia-oxidizing activity by ATU at 86 μmol l−1, a concentration known to completely inhibit cultivated AOB (Hooper and Terry, 1973) and bacteria-rich environmental samples (Ginestet et al., 1998). Near complete inhibition of ammonia oxidation in CN25 was observed at 860 μmol l−1 ATU. The mechanism of ATU inhibition in AOB is thought to be chelation of the Cu active site in the Amo protein (Bedard and Knowles, 1989). Our results are consistent with observations of N. gargensis (Hatzenpichler et al., 2008) and support the idea that ATU-inhibited nitrification rates may represent conservative estimates of AOA-only activity in the field. Further, these results suggest that ATU should not be relied on for complete inhibition of nitrifying activity in environmental samples. For example, nitrification rate measurements in the California Current showed variable (0–90%) but on average low levels of inhibition by ATU (mean 35%, n=11) consistent with other lines of evidence supporting AOA activity in that study (Santoro et al., 2010). On the other hand, Lam et al., 2009 reported complete inhibition of ammonium oxidation in the presence of 86 μmol l−1 ATU in the Eastern Tropical South Pacific oxygen minimum zone. The physiology behind the differential responses of AOA and AOB and different environmental communities to this inhibitor is unknown, but could be the result of different metal active sites for enzymes in the ammonia oxidation pathway between the two groups of organisms or differences in metal active sites or affinities of enzymes, or differences in trace metal availability in different field locations.
Nitrogen isotopic fractionation
Multiple experiments with the three archaeal enrichments suggest a 15ɛNH3 for archaeal ammonia oxidation of approximately 22%. This value falls within the reported 15ɛNH3 for bacterial ammonia oxidation of 14% to 42% (Mariotti et al., 1981; Casciotti et al., 2003, 2010). Thus, despite genome-inferred differences in the ammonia oxidation pathways of AOA and AOB (Walker et al., 2010), a difference in the magnitude of 15ɛNH3 between these two groups was not apparent.
The physiological reasons for the large range in 15ɛNH3 for both AOA and AOB are unknown, but could result from changes in growth state of the organisms or violations of assumptions associated with using the Rayleigh accumulated product equation to estimate 15ɛNH3 from δ15NNO2 measurements. Factors to consider include variations in substrate concentration, ammonia oxidation rate and loss of N to unmeasured pools. Furthermore, unbalanced growth could lead to accumulation of intermediate products in the multi-step oxidation pathway, leading to larger errors involved with characterization of ammonia oxidation as a single-step unidirectional reaction. At high starting [NH4+], expressed 15ɛNH3 values were consistently around 20–22‰, whereas at low starting [NH4+], expressed 15ɛNH3 values were more variable (both higher and lower) and greater uncertainty was involved in their estimation (Table 2). Therefore, no direct relationship can be drawn between [NH4+] and 15ɛNH3. Similarly, there is no direct relationship between ammonia oxidation rate (or oxidation rate/[NH4+]) and 15ɛNH3 (Table 2).
Although dissolved inorganic nitrogen conservation ([NH4+ + NO2−]) was not 100% in all experiments, the majority of experiments had NO2− recoveries over 90% (Table 2), which is within the range reported for pure cultures of AOB (Mariotti et al., 1981; Casciotti et al., 2002). There is likely to be some loss of NH4+ because of uptake for anabolic metabolism by AOA and the bacteria in the enrichments, but this sink should be small and consistent across experiments. This process would alter observed 15ɛNH3 values in proportion to the amount of NH4+ assimilated and the isotope effect for NH4+ assimilation (4–27‰ (Hoch et al., 1992)). However, we emphasize that this effect is likely to be small and note that relatively high estimates of 15ɛNH3, not low ones, were observed where recovery was low (Table 2). We did not make measurements of [NO3−] in the experiments described here, but we have not observed NO2− loss over time and have no evidence that NO2− is further oxidized to NO3− in any of the enrichments.
