Abstract
Macroautophagy (referred to hereafter as autophagy) is a highly regulated cellular process that serves to remove damaged proteins and organelles from the cell. Autophagy contributes to an array of normal and pathological processes, and has recently emerged as a key regulator of multiple aspects of cancer biology. The role of autophagy in cancer is complex and is likely dependent on tumor type, stage, and genetic context. This complexity is illustrated by the identification of settings where autophagy acts potently to either promote or inhibit tumorigenesis. In this review, I discuss the underlying basis for these opposing functions and propose a model suggesting a dynamic role for autophagy in malignancy. Collectively, the data point to autophagy as serving as a barrier to limit tumor initiation. Once neoplastic lesions are established, it appears that adaptive changes occur that now result in positive roles for autophagy in malignant progression and in subsequent tumor maintenance. Remarkably, constitutive activation of autophagy is critical for continued growth of some tumors, serving to both reduce oxidative stress and provide key intermediates to sustain cell metabolism. Autophagy is also induced in response to cancer therapies where it can function as a survival mechanism that limits drug efficacy. These findings have inspired significant interest in applying anti-autophagy therapies as an entirely new approach to cancer treatment. It is now apparent that aberrant control of autophagy is among the key hallmarks of cancer. While much needs to be learned about the regulation and context-dependent biological functions of autophagy, it seems clear that modulation of this process will be an attractive avenue for future cancer therapeutic approaches.
Keywords: autophagy, cancer, tumor, metabolism, therapy
The evolution of advanced cancers from normal cells
As advanced cancers evolve, many fundamental changes in basic cellular processes are required for the initiation and maintenance of the transformed state. Discoveries in cancer biology over the past 20 years have identified an increasing list of these essential hallmarks of cancer. These include modulation of key aspects of cancer cell physiology, such as apoptosis and cell cycle checkpoint control, as well as interactions with the surrounding microenvironment (immune modulation and angiogenesis) (Hanahan and Weinberg 2011). Recent discoveries have pointed to deregulation of autophagy as a novel feature that is central to the pathogenesis of human malignancy. Importantly, autophagy intersects at multiple levels with cell metabolism, protein and organelle turnover, cell survival, and other cellular functions, suggesting that the role of autophagy in cancer is both dynamic and highly complex.
Autophagy: the process and its machinery
The capacity to turn over proteins and higher-order structures, such as organelles, is central to the homeostasis of cells. For example, inflammatory, developmental, and proliferative signals are linked to protein degradation as a means to acutely turn on and off signal transduction pathways that alter the cell state. These rapid changes are mainly regulated by the well-known ubiquitin/proteasome system (Ciechanover 2005). Long-lived proteins and organelles also need to be periodically degraded, as they can accumulate damage (e.g., oxidative changes) that render them nonfunctional or toxic to the cell. Macroautophagy (referred to hereafter as autophagy) is a regulated and evolutionarily conserved catabolic process that serves to degrade these structures and recycle their biochemical components for use in energy production and other biosynthetic reactions (Levine and Kroemer 2008; Yang and Klionsky 2010b). Autophagy was first discovered >40 years ago in mammalian cells as a process of self-digestion and was initially thought to be a nonspecific bulk degradation process (De Duve and Wattiaux 1966; Klionsky 2008; Yang and Klionsky 2010a). The relevance to stress responses has been borne out in subsequent studies, and it is now apparent that autophagy can serve a multitude of critical functions, including cellular quality control, tissue homeostasis, and energy production (Mizushima et al. 2008). Additionally, it is clear that there is some cargo specificity, in particular for organelles such as the mitochondria and endoplasmic reticulum (ER). In contrast, the ubiquitin/proteasome system typically mediates the regulated degradation of soluble and short-lived proteins, does not degrade organelles, and does not involve the lysosome. Recent work has elucidated significant cross-talk between the two systems, and this is an active area of research (Wong and Cuervo 2010).
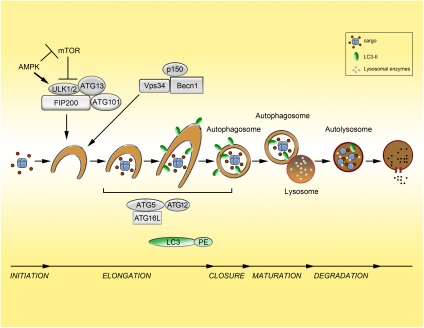
Autophagy begins with the formation of a double-membrane vesicle (known as an autophagosome) around the cargo macromolecules. The cargo can consist of entire organelles, protein aggregates, nucleic acids, and lipids, as well as pathogens such as Salmonella and Mycobacterium. Autophagosomes fuse with lysosomes to form autolysosomes where lysosomal enzymes degrade the cargo (Fig. 1). The end products of this digestive process are basic molecular building blocks such as amino acids, fatty acids, and nucleotides, which are released back into the cytoplasm by lysosomal permeases. The formed autophagosome can also fuse to the endosomal compartment (early and late endosomes; multivesicular bodies) prior to fusing to lysosomes, adding further complexity to the process (Liou et al. 1997; Razi et al. 2009). In addition to macroautophagy, which is the subject of this review, there are also two additional types of autophagy that differ in function as well as in how cargo is delivered to the lysosome:microautophagy (Mijaljica et al. 2011) and chaperone-mediated autophagy (Arias and Cuervo 2011), which will not be discussed further and have been reviewed elsewhere.
Figure 1.
The process and regulation of autophagy. The stages of autophagy (initiation, elongation, closure, maturation, and degradation) are depicted. Cargo is sequestered in a double-membrane vesicle that eventually forms the autophagosome. This fuses to the lysosome (autolysosome), where the cargo is degraded by lysosomal enzymes and degradation products are recycled back into the cytosol by lysosomal permeases. mTOR is a key regulator of autophagy in response to changes in nutrient availability. During nutrient-replete conditions, mTOR is activated and autophagy is inhibited through repression of ULK1/2 (the mammalian homologs of ATG1). Upon nutrient depletion, ULK1/2 is activated and can promote autophagy initiation. ULK is also activated in states of low energy (increased AMP/ATP ratio) by phosphorylation by AMPK as well as repression of mTORC1. Also critical to autophagy initiation is the production of phosphatidylinositol-3-phosphate (PI3P) by the class III PI3K Vps34, which is in a complex with ATG6/Beclin1 and p150 (Vps15). This complex is an additional level of regulation and, depending on the particular proteins bound, can activate or repress Vps34 activity. The ATG5–ATG12 complex as well as LC3 conjugated to PE, known as LC3-II, act downstream from Vps34 and ULK1/2 and, along with other proteins, have roles in autophagosome membrane elongation. LC3 acts downstream from the ATG5–ATG12 system and is present on the outer and inner surfaces of the autophagosome (depicted as a green oval). LC3 is a commonly used marker to monitor autophagosomes.
The autophagic process and machinery are complex and are the subject of multiple excellent reviews to which I direct the readers for reference (He and Klionsky 2009; Klionsky et al. 2010; Yang and Klionsky 2010b). In brief, the process consists of five basic phases: initiation, elongation, closure, maturation, and degradation (Fig. 1). Much of the core molecular machinery responsible for autophagosome formation has been identified over the past decade. The pathway shows strong evolutionary conservation from yeast to humans. A key set of autophagy-related genes (ATGs), which are essential for the process, were identified in a series of seminal studies in the yeast system and have moved the field into the molecular era (Nakatogawa et al. 2009). Subsequent studies have identified many of the mammalian orthologs, allowing for detailed genetic and biochemical studies in higher eukaryotes (Mizushima et al. 1998; Kabeya et al. 2000).
The core machinery can be divided into four subgroups (Yang and Klionsky 2010b) that regulate successive steps of the autophagic process. The ATG1/unc-51-like kinase (ULK) complex, including ATG13 and the scaffold FIP200 (presumed ortholog of yeast ATG17), and a second subgroup containing a complex comprised of Vps34, a class III phosphatidylinositol-3-kinase (PI3K), and ATG6/Beclin1 (Simonsen and Tooze 2009) both regulate the initiation phases of autophagy. A third subgroup regulates later steps of autophagy, autophagosome elongation, and expansion, and is characterized by two ubiquitin-like proteins: LC3 (also known as ATG8/microtubule-associated protein 1 light chain 3) and ATG12 (Geng and Klionsky 2008). LC3 becomes inserted into the inner and outer membranes of the autophagosome and has been used extensively to monitor autophagy (see below). ATG12 is conjugated to ATG5 by the ATG7 and ATG10 proteins that resemble the E1 and E2 components of the ubiquitin system, respectively (Ichimura et al. 2000). This ATG5–ATG12 conjugate interacts with ATG16L, and this complex then promotes LC3 lipidation (Hanada et al. 2007; Fujita et al. 2008). Last, the fourth subgroup contains transmembrane proteins such as ATG9 and VMP1 (Vacuole membrane protein 1) and is also critical for autophagy. The function of ATG9 in mammalian autophagy is not entirely defined, but it may have a role in the delivery of membrane to the autophagosome (Yang and Klionsky 2010b). VMP1 has been shown to interact with Becn1 and is required for autophagy, while its overexpression can actually induce autophagy (Ropolo et al. 2007).
Levels of autophagy are under the tight control of multiple signal transduction pathways that link this process to a variety of environmental cues (Levine and Kroemer 2008; Kroemer et al. 2010; Levine et al. 2011). Typically, autophagy is present at low levels in cells and is important for housekeeping functions such as protein and organelle quality control. Nutrient deprivation is among the best-characterized inducers of autophagy. This is largely dependent on mTOR (mammalian target of rapamycin) signaling (Neufeld 2010), in particular on mTORC1 (mTOR complex 1). mTORC1 is an important nutrient sensor whose kinase activity drives biosynthetic pathways and cell growth when cellular energy and amino acid levels are plentiful. In such a high-nutrient state, mTOR binds and phosphorylates the ULK1 complex, blocking its kinase activity and thereby inhibiting autophagy initiation (Fig. 1; Jung et al. 2010). Interestingly, there is additional spatial regulation of mTOR, as it is recruited to the lysosomal surface upon activation as well as to compartments containing autolysosomes (Sancak et al. 2010; Narita et al. 2011). Energy charge (ATP:AMP ratio) has also been shown to be a further level of autophagy regulation. Specifically, the AMP-activated protein kinase (AMPK), which is stimulated by energy stress, promotes autophagy by both acting as a negative regulator of mTORC1 (Gwinn et al. 2008) and directly phosphorylating ULK1 at sites that lead to enhanced catalytic activity (Egan et al. 2011; J Kim et al. 2011).
An additional level of autophagy regulation occurs at the Beclin1/Vps34 complex, of which several distinct forms exist (Sinha and Levine 2008; Simonsen and Tooze 2009). When AMBRA (activating molecule in Beclin-1-regulated autophagy) and ATG14L (Barkor) are bound to the complex—or, alternatively, UVRAG (ultraviolet irradiation resistance-associated gene) is bound—autophagy is stimulated. However, if Rubicon (RUN domain and cysteine-rich domain containing Beclin-1-interacting protein) is bound in the complex with UVRAG, autophagy is inhibited.
Other stimuli, in addition to nutrient deprivation, have been shown to activate autophagy, such as anti-cancer therapies (chemotherapy, radiation therapy, targeted agents) (Amaravadi et al. 2011), reactive oxygen species (ROS) (Scherz-Shouval and Elazar 2007, 2011), ER stress, the unfolded protein response (Buchberger et al. 2010), and ammonia (Eng et al. 2010; Cheong et al. 2011). Whether the autophagy induced by each of these stimuli is somehow distinct is not clear. In this regard, an important area of future investigation will be to determine whether cargo selection, engagement of specific autophagic machinery, or other aspects of the process differ depending on the particular stimulus.
Measuring autophagy
The ability to accurately measure autophagy is essential for efforts to define the contributions of this process to normal cell physiology as well as in cancer. The measurement of autophagy has been a topic of intense discussion, and there are several comprehensive reviews on the subject as well a variety of published guidelines (Mizushima 2004; Klionsky et al. 2008; Mizushima et al. 2010). As is the case for any dynamic process, the difficulty occurs in the attempt to develop static or indirect measurements as accurate surrogates for true pathway activity, which can be defined as delivery of a substrate to and degradation within the lysosome. Classically, electron microscopy (EM) has been the standard approach to detect autophagy by the morphological identification of autophagosomes. Although well established, it is quite costly and not readily accessible. Many newer assays are based on LC3 (ATG8). This protein is first cleaved by ATG4 to generate LC3-I and then lipidated to produce LC3-II (Ichimura et al. 2000). The conjugation to phosphatidylethanolamine (PE) occurs at a C-terminal glycine and is mediated by the E1-like ATG7 and E2-like ATG3 (Geng and Klionsky 2008). The lipidated (LC3-II) form can be detected as a faster-migrating band by immunoblotting (Mizushima and Yoshimori 2007). As LC3-II is incorporated into the inner and outer surfaces of autophagosomes, the expression of a green fluorescence protein (GFP) LC3 fusion protein can be used to identify GFP puncta or dots representing autophagosomes. Each of these approaches suffers from the limitation of a static measurement, rather than a measurement of the process (i.e., autophagosome degradation). For example, an elevation of GFP-LC3 puncta or LC3-II levels can be due to either a true increase in autophagy or, alternatively, a block late in the process at the level of degradation. Thus, while these assays provide useful information, additional data are needed to confirm autophagy activation. This is true even of using EM to identify the presence of autophagosomes, which, as mentioned previously, has been a standard approach.