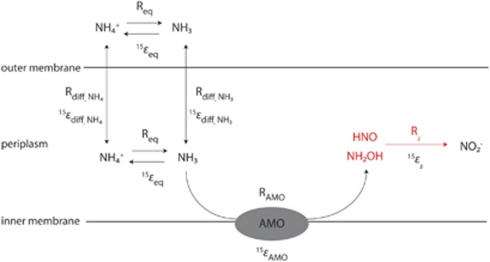
In addition to variation between experiments, we found evidence for variable 15ɛNH3 during archaeal ammonia oxidation within some experiments, with the greatest 15ɛNH3 expressed at the early stages of growth. This was particularly apparent for CN25 and CN75 enrichments grown at low [NH4+] (<20 μ), which exhibited a considerable lag phase before the commencement of ammonia oxidation. Variable ɛ values have been observed in many other organisms, including denitrifiers (Granger et al., 2008), methane oxidizers (Templeton et al., 2006), sulfate reducers (Habicht et al., 2005) and nitrite oxidizers (Casciotti, 2009; Buchwald and Casciotti, 2010), and during assimilation of NH4+ by heterotrophic bacteria (Hoch et al., 1992) but has not previously been reported in ammonia-oxidizing organisms. We explore two hypotheses that could explain a large apparent isotope effect at the early stages of growth (Figure 6).
Figure 6.
Potential pathways of NH3 acquisition and oxidation by AOA that could lead to a variable kinetic isotope effect (15ɛNH3) (Walker et al., 2010). Req and 15ɛeq are the rate and equilibrium isotope effect for NH3/NH4+ equilibration, Rdiff,NH4 and 15ɛdiff,NH4 are the rate and kinetic isotope effect for NH4+ diffusion, Rdiff,NH3 and 15ɛdiff,NH3 are the rate and kinetic isotope effect for NH3 diffusion, RAMO and 15ɛAMO are the rate and kinetic isotope effect for ammonia oxidation by ammonia monooxygenase (Amo), and R2 and 15ɛ2 are the rate and kinetic isotope effect for the second step of the ammonia oxidation pathway. Major uncertainties in the archaeal oxidation pathway are shown in red.
The first hypothesis is that, in the early stages of growth in batch culture, diffusion of NH3 and/or NH4+ between the growth medium and the periplasm is rapid relative to the oxidation rate (Rdiff,NH3 or Rdiff,NH4 > RAMO) and enzymatic ammonia oxidation is the rate-limiting step. In this scenario, the enzyme-level isotope effect (15ɛAMO), plus the equilibrium isotope effect between NH3 and NH4+ (15ɛeq = 19‰ (Hermes et al., 1985), should dominate the observed 15ɛNH3 early in the experiment. In the later stages of growth, the reaction may become diffusion limited (Rdiff <RAMO) and the isotope effect for diffusion (15ɛdiff) would dominate. A similar mechanism has been examined for variations in expressed isotopic fractionation during NH4+ assimilation by bacteria (Hoch et al., 1992) and marine algae (Pennock et al., 1996), as well as CO2 uptake by algae (Laws et al., 1997) and plants (O'Leary, 1981). Diffusion of NH3 in aqueous solution is estimated to have an isotope effect of 20‰ (Hoch et al., 1992), which is similar to the late-stage 15ɛNH3 values we observed. However, this scenario would predict a direct relationship between [NH4+] and 15ɛNH3 in experiments starting at different initial NH4+ concentrations, which is not observed in our data (Table 2). If anything, we observe larger 15ɛNH3 values at low initial [NH4+]. Therefore, we reject the hypothesis of diffusion limitation in controlling 15ɛNH3 in these experiments.
It has been assumed here that the Amo-catalyzed reaction occurs in the periplasm, as it does in AOB, and that active transport of NH3 or NH4+ into the periplasm from the medium does not occur. Genes for putative Amt transporters have been identified in the genomes of both AOB (Arp et al., 2007) and AOA (Hallam et al., 2006; Walker et al., 2010; Blainey et al., 2011) and expression of these genes in environmental samples appears proportional to expression of amo genes (Stewart et al., 2011). At this time it is uncertain whether Amt supplies N for anabolic metabolism serves a regulatory function (Arp et al., 2007), or supplies substrate to the Amo enzyme. However, if Amt is localized for transport across the cytoplasmic membrane (Andrade et al., 2005), it would be difficult to envision a role for Amt in the 15ɛNH3 variations observed here.