True determination of autophagic activity is obtained by assessment of autophagic flux, a dynamic measurement of the passage of substrates through the autophagic pathway. Long-term protein degradation has been used classically as an indirect measurement of autophagy, as long-lived proteins are degraded by autophagy. The results from this assay must be interpreted with caution, as there are other factors that can influence the degradation of proteins in addition to autophagy. Thus, it is most useful as an estimate for relative flux when comparing the impact of perturbation within a single cell type. Flux can also be measured by blocking lysosomal degradation of autophagosomes—the last stage of autophagy—using drugs such as chloroquine (CQ), Bafilomycin A1, or lysosomal protease inhibitors. This results in a buildup of autophagy intermediates; the increase in inhibited cells compared with basal levels reflects flux. This concept can be applied via immunoblotting for LC3-II or immunofluorescence detection of GFP-LC3 puncta under basal and inhibited conditions. p62/SQSTM1 has multiple critical functions and is ultimately degraded by autophagy, so serially monitoring levels of p62 may also be considered a relative measure of flux (Bjorkoy et al. 2009). Several novel LC3-based reporters have been developed to measure flux, including the tandem fluorescence LC3 reporter. This reporter has GFP–RFP fused to the LC3 protein (Kimura et al. 2007). Because GFP fluorescence is pH-labile, autolysosomes will contain only red puncta, while autophagosomes will have both colors and appear yellow. The ratio of red to yellow puncta is a measure of flux.
Thus, unless flux is measured, it is difficult to make definitive conclusions about the levels of autophagy in a particular system. Unfortunately, it is often a challenge to assess autophagic flux in vivo. This is highly relevant in the clinical application of drugs that target autophagy, since accurate determination of autophagy levels are likely to be important as a biomarker for patient selection as well as evaluation of biochemical effects (pharmacodynamics).
Autophagy and early tumorigenesis
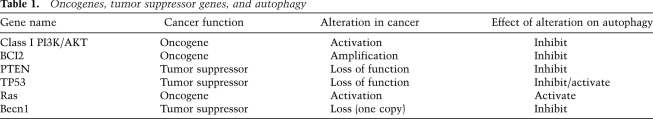
Autophagy was initially considered a process that suppressed malignant transformation. There is indirect evidence of a tumor-suppressive role for autophagy when considering the spectrum of oncogene and tumor suppressor gene alterations in human malignancy (Table 1; Maiuri et al. 2009). Activation of the PI3K/Akt pathway via activating PI3K mutations, AKT amplifications, or Pten loss leads to decreased autophagy in many settings largely through mTOR activation (Guertin and Sabatini 2007; Diaz-Troya et al. 2008). Other common alterations in human tumors that potentially inhibit autophagy include Bcl-2 amplification/overexpression. In addition to inhibiting apoptosis, Bcl-2 has been reported in some circumstances to inhibit autophagy through an inhibitory binding to Becn1 (Maiuri et al. 2007; Sinha and Levine 2008). The tumor suppressor p53, one of the most commonly altered genes in human malignancy, has what appears to be opposing roles in autophagy. Activation of p53 by nutrient deprivation or genotoxic stress leads to activation of autophagy (Balaburski et al. 2010). This can be through inhibition of mTOR (Feng et al. 2005) or by activation of DRAM (damage-regulated autophagy modulator) (Crighton et al. 2006). Functional loss of p53 would therefore be expected to lead to decreased autophagy, consistent with the role of autophagy as tumor suppressor. In contrast, evidence from the Kroemer group (Tasdemir et al. 2008) also suggests that p53 can suppress autophagy in the basal state. In this case, the suppression of basal autophagy was dependent on the cytoplasmic, not the nuclear, pool of p53 (Tasdemir et al. 2008).
Table 1.
Oncogenes, tumor suppressor genes, and autophagy
More direct evidence of the tumor-suppressing properties of autophagy was provided through the work of the Levine laboratory (Liang et al. 1999). The identification of Beclin1 as the mammalian ortholog of yeast ATG6 and subsequent discovery that a large fraction of breast and ovarian cancers have single copy loss of this gene suggested that intact autophagy may be constraining tumor initiation (Liang et al. 1999). The functional importance of this loss was demonstrated in the Becn1 knockout mouse, where the heterozygotes were predisposed to a variety of tumors (Qu et al. 2003; Yue et al. 2003). The wild-type Becn1 allele was typically intact in both the mouse and human tumors, confirming that is a haploinsufficient tumor suppressor gene. In fact, while cells from the heterozygous mice have significantly decreased levels of autophagy compared with wild-type mice, they still have appreciable autophagic activity. These mice developed a mixture of benign as well as malignant tumors, indicating that in the context of Becn1 heterozygosity with reduced autophagy, tumors can fully progress. In a related fashion, the Mizushima laboratory (Takamura et al. 2011) has recently created a mouse with mosaic deletion of Atg5 in all tissues. This predisposed homozygous mice to liver tumors with high penetrance, with no tumors detected in other organs. Conditional deletion of Atg7 in the liver also resulted in benign liver tumors. The Komatsu laboratory (Inami et al. 2011) had similar results with conditional deletion of Atg7 in the liver. Importantly, tumors in both studies were benign hepatic adenomas and did not progress to frank cancer, which suggests that loss of autophagy may be sufficient for the initiation of tumorigenesis, but not for the progression to advanced cancers. This is consistent with the fact that homozygous deletion of Atg5 or Atg7 more completely abrogates autophagy than single copy loss of Becn1.
The White laboratory (Degenhardt et al. 2006; Karantza-Wadsworth et al. 2007; Mathew et al. 2007) has also provided mechanistic insights into the tumor-suppressive functions of autophagy. Their studies have shown both cell-intrinsic and -extrinsic effects of autophagy inhibition that promote tumorigenesis. In the setting of autophagy- and apoptosis-incompetent cells, tumorigenesis is accelerated in vivo. There was an increase in necrosis and inflammation that was likely at least partially responsible for the increased tumor growth by providing a protumorigenic inflammatory microenvironment. Further studies from the White laboratory (Degenhardt et al. 2006; Karantza-Wadsworth et al. 2007; Mathew et al. 2007) have shown that loss of autophagy leads to genomic instability and aneuploidy, which promotes tumorigenesis. Additionally, the overexpression of p62 as a result of autophagy inhibition was shown to be important in the promotion of tumorigenesis through a variety of mechanisms, including deregulation of NF-κB signaling, accumulation of ROS, and increased DNA damage (Mathew et al. 2009). p62 contains a UBA domain, allowing it to bind to ubiquitinated proteins, as well as an LIR (LC3-interacting region), conferring the ability to deliver targeted proteins to the autophagosome (Bjorkoy et al. 2006). Another link of elevated p62 to tumorigenesis was provided by two groups (Komatsu et al. 2010; Lau et al. 2010) who demonstrated that p62 binds to Keap1, leading to the up-regulation of NRF2, a transcription factor whose functions include the coordination of an antioxidant defense. The exact mechanism of how this may facilitate tumorigenesis has not been completely elucidated, but persistent NRF2 activation appears to be critical for anchorage-independent growth of hepatocellular carcinoma cells in the context of p62 overexpression (Inami et al. 2011).