A second hypothesis to explain the within-experiment variations in 15ɛNH3 is that during early exponential phase growth, the first step of ammonia oxidation proceeds more quickly than the second step (RAMO>R2) and an as-yet unknown intermediate (NH2OH or HNO) accumulates. This intermediate pool would be isotopically depleted relative to NH4+ outside the cell, and a second isotope effect would be expressed (15ɛ2; Figure 6) resulting in the production of NO2− with a large apparent isotope effect. If the intermediate pool is eventually used up and ammonia oxidation proceeds as a pseudo one-step reaction, the apparent isotope effect should decrease to 15ɛAMO (plus 15ɛeq) late in the experiment. This hypothesis is consistent with the fact that curvature in the Rayleigh plots was most prominent where long lag phases preceded ammonia oxidation, which may be associated with imbalanced growth early in the experiment. Interestingly, if this hypothesis is correct, the similarity between average 15ɛNH3 (22‰) and 15ɛeq (19‰); (Hermes et al., 1985) may indicate a small kinetic isotope effect for Amo. Use of the Rayleigh distillation equation with measurements of δ15NNH4 would get around the potential problem from accumulation of intermediates, although it would still be affected by NH3/NH4+ equilibration and NH4+ transport effects discussed above. At this time there are many additional uncertainties in the archaeal ammonia oxidation pathway (Klotz and Stein, 2008; Walker et al., 2010) including the exact chemical intermediates, the localization of the respiratory enzymes within the cell, and the role of active NH3/NH4+ transport that must be resolved before the mechanisms causing variable 15ɛNH3 can be fully explained.
The ultimate goal of determining species-level isotope effects is to better interpret natural abundance stable isotope ratios (δ15NNO3 and δ18ONO3) in the environment. The average δ15N of marine NO3− is set by the balance of N2 fixation and N removal by denitrification and anammox (Brandes and Devol, 2002), although nitrification can affect the partitioning of 15N between dissolved and particulate N (Wankel et al., 2007). The potential for variable 15ɛNH3 adds an extra level of complexity to the interpretations of δ15NNO3 in the environment.
Nitrification is also the main determinant in setting the average δ18O of marine NO3− (Casciotti et al., 2002; Sigman et al., 2009) and N2O (Ostrom et al., 2000; Popp et al., 2002). Determining the oxygen isotope effects for archaeal ammonia oxidation will therefore be an important next step in advancing the use of the dual isotope signatures of NO3− and N2O to understand the marine nitrogen cycle.
Acknowledgments
This work was supported by a Woods Hole Oceanographic Institution (WHOI) Postdoctoral Scholar fellowship to AES and the WHOI Ocean Life Institute. We thank Josh Plant and Ken Johnson for the NH4+ concentration data from cruise CN107, and Christopher Francis for logistical support during that cruise. Matt McIlvin is acknowledged for expert technical assistance with mass spectrometry. Cornelia Wuchter is acknowledged for assistance with the CARD-FISH procedure. Louis Kerr at the Marine Biological Laboratory microscopy facility assisted with the fluorescence micrographs. We thank Stefan Sievert and Matt Johnson for access to equipment in their laboratories. Carolyn Buchwald, Edward Leadbetter and three anonymous reviewers provided invaluable comments on the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andrade SLA, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2005;102:14994. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arp DJ, Chain PSG, Klotz MG. The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Microbiology. 2007;61:503. doi: 10.1146/annurev.micro.61.080706.093449. [DOI] [PubMed] [Google Scholar]
- Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens—re-evaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard C, Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Mol Biol Rev. 1989;53:68. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- Beman JM, Sachdeva R, Fuhrman JA. Population ecology of nitrifying Archaea and Bacteria in the Southern California Bight. Environ Microbiol. 2010;12:1282–1292. doi: 10.1111/j.1462-2920.2010.02172.x. [DOI] [PubMed] [Google Scholar]