Perhaps another mechanism by which autophagy has tumor-suppressive functions is its role in senescence. Young et al. (2009) have shown that autophagy is activated during oncogene-induced senescence by oncogenic Ras in fibroblasts. Inhibition of autophagy in this setting resulted in delayed onset of senescence. Given that senescence is thought to be a barrier to malignant transformation, the loss of autophagy may allow the escape of cells that have developed an oncogenic mutation that would normally undergo senescence and stop cycling.
Autophagy in advanced cancer—response to therapy
In addition to the well-established role of autophagy as a tumor suppressor, there is significant evidence that autophagy has a potentiating role in established cancers. Work from multiple groups has shown that autophagy can promote survival in cells under a variety of stresses (Kroemer et al. 2010). For example, evolving tumors develop regions of hypoxia and nutrient limitation, where elevated autophagy activation has been found previously to promote tumor survival (Degenhardt et al. 2006). Thus, it is easy to conceive that in these select regions of a tumor, autophagy is protumorigenic. However, it appears that the protumorigenic role of autophagy extends beyond the hypoxic/nutrient-deprived regions of a tumor. For example, autophagy has been shown to mediate therapeutic resistance in a variety of situations. The Thompson laboratory (Amaravadi et al. 2007) has demonstrated that autophagy is induced by restoration of p53 expression or by alkalating chemotherapy in a mouse lymphoma model. Inhibition of the autophagy by CQ or RNAi to ATG genes resulted in a profound synergy in cell kill. CQ acts at the level of the lysosome by interfering with lysosomal pH and thus autophagosome degradation (discussed below; Rubinsztein et al. 2007). Subsequently, a series of studies have shown synergy of autophagy inhibition with a variety of cytotoxic, targeted agents and radiotherapy (Amaravadi et al. 2007, 2011; Carew et al. 2007; Apel et al. 2008; Degtyarev et al. 2008).
While many studies have shown a protective role of autophagy in tumor cells, there do appear to be situations in which autophagy may contribute to the cell killing (Chen and Karantza 2011; Levy and Thorburn 2011). In fact, while autophagy can promote survival in many settings, it has also been described as type II programmed cell death (Debnath et al. 2005). The evidence used to suggest the importance of autophagy in the cell death by a particular agent is typically the increased presence of markers of elevated autophagy (autophagosomes or LC3-II expression) upon treatment and a mitigation of the agent's efficacy upon autophagy inhibition, often by RNAi to essential autophagy genes. While this may be a true phenomenon, the presence of elevated autophagy does not necessarily imply that it is the cause of death, but instead could signify a failed survival response. In fact, recent data from the Kroemer laboratory (Shen et al. 2011) using high-throughput chemical screens identified many compounds that activated autophagy in mammalian cells, but failed to demonstrate that any of these compounds killed cells via autophagy. Additionally, it is possible that autophagy genes may have other functions involved in cell death that are unrelated to autophagy, which could influence the results of such studies. One must also be cautious in these cases, as while there are data for autophagy as a cell death mechanism in organisms such as Drosophila melanogaster and Caenorhabditis elegans, in vivo data in mammalian systems are lacking (Debnath et al. 2005; Kroemer and Levine 2008). The use of mouse models with autophagy defects may be helpful in these situations.
Also relevant to the use of appropriate models is the fact that autophagy has a critical role in the host immune response. Indeed, autophagy is important in the proper functioning of multiple immune cell types, including T cells and macrophages (Pua et al. 2007; Deretic 2011; Levine et al. 2011). Given the known importance of the immune system in cancer development and progression (Schreiber et al. 2011), the response of tumors to autophagy inhibition must also take the immune system into account. This also applies to assessing combinations of therapies with autophagy inhibition. It is possible that the suppressive effects that autophagy inhibition has on the immune system may actually blunt some of the response of the tumor to anti-autophagy therapies. For example, elevated autophagy in response to particular therapies can promote the extracellular release of the high-mobility group box 1 protein (HMGB1) (Thorburn et al. 2009a). This HMGB1 can bind to and activate Toll-like receptors on dendritic cells, which can increase anti-tumor T-cell responses (Apetoh et al. 2007). The release of HMGB1 is diminished when autophagy is inhibited. Thus, while inhibition of autophagy in glioblastoma cells resulted in increased cell death by an EGF-conjugated toxin in vitro, there was actually a decrease in extracellular HMGB1 release (Thorburn et al. 2009b). Thorburn et al. (2009b) concluded that the increased cell kill with autophagy inhibition needs to be weighed against the potential decrease in tumor immune response. Unfortunately, most commonly used cell culture or xenograft models in immunocompromised mice do not allow one to assess the interplay with the immune response. Syngeneic tumor transplant models or autochthonous models are necessary for such studies.
Autophagy and tumor maintenance
In addition to its role as a reactive survival mechanism, autophagy appears to have an integral role in tumor maintenance in certain settings. Autophagy is activated in rapidly growing tumors in regions of hypoxia and/or nutrient deprivation, allowing survival. This cell-extrinsic role of autophagy has been well described (Degenhardt et al. 2006). In fact, several of the consequences of autophagy loss mentioned previously that have been shown to promote tumorigenesis (e.g., inflammation/ROS and genomic instability/DNA damage) are actually detrimental to tumor growth at high levels. Therefore, autophagy may be reactivated or up-regulated in tumors to allow continued growth by mitigating this damage. For example, autophagy is activated by ROS via several mechanisms, including direct modification of ATG4 as well as increasing HMGB1 expression and extracellular release (Kang et al. 2011; Scherz-Shouval and Elazar 2011). Once activated, autophagy can mitigate ROS-induced damage by degrading oxidized proteins or, alternatively, degrading damaged mitochondria (mitophagy), which are a primary source of intracellular ROS (Scherz-Shouval and Elazar 2011).