- Bernhard AE, Landry ZC, Blevins A, de la Torre JR, Giblin AE, Stahl DA. Abundance of ammonia-oxidizing archaea and bacteria along an estuarine salinity gradient in relationship to potential nitrification rates. Appl Environ Microbiol. 2010;76:1285–1289. doi: 10.1128/AEM.02018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR, Gilbert J. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One. 2011;6:148–149. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes JA, Devol AH. A global marine-fixed nitrogen isotopic budget: implications for Holocene nitrogen cycling. Global Biogeochem Cycles. 2002;16:1120. [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- Buchwald C, Casciotti KL. Oxygen isotopic fractionation and exchange during bacterial nitrite oxidation. Limnol Oceanogr. 2010;55:1064–1074. [Google Scholar]
- Casciotti KL. Inverse kinetic isotope fractionation during bacterial nitrite oxidation. Geochimica Et Cosmochimica Acta. 2009;73:2061–2076. [Google Scholar]
- Casciotti KL, Bohlke JK, McIlvin MR, Mroczkowski SJ, Hannon JE. Oxygen isotopes in nitrite: analysis, calibration, and equilibration. Anal Chem. 2007;79:2427–2436. doi: 10.1021/ac061598h. [DOI] [PubMed] [Google Scholar]
- Casciotti KL, Buchwald C, Santoro AE, Frame C. Assessment of nitrogen and oxygen isotope fraction during nitrification and its expression in the marine environment. Methods Enzymol. 2011;486:253–280. doi: 10.1016/B978-0-12-381294-0.00011-0. [DOI] [PubMed] [Google Scholar]
- Casciotti KL, McIlvin MR, Buchwald C. Oxygen isotopic exchange and fractionation during bacterial ammonia oxidation. Limnol Oceanogr. 2010;55:753–762. [Google Scholar]
- Casciotti KL, Sigman DM, Hastings MG, Bohlke JK, Hilkert A. Measurement of the oxygen isotopic composition of nitrate in seawater and freshwater using the denitrifier method. Anal Chem. 2002;74:4905–4912. doi: 10.1021/ac020113w. [DOI] [PubMed] [Google Scholar]
- Casciotti KL, Sigman DM, Ward BB. Linking diversity and stable isotope fractionation in ammonia-oxidizing bacteria. Geomicrobiology Journal. 2003;20:335–353. [Google Scholar]
- Casciotti KL, Trull TW, Glover DM, Davies D. Constraints on nitrogen cycling at the subtropical North Pacific Station ALOHA from isotopic measurements of nitrate and particulate nitrogen. Deep Sea Res. 2008;55:1661–1672. [Google Scholar]
- Church MJ, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DR, Rees AP, Joint I. Ammonium regeneration and nitrification rates in the oligotrophic Atlantic Ocean: Implications for new production estimates. Limnol Oceanogr. 2008;53:52–62. [Google Scholar]
- Coolen MJL, Abbas B, van Bleijswijk J, Hopmans EC, Kuypers MMM, Wakeham SG, et al. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ Microbiol. 2007;9:1001–1016. doi: 10.1111/j.1462-2920.2006.01227.x. [DOI] [PubMed] [Google Scholar]
- de la Torre JR, Walker CB, Ingalls AE, Koenneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- Delong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiore PJ, Sigman DM, Dunbar RB. Upper ocean nitrogen fluxes in the Polar Antarctic Zone: constraints from the nitrogen and oxygen isotopes of nitrate. Geochemistry Geophysics Geosystems. 2009;10:Q11016. [Google Scholar]
- Eppley RW, Peterson BJ. Particulate organic matter flux and planktonic new production in the deep ocean. Nature. 1979;282:677. [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestet P, Audic JM, Urbain V, Block JC. Estimation of nitrifying bacterial activities by measuring oxygen uptake in the presence of the metabolic inhibitors allylthiourea and azide. Appl Environ Microbiol. 1998;64:2266–2268. doi: 10.1128/aem.64.6.2266-2268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger J, Sigman DM, Lehmann MF, Tortell PD. Nitrogen and oxygen isotope fractionation during dissimilatory nitrate reduction by denitrifying bacteria. Limnol Oceanogr. 2008;53:2533–2545. [Google Scholar]
- Granger J, Sigman DM, Needoba JA, Harrison PJ. Coupled nitrogen and oxygen isotope fractionation of nitrate during assimilation by cultures of marine phytoplankton. Limnol Oceanogr. 2004;49:1763–1773. [Google Scholar]