More recently, the concept that there may be cell-autonomous roles for autophagy in tumor maintenance has been proposed. In this case, the tumor cells have evolved (through either genetic or epigenetic changes) to require autophagy under basal conditions. For example, my group (Yang and Kimmelman 2011; Yang et al. 2011) has shown that pancreatic cancers have elevated basal autophagy in a cell-autonomous manner and require this autophagy for continued growth. In fact, inhibiting autophagy in these cells by CQ or RNAi leads to decreased in vitro growth as well as significant responses in a variety of mouse models of the disease. The cellular consequences of autophagy inhibition in pancreatic cancer cells include profound metabolic changes. Oxidative phosphorylation was markedly decreased, as was ATP production. In this setting, autophagy is likely providing critical metabolic intermediates that the cells require. Using a different system, White and colleagues (Guo et al. 2011) had similar findings. They showed that Ras-driven tumorigenesis resulted in elevated basal autophagy. Using cells derived from a variety of genetically engineered mice with autophagy defects, they demonstrated that tumorigenesis was attenuated in the context of inhibited autophagy. Additionally, they showed that several human tumor cell lines with Ras mutations from various tissues were dependent on autophagy for growth. Consistent with our results (Yang and Kimmelman 2011; Yang et al. 2011), White and colleagues (Guo et al. 2011) identified that Ras-transformed epithelial cells had decreased oxidative phosphorylation in the context of genetic inhibition of autophagy. Interestingly, the autophagy-incompetent cells in their system demonstrated an accumulation of damaged mitochondria due to impaired mitophagy. This was not seen in pancreatic cancer cells, which may reflect the difference between inhibiting autophagy acutely in a fully transformed pancreatic cancer cell versus transforming a chronically autophagy-incompetent epithelial cell. Alternatively, it may reflect cell type-specific differences.
The data imply that the dependence on autophagy may be a property of Ras-driven tumors. Pancreatic cancers almost universally possess activating Kras mutations (Hezel et al. 2006). In further support of this concept, the Debnath laboatory (Lock et al. 2011) has shown that transformation of mammary epithelial cells by oncogenic H-Ras requires autophagy. MJ Kim et al. (2011) had similar results using Kras in mammary epithelial cells. While the systems and some of the results differed between these studies, the importance of autophagy during Ras-induced transformation was consistent. Interestingly, tumors driven by other oncogenic events may also depend on autophagy. For example, inhibition of autophagy in mammary tissue using a conditional FIP200 (probable ATG17 homolog) knockout mouse significantly attenuated mammary tumorigenesis by the polyoma middle T (PyMT) oncogene (Wei et al. 2011).
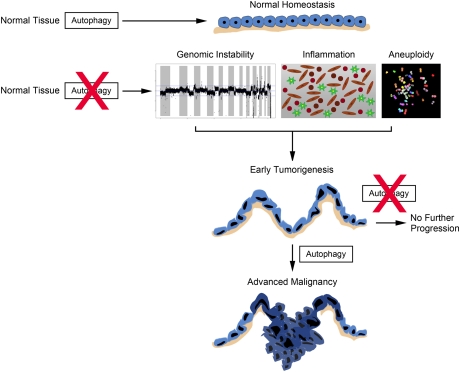
While much of the data have demonstrated that Ras-transformed cells have elevated basal autophagy, the level of Ras expression and genetic constellation of a particular cell may dictate the ultimate biological outcome of the autophagy activation. For example, in human ovarian surface epithelial (HOSE) cells, where high levels of H-Ras activation lead to growth arrest followed by cell death, the elevated autophagy caused by oncogenic Ras appears to contribute to cell death (Elgendy et al. 2011). Knockdown of autophagy genes attenuated the death, implicating autophagy or at least particular autophagy genes in the cell death process. These seemingly disparate results from previous studies can be explained by several factors. First, the high levels of Ras overexpression and the acute induction of Ras are distinct from previous studies where Ras was stably expressed and functioned to promote tumorigenesis or, in the case of pancreatic cancers, an endogenous mutation. There is a wealth of literature demonstrating the anti-proliferative effects of high levels of oncogenic Ras expression in primary cells as a barrier to transformation (Collado and Serrano 2010; Kuilman et al. 2010), and, perhaps in the initial stages of transformation, autophagy provides a barrier. This is consistent with the concept that autophagy has a tumor suppressor function. However, it is possible that once a cell has committed to a program of malignant transformation, autophagy may become critical for its continued growth. Indeed, the emerging role of autophagy in established tumors, particularly those driven by oncogenic Ras, is quite compelling. Taken together, the data suggest that the role of autophagy in cancer is likely dynamic, suppressing tumor growth in the initial phases, but serving critical metabolic and survival functions in advanced cancers (Fig. 2).
Figure 2.
Proposed model of the role of autophagy in cancer development and progression. In normal tissue, autophagy performs homeostatic functions such as organelle and protein quality control. If autophagy is suppressed in tissues, normal homeostasis is disrupted. Consequences of this include increased inflammation, genomic instability, and aneuploidy. Together, these changes can promote tumor initiation and lead to early tumorigeneisis (the schematic depicts an early lesion with increased nuclear atypia and loss of cell polarity, but an intact basement membrane in tan). However, if autophagy continues to be suppressed, then tumor progression will not proceed. Alternatively, if autophagy is activated at this stage, cells can keep up with their metabolic demand as well as regulate oxidative stress. This allows progression to more advanced malignancy (tumor breaks through the basement membrane) as well as continued tumor growth. Genomic instability is depicted as a recurrence plot from array comparative genomic hybridization of a pancreatic cancer cell line showing multiple high-amplitude genomic changes. Aneuploidy is shown by a spectral karyotype of a tumor cell line demonstrating aneuplody and chromosomal rearrangements.
There may also be a role for autophagy inhibition in tumor prevention. Using mouse models of lymphoma, the Kastan group (Maclean et al. 2008) showed that CQ treatment was able to impair lymphoma development in Atm knockout or Myc overexpression models. This prevention was p53-dependent, as CQ did not affect lymphoma development in a p53-null model. Therefore, there may be a role for such chemoprevention in families with inherited tumor syndromes or at high risk for development of a particular tumor type. One must use caution in these cases, given the aforementioned tumor suppressor role of autophagy, particularly in the early phases of tumorigenesis (Chen and White 2011). These patients often have genetic mutations predisposing them to malignancy that may cooperate with autophagy inhibition. Thus, a complete understanding of the role of autophagy in a particular tumor type as well as in its various stages of development will be essential.
Elevated autophagy in tumors—meeting metabolic demand
As has been shown in pancreatic cancers and other systems, some tumors have elevated autophagy even in nutrient-replete conditions. While the data suggest that autophagy is required for growth, it is conceptually interesting why these cells have evolved to require autophagy as an essential part of their cellular metabolism, as well as how they have this “self-eating” program on all of the time yet maintain their size and organelle content. As far as the metabolic aspects, a rapidly dividing tumor cell needs energy as well as an abundance of proteins, sugars, lipids, and nucleic acids, which autophagy can help provide (Rabinowitz and White 2010; Eng and Abraham 2011). The question of how the cell maintains high autophagic flux at all times may rely on the idea that basal autophagy is different from autophagy induced by starvation or other stimuli; perhaps it is more self-limited and restricted to specific substrates. For example, pancreatic cancer cells display minimal mitophagy (Yang et al. 2011). There may be more selectivity in this constitutive autophagy, similar to the highly selective ubiquitin–proteasome pathway, rather than a general degradation of long-lived proteins.
What has been fairly consistent in many of the recent studies is that the elevated basal autophagy seen in tumors has become critical for the cellular metabolism of these cancers. Indeed, this concept of altered tumor metabolism is an area of active research, and efforts are under way to exploit this for therapeutic gain (Wise and Thompson 2010). Tumor cells require elevated energy production as well as increased protein, lipid, and nucleic acid synthesis to increase biomass. Many tumors rely heavily on glucose metabolism via glycolysis for energy production (Vander Heiden et al. 2009; Levine and Puzio-Kuter 2010), while tumors with Myc overexpression or activation of Ras have been shown to require glutamine to fuel the TCA cycle (Wise et al. 2008; Weinberg et al. 2010; Wise and Thompson 2010). Autophagy, at the most basic level, provides raw materials for multiple metabolic pathways. Through the degradation of proteins, it can provide a pool of amino acids that can be used for anabolic reactions or energy production. Amino acids can fuel the TCA cycle via conversion to α-ketoglutarate (αKG), pyruvate, or other TCA cycle intermediates (Rabinowitz and White 2010). Additionally, fatty acids produced by lipid autophagy (lipophagy) can undergo β-oxidation, resulting in acetyl-CoA, which can enter the TCA cycle. Breakdown of RNA to nucleosides and, subsequently, to ribose-phosphate can also result in ATP production.
In addition to decreased oxidative phosphorylation, the metabolic consequences of autophagy inhibition in Ras-transformed cells may also include decreased glycolysis in certain contexts. The Debnath laboratory (Lock et al. 2011) has shown that glycolysis is impaired in oncogenic Ras-expressing Atg5-null mouse embryonic fibroblasts (MEFs) compared with wild-type MEFs expressing Ras. Similar results were obtained in autophagy-incompetent FIP200-null MEFs expressing oncogenic Ras (Wei et al. 2011). In contrast, inhibition of autophagy in pancreatic cancer lines actually leads to a compensatory increase in glycolysis (Yang et al. 2011). This may reflect cell type differences or, as mentioned previously, the distinction of inhibiting autophagy acutely in a chronically transformed cell (pancreatic cancer lines) versus transforming a chronically autophagy-incompetent cell (Atg5-null MEFs). Interestingly, the Sabatini group (Sheen et al. 2011) has shown that melanomas with activated Ras–ERK pathways also have a distinct metabolic phenotype involving autophagy. These tumors are sensitive to deprivation of leucine due to the inability to repress mTOR and activate autophagy (Sheen et al. 2011). Furthermore, there was a synergestic anti-tumor effect with both leucine deprivation and autophagy inhibition.
Taken together, the data support the concept that autophagy has the potential to intersect both normal and tumor metabolism at many levels. A comprehensive assessment of the role of autophagy in the metabolism of particular tumor types will be critical, as it may differ depending on a given histology, stage, or genetic context.
Clinical trials—concepts and obstacles
Currently, there are nearly 20 clinical trials registered with the National Cancer Institute (http://www.cancer.gov/clinicaltrials) exploring anti-autophagy strategies in a variety of human cancers. Most of these trials are ongoing, with minimal published results available, and nearly all use the CQ derivative hydroxychloroquine (HCQ). These lysosomotropic agents act at the level of the lysosome by inhibiting acidification, thereby impairing autophagosome degradation (Rubinsztein et al. 2007). The majority of these trials employ a combination with cytotoxic chemotherapies or targeted agents. Much of the rationale has been based on the aforementioned data that autophagy is induced in a variety of tumor cell lines and preclinical models by several types of chemotherapeutic agents as a reactive survival mechanism. Therefore, combining autophagy inhibition with traditional and targeted agents may be synergistic. Based on the importance of autophagy in pancreatic cancer biology, we and others have opened trials using HCQ as part of the therapeutic regimen. It is possible that these findings will be extended to a variety of Ras-driven cancers as well. Given that many centers are screening for Kras mutations as part of routine care, such trials are certainly feasible. While we know certain contexts in which tumors are dependent on autophagy, there is much work to be done to truly define these tumors a priori. It is likely that oncogenic Ras is only part of the story and much more will be discovered in this area in the coming years. In the interim, it is tempting to speculate that elevated basal autophagy, such as that seen in cancer cells that respond to autophagy inhibition (Guo et al. 2011; Yang et al. 2011), may predict response to autophagy inhibition in patients. A more comprehensive assessment of basal autophagy levels and sensitivity to autophagy inhibition using a large panel of tumor cell lines needs to be performed before definitive conclusions can be made.
While the above preclinical data are generally supportive of incorporating anti-autophagy therapies in cancer treatment trials, there are many critical questions that remain. Initially, it will be important to identify which cancers will benefit from anti-autophagy therapies. While preclinical data have provided some insights, these must be tested in clinical trials to validate the approach. Additionally, the proper combinations of therapies will be critical for success in patients. A significant concern is that there have been certain therapies in particular tumor lines where inhibition of autophagy decreases therapeutic efficacy. Understanding the circumstances in which autophagy inhibition impairs the therapeutic effect will be of great importance.
Importantly, while CQ and HCQ are effective inhibitors of autophagy in vitro, whether they will do so at doses used in current clinical trials is still uncertain. Part of the concern is the micromolar concentration that is required to inhibit autophagy and show anti-tumor efficacy in preclinical models. While this is theoretically achievable at tolerated doses after prolonged dosing, it is not ideal (Tett et al. 1993; Munster et al. 2002). Furthermore, given the aforementioned difficulties in measuring autophagy, particularly in fixed tissue such as tumor biopsies, it will be difficult to measure the basal levels of autophagy in patients' tumors to select candidates for trials. In particular, an accurate method to measure autophagic flux in biopsy specimens will be essential. These assays will also be necessary for measuring whether autophagy is inhibited in tumors during treatments as a pharmacodynamic assay. Many of the current trials will have some form of pharmacodynamic measurements built in. For example, several trials are using serial blood draws to isolate peripheral white blood cells (WBCs). These can be monitored by Western blot for p62 or LC3-II or by EM to assess for autophagy inhibition. Whether these measurements will (1) be reliable and (2) accurately reflect what is happening in tumors is still unresolved. The preclinical data in pancreatic cancer demonstrate that glycolysis is increased upon inhibition of autophagy (Yang et al. 2011). Because of this, there is potential to use positron emission tomography (PET) scans as a pharmacodynamic marker in pancreatic cancer trials using HCQ, as this can give a relative measure of glucose use by monitoring the FDG (fluorodeoxyglucose) uptake in the tumor (Vander Heiden et al. 2009).