- Habicht K, Salling L, Thamdrup B, Canfield D. Effect of low sulfate concentrations on lactate oxidation and isotope fractionation during sulfate reduction by Archaeoglobus fulgidus strain Z. Appl Environ Microbiol. 2005;71:3770. doi: 10.1128/AEM.71.7.3770-3777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:520–536. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R, Lebecleva EV, Spieck E, Stoecker K, Richter A, Daims H, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes JD, Weiss PM, Cleland W. Use of nitrogen-15 and deuterium isotope effects to determine the chemical mechanism of phenylalanine ammonia-lyase. Biochemistry. 1985;24:2959–2967. doi: 10.1021/bi00333a023. [DOI] [PubMed] [Google Scholar]
- Hoch MP, Fogel ML, Kirchman DL. Isotope fractionation associated with ammonium uptake by a marine bacterium. Limnol Oceanogr. 1992;37:1447–1459. [Google Scholar]
- Holmes RM, Aminot A, Kèrouel R, Hooker BA, Peterson BJ. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J Fish Aquat Sci. 1999;56:1801–1808. [Google Scholar]
- Hooper AB, Terry KR. Specific inhibitors of ammonia oxidation in Nitrosomonas. J Bacteriol. 1973;115:480. doi: 10.1128/jb.115.2.480-485.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman MR, Wood P. Suicidal inactivation and labelling of ammonia mono-oxygenase by acetylene. Biochem J. 1985;227:719. doi: 10.1042/bj2270719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KS, Boyle E, Bruland K, Coale K, Measures C, Moffett J, et al. The SAFe iron intercomparison cruise: an international collaboration to develop dissolved iron in seawater standards. EOS Trans Am Geophysical Union. 2007;88:131–132. [Google Scholar]
- Klotz MG, Stein LY. Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol Lett. 2008;278:146–156. doi: 10.1111/j.1574-6968.2007.00970.x. [DOI] [PubMed] [Google Scholar]
- Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA. 2007;104:7104–7109. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Lavik G, Jensen MM, van de Vossenberg J, Schmid M, Woebken D, et al. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci USA. 2009;106:4752. doi: 10.1073/pnas.0812444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws EA, Bidigare RR, Popp BN. Effect of growth rate and CO 2 concentration on carbon isotopic fractionation by the marine diatom Phaeodactylum tricornutum. Limnol Oceanogr. 1997;42:1552–1560. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar , et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti A, Germon JC, Hubert P, Kaiser P, Letolle R, Tardieux A, et al. Experimental determination of nitrogen kinetic isotope fractionation: some principles; illustration for the denitrification and nitrification processes. Plant Soil. 1981;62:413–430. [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–981. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- McIlvin MR, Altabet MA. Chemical conversion of nitrate and nitrite to nitrous oxide for nitrogen and oxygen isotopic analysis in freshwater and seawater. Anal Chem. 2005;77:5589–5595. doi: 10.1021/ac050528s. [DOI] [PubMed] [Google Scholar]
- McIlvin MR, Casciotti KL. Fully automated system for stable isotopic analysis of dissolved nitrous oxide at natural abundance levels. Limnol Oceanograph Methods. 2010;8:54–66. [Google Scholar]
- McIlvin MR, Casciotti KL. Technical updates to the bacterial method for nitrate isotopic analyses. Anal Chem. 2011;83:1850–1856. doi: 10.1021/ac1028984. [DOI] [PubMed] [Google Scholar]
- Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by ‘helper' heterotrophic bacteria. Appl Environ Microbiol. 2008;74:4530. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosier AC, Francis CA. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol. 2008;10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- Needoba JA, Waser NA, Harrison PJ, Calvert SE. Nitrogen isotope fractionation in 12 species of marine phytoplankton during growth on nitrate. Marine Ecol Prog Ser. 2003;255:81–91. [Google Scholar]
- O'Leary MH. Carbon isotope fractionation in plants. Phytochemistry. 1981;20:553–567. [Google Scholar]
- Olson RJ. 15 N tracer studies of the primary nitrite maximum. J Mar Res. 1981;39:203–226. [Google Scholar]