Trials combining HCQ in the neoadjuvant setting (prior to surgical resection) will have tumor tissue available for analysis both before and after HCQ treatment. The interpretation of these assays may still prove difficult, as HCQ is often combined with other therapies that are known to modulate autophagy (e.g., chemotherapy and radiation). Alternative biomarkers to predict for autophagy activation as well as autophagy dependence are an area of active investigation in multiple laboratories and will be important to allow for continued refinement of the ideal therapeutic approach as well as for proper patient selection.
Novel autophagy inhibitors
While CQ and HCQ have the advantage of having a great deal of preclinical evidence behind them as well as a long track record of use in human patients, they are not ideal in terms of their pharmacology due to needing micromolar levels for activity. Because of this, there is a great deal of interest in developing new inhibitors. CQ, as mentioned previously, inhibits the last step of autophagy at the level of the lysosome. As it impacts lysosomal function, its affects are not entirely specific to autophagy. However, it is possible that there may be a benefit to having an accumulation of autophagosomes (which itself is potentially toxic), as seen with CQ, compared with inhibiting autophagy at early stages and preventing autophagosome formation. Because of the long-term use at relatively high doses in patients with a variety of rheumatologic conditions, there is reason to believe that inhibition (or partial inhibition) of autophagy in an adult patient is not detrimental. Conversely, there is little evidence (as mentioned above) that we are actually inhibiting autophagy in patients at these doses. As many of the ongoing clinical trials using HCQ are attempting to perform pharmacodynamic studies, we may have an answer to these questions relatively soon. Quinacrine, which also has been used in patients as an anti-malarial in the past, has been shown to inhibit autophagy similarly to CQ. In fact, quinacrine showed greater cytotoxicity in gastrointestinal stromal tumor (GIST) cell lines treated with imatinib than CQ (Gupta et al. 2010), and therefore this may be a promising anti-autophagy agent for future clinical trials.
Given the complexity of the autophagic process, there are multiple additional points that are potential targets for therapeutic intervention. Because of the past success of developing kinase inhibitors for clinical use, it is likely that kinases would be prime candidates for inhibition. Vps34, a class III PI3K, has a critical early role in autophagosome development and is one such potential target. This is particularly attractive, as there has been significant success in designing effective class I PI3K inhibitors (Wong et al. 2010). However, one potential issue is that Vps34 has roles in other aspects of endosome trafficking, and this may lead to unwanted effects and toxicity (Backer 2008). ULK1/2 are the mammalian orthologs of yeast ATG1. Recent evidence has shown that they are critical for autophagy activation, acting downstream from AMPK and the TOR complex (Hara et al. 2008; Egan et al. 2011; J Kim et al. 2011). Thus, these may make effective targets for autophagy inhibitors as well. Because there is some redundancy between ULK1 and ULK2 in terms of requirements for autophagy, an inhibitor may need to block activation of both. Alternatively, if one could identify tumor-selective roles of ULK1 versus ULK2, then a more specific inhibitor would be desirable. Given the parallels of autophagy and ubiquitin pathways, novel inhibitors to key protein interactions may provide additional selectivity. Another potential target would be LC3 proteases such as ATG4b, which are necessary for LC3 processing. Whichever approach is taken, the delicate balance between potency and toxicity must be determined to achieve a clinical success.
Conclusions
There has been a tremendous amount of progress in our understanding of the role of autophagy in cancer. Overall, the data support a dynamic role of autophagy in cancer—both as a tumor suppressor early in progression and later as a protumorigenic process, critical for tumor maintenance and therapeutic resistance (Fig. 2). However, there are still many critical questions that remain. If the role of autophagy is indeed dynamic in human cancer, the regulation of this switch from low levels to elevated levels has not been elucidated and will need to be explored for a complete understanding of this process. Given the complexities of the tumor's interaction with the microenvironment, these experiments will likely need to be performed in vivo with relevant tumor models. To accomplish this, it will be necessary to further refine our mouse models. In particular, we will need to genetically modulate autophagy, not only in a spatial fashion (to a particular organ), but also temporally (at a particular stage of tumor development). Such models are possible with currently available tools; e.g., inducible alleles and different recombinase systems. Last, the delineation of the autophagic cargo in tumors with elevated autophagy will allow for a greater understanding of how this process is integrated into the metabolism of the cell. Most likely, there will be differences in the role of autophagy in normal metabolism versus that of its tumorgenic counterpart. In fact, this may differ between tumor types or for a particular constellation of genetic events. Given the progress that has been made in proteomic and metabolomic approaches, this type of analysis is certainly feasible.
While there are still uncertainties of how autophagy inhibition will fare as an anti-cancer therapy, the preclinical data generally support the approach. The current clinical trials will hopefully provide insight into whether this will be a viable therapeutic paradigm. Importantly, pharmacodynamic studies may enable the determination of whether HCQ is actually inhibiting autophagy in patients. Correlative studies by multiple groups have the potential to define subgroups of patients who will most benefit and will guide future efforts.
Acknowledgments
I apologize for any omission of primary citations. I thank Dr. Alexei Protopopov for the SKY images and Drs. Jay Debnath, Nabeel Bardeesy, Brian Alexander, and Harvey Cantor for informative discussion and critical reading of the manuscript. I am supported by the Dana Farber Cancer Institute, National Cancer Institute Grant R01CA157490, Kimmel Scholar Award, and AACR-PanCAN Career Development Award.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17558811.
References
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB 2007. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 117: 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E 2011. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 17: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A 2008. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res 68: 1485–1494 [DOI] [PubMed] [Google Scholar]
- Apetoh L, Ghiringhelli F, Tesniere A, Criollo A, Ortiz C, Lidereau R, Mariette C, Chaput N, Mira JP, Delaloge S, et al. 2007. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunol Rev 220: 47–59 [DOI] [PubMed] [Google Scholar]
- Arias E, Cuervo AM 2011. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol 23: 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer JM 2008. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J 410: 1–17 [DOI] [PubMed] [Google Scholar]
- Balaburski GM, Hontz RD, Murphy ME 2010. p53 and ARF: unexpected players in autophagy. Trends Cell Biol 20: 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Johansen T 2006. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2: 138–139 [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T 2009. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 452: 181–197 [DOI] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T 2010. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell 40: 238–252 [DOI] [PubMed] [Google Scholar]
- Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL 2007. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood 110: 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Karantza V 2011. Autophagy as a therapeutic target in cancer. Cancer Biol Ther 11: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, White E 2011. Role of autophagy in cancer prevention. Cancer Prev Res (Phila) 4: 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Lindsten T, Wu J, Lu C, Thompson CB 2011. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci 108: 11121–11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A 2005. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin–proteasome system and onto human diseases and drug targeting. Cell Death Differ 12: 1178–1190 [DOI] [PubMed] [Google Scholar]
- Collado M, Serrano M 2010. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer 10: 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM 2006. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126: 121–134 [DOI] [PubMed] [Google Scholar]
- Debnath J, Baehrecke EH, Kroemer G 2005. Does autophagy contribute to cell death? Autophagy 1: 66–74 [DOI] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R 1966. Functions of lysosomes. Annu Rev Physiol 28: 435–492 [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC, et al. 2008. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol 183: 101–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V 2011. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev 240: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL 2008. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4: 851–865 [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgendy M, Sheridan C, Brumatti G, Martin SJ 2011. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell 42: 23–35 [DOI] [PubMed] [Google Scholar]
- Eng CH, Abraham RT 2011. The autophagy conundrum in cancer: influence of tumorigenic metabolic reprogramming. Oncogene. doi: 10.1038/onc.2011.220 [DOI] [PubMed] [Google Scholar]
- Eng CH, Yu K, Lucas J, White E, Abraham RT 2010. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal 3: ra31 doi: 10.1126/scisignal.2000911 [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S 2005. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci 102: 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T 2008. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ 2008. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects' review series. EMBO Rep 9: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM 2007. Defining the role of mTOR in cancer. Cancer Cell 12: 9–22 [DOI] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. 2011. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25: 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Roy S, Lazar AJ, Wang WL, McAuliffe JC, Reynoso D, McMahon J, Taguchi T, Floris G, Debiec-Rychter M, et al. 2010. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST). Proc Natl Acad Sci 107: 14333–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y 2007. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282: 37298–37302 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ 2009. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA 2006. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 20: 1218–1249 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, et al. 2011. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 193: 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH 2010. mTOR regulation of autophagy. FEBS Lett 584: 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Livesey KM, Zeh HJ III, Lotze MT, Tang D 2011. HMGB1 as an autophagy sensor in oxidative stress. Autophagy 7: 904–906 [DOI] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E 2007. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 21: 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Woo SJ, Yoon CH, Lee JS, An S, Choi YH, Hwang SG, Yoon G, Lee SJ 2011. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem 286: 12924–12932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452–460 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ 2008. Autophagy revisited: a conversation with Christian de Duve. Autophagy 4: 740–743 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, Gewirtz DA, Kroemer G, Levine B, Mizushima N, et al. 2010. A comprehensive glossary of autophagy-related molecules and processes. Autophagy 6: 438–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223 [DOI] [PubMed] [Google Scholar]
- Kroemer G, Levine B 2008. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol 9: 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B 2010. Autophagy and the integrated stress response. Mol Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS 2010. The essence of senescence. Genes Dev 24: 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD 2010. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 30: 3275–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Puzio-Kuter AM 2010. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330: 1340–1344 [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW 2011. Autophagy in immunity and inflammation. Nature 469: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JM, Thorburn A 2011. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther 131: 130–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676 [DOI] [PubMed] [Google Scholar]
- Liou W, Geuze HJ, Geelen MJ, Slot JW 1997. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol 136: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J 2011. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell 22: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean KH, Dorsey FC, Cleveland JL, Kastan MB 2008. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest 118: 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. 2007. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 26: 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, Kroemer G 2009. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ 16: 87–93 [DOI] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E 2007. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 21: 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijaljica D, Prescott M, Devenish RJ 2011. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 7: 673–682 [DOI] [PubMed] [Google Scholar]
- Mizushima N 2004. Methods for monitoring autophagy. Int J Biochem Cell Biol 36: 2491–2502 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T 2007. How to interpret LC3 immunoblotting. Autophagy 3: 542–545 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y 1998. A protein conjugation system essential for autophagy. Nature 395: 395–398 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B 2010. Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster T, Gibbs JP, Shen D, Baethge BA, Botstein GR, Caldwell J, Dietz F, Ettlinger R, Golden HE, Lindsley H, et al. 2002. Hydroxychloroquine concentration–response relationships in patients with rheumatoid arthritis. Arthritis Rheum 46: 1460–1469 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, et al. 2011. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332: 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP 2010. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol 22: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW 2007. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med 204: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112: 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E 2010. Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M, Chan EY, Tooze SA 2009. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol 185: 305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Lo Re A, Seux M, Nowak J, Gonzalez CD, Iovanna JL, et al. 2007. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem 282: 37124–37133 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ 2007. Potential therapeutic applications of autophagy. Nat Rev Drug Discov 6: 304–312 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Elazar Z 2007. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol 17: 422–427 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Elazar Z 2011. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci 36: 30–38 [DOI] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ 2011. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 331: 1565–1570 [DOI] [PubMed] [Google Scholar]
- Sheen JH, Zoncu R, Kim D, Sabatini DM 2011. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell 19: 613–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Kepp O, Michaud M, Martins I, Minoux H, Metivier D, Maiuri MC, Kroemer RT, Kroemer G 2011. Association and dissociation of autophagy, apoptosis and necrosis by systematic chemical study. Oncogene. doi: 10.1038/onc.2011.168 [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 186: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Levine B 2008. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene 27: S137–S148 doi: 10.1038/onc.2009.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N 2011. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25: 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. 2008. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 10: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tett SE, Day RO, Cutler DJ 1993. Concentration-effect relationship of hydroxychloroquine in rheumatoid arthritis—a cross sectional study. J Rheumatol 20: 1874–1879 [PubMed] [Google Scholar]
- Thorburn J, Frankel AE, Thorburn A 2009a. Regulation of HMGB1 release by autophagy. Autophagy 5: 247–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn J, Horita H, Redzic J, Hansen K, Frankel AE, Thorburn A 2009b. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Differ 16: 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL 2011. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 25: 1510–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS 2010. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci 107: 8788–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB 2010. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci 35: 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci 105: 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM 2010. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol 2: a006734 doi: 10.1101.cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Engelman JA, Cantley LC 2010. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev 20: 87–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Kimmelman AC 2011. A critical role for autophagy in pancreatic cancer. Autophagy 7: 912–913 [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ 2010a. Eaten alive: a history of macroautophagy. Nat Cell Biol 12: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ 2010b. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 22: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, et al. 2011. Pancreatic cancers require autophagy for tumor growth. Genes Dev 25: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM 2009. Autophagy mediates the mitotic senescence transition. Genes Dev 23: 798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci 100: 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]