- Ostrom NE, Russ ME, Popp BN, Rust TM, Karl DM. Mechanisms of nitrous oxide production in the subtropical North Pacific based on determinations of the isotopic abundances of nitrous oxide and di-oxygen. Chemosphere-Global Change Sci. 2000;2:281–290. [Google Scholar]
- Pennock JR, Velinsky DJ, Ludlam JM, Sharp JH, Fogel ML. Isotopic fractionation of ammonium and nitrate during uptake by Skeletonema costatum: implications for 15N dynamics under bloom conditions. Limnol Oceanogr. 1996;41:451–459. [Google Scholar]
- Popp BN, Westley MB, Toyoda S, Miwa T, Dore JE, Yoshida N, et al. Nitrogen and oxygen isotopomeric constraints on the origins and sea-to-air flux of N2O in the oligotrophic subtropical North Pacific gyre. Global Biogeochem Cycles. 2002;16:1064. [Google Scholar]
- Prosser JI. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1990;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through Eastern Tropical Pacific. Plos Biology. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro AE, Casciotti KL, Francis CA. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol. 2010;12:1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- Santoro AE, Francis CA, de Sieyes NR, Boehm AB. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ Microbiol. 2008;10:1068–1079. doi: 10.1111/j.1462-2920.2007.01547.x. [DOI] [PubMed] [Google Scholar]
- Sigman DM, DiFiore PJ, Hain MP, Deutsch C, Wang Y, Karl DM, et al. The dual isotopes of deep nitrate as a constraint on the cycle and budget of oceanic fixed nitrogen. Deep Sea Research Part I: Oceanographic Research Papers. 2009;56:1419–1439. [Google Scholar]
- Sigman DM, Granger J, DiFiore PJ, Lehmann MF, Ho R, Cane G, et al. Coupled nitrogen and oxygen isotope measurements of nitrate along the eastern North Pacific margin. Global Biogeochem Cycles. 2005;19:GB4022. [Google Scholar]
- Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Stein JL, Marsh TL, Wu KY, Shizuya H, DeLong EF. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996;178:591. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen JR, Chang YJ, Macnaughton SJ, Kowalchuk GA, Leung KT, Flemming CA, et al. Effect of toxic metals on indigenous soil beta-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol. 1999;65:95–101. doi: 10.1128/aem.65.1.95-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart FJ, Ulloa O, DeLong EF.2011Microbial metatranscriptomics in a permanent marine oxygen minimum zone Environ Microbioldoi: 10.1111/j.1462-2920.2010.02400 [DOI] [PubMed]
- Strickland J, Parsons T. A practical handbook of seawater analysis. Fish Res Board Can Bull. 1968;167:71–75. [Google Scholar]
- Teira E, Reinthaler T, Pernthaler A, Pernthaler J, Herndl GJ. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl Environ Microbiol. 2004;70:4411. doi: 10.1128/AEM.70.7.4411-4414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AS, Chu KH, Alvarez-Cohen L, Conrad ME. Variable carbon isotope fractionation expressed by aerobic CH4-oxidizing bacteria. Geochimica Et Cosmochimica Acta. 2006;70:1739–1752. [Google Scholar]
- Urakawa H, Martens-Habbena W, Stahl DA. High abundance of ammonia-oxidizing archaea in coastal waters, determined using a modified DNA extraction method. Appl Environ Microbiol. 2010;76:2129. doi: 10.1128/AEM.02692-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wankel SD, Kendall C, Pennington JT, Chavez FP, Paytan A. Nitrification in the euphotic zone as evidenced by nitrate dual isotopic composition: Observations from Monterey Bay, California. Global Biogeochemical Cycles. 2007;21:GB2009. [Google Scholar]
- Ward BB.2002Nitrification in aquatic environmentsIn: Capone DG (ed).Encyclopedia of Environmental Microbiology John Wiley & Sons: New York; 2144–2167. [Google Scholar]
- Ward BB, Olson RJ, Perry MJ. Microbial nitrification rates in the primary nitrite maximum off Southern-California. Deep-Sea Res A-Oceanograph Res Papers. 1982;29:247–255. [Google Scholar]
- Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yool A, Martin AP, Fernandez C, Clark DR. The significance of nitrification for oceanic new production. Nature. 2007;447:999–1002. doi: 10.1038/nature05885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.