Abstract
DNA-dependent protein kinase (DNA-PK) is a central regulator of DNA double-strand break (DSB) repair; however, the identity of relevant DNA-PK substrates has remained elusive. NR4A nuclear orphan receptors function as sequence-specific DNA-binding transcription factors that participate in adaptive and stress-related cell responses. We show here that NR4A proteins interact with the DNA-PK catalytic subunit and, upon exposure to DNA damage, translocate to DSB foci by a mechanism requiring the activity of poly(ADP-ribose) polymerase-1 (PARP-1). At DNA repair foci, NR4A is phosphorylated by DNA-PK and promotes DSB repair. Notably, NR4A transcriptional activity is entirely dispensable in this function, and core components of the DNA repair machinery are not transcriptionally regulated by NR4A. Instead, NR4A functions directly at DNA repair sites by a process that requires phosphorylation by DNA-PK. Furthermore, a severe combined immunodeficiency (SCID)-causing mutation in the human gene encoding the DNA-PK catalytic subunit impairs the interaction and phosphorylation of NR4A at DSBs. Thus, NR4As represent an entirely novel component of DNA damage response and are substrates of DNA-PK in the process of DSB repair.
Keywords: NR4A, Nurr1, Nur77, DNA-PK, PARP-1, DNA repair
Nur77 (NR4A1), Nurr1 (NR4A2), and Nor1 (NR4A3) are orphan members of the nuclear receptor family that have been suggested to function as ligand-independent transcription factors (Baker et al. 2003; Wang et al. 2003). A distinguishing feature of the NR4A family of nuclear receptors is that they are rapidly induced by various acute stimuli and are functioning in adaptive and stress-responsive physiological functions, in addition to important roles in cellular differentiation into midbrain dopamine neurons and cells of the hematopoietic lineage (Zetterström et al. 1997; Castillo et al. 1998; Saucedo-Cardenas et al. 1998; Ponnio et al. 2002; Mullican et al. 2007; Pearen and Muscat 2010; Sirin et al. 2010; Zhao and Bruemmer 2010). Moreover, NR4A proteins have recently been identified as tumor suppressors in myeloid cells, and NR4A loss of function results in acute myeloid leukemia (Mullican et al. 2007; Ramirez-Herrick et al. 2011). Interestingly, loss of function of NR4As has also been associated with increased DNA damage in myeloid and other cell types (Smith et al. 2008; Ramirez-Herrick et al. 2011). The mechanism whereby these proteins promote DNA repair has remained unclear; however, since NR4A receptors can function as conventional transcription factors, it has seemed likely that their participation in DNA repair is indirect and occurs via target gene transcriptional regulation.
DNA double-strand breaks (DSBs) belong to the most toxic DNA lesions and are typically repaired via either homologous recombination or nonhomologous end-joining (NHEJ) pathways. NHEJ is considered the main pathway for DSB repair in mammalian cells, as it can operate in any phase of the cell cycle and, in contrast to homologous recombination, does not require a sister chromatid for completion of the repair (Jackson and Bartek 2009). NHEJ is initiated by binding of DNA-dependent protein kinase (DNA-PK) regulatory subunits (Ku70/Ku80 heterodimer) to free DNA ends, followed by recruitment of the DNA-dependent kinase catalytic subunit protein (DNA-PKcs) to DSBs. This assembly results in DNA-PK kinase activation. The DNA-PK complex (Ku70/Ku80/DNA-PKcs) serves as a platform that holds both DNA ends together and orchestrates DNA processing and ligation. The latter steps of NHEJ require additional proteins, including Artemis (end-processing nuclease), XLF/Cerrunos, and the XRCC4/ligIV complex (ligase) (Jackson and Bartek 2009). More recent data on NHEJ assembly during DNA repair argue for a more complex model in which cooperative interactions between various NHEJ components orchestrate a precise architecture (Yano et al. 2008). It has been shown that DNA-PK is autophosphorylated on DNA-PKcs at multiple residues, and such autophosphorylation is important for the completion of DNA repair (Meek et al. 2008). While the precise function of DNA-PKcs autophosphorylation is still under intense investigation, it appears that it controls access of DNA repair accessory factors to DNA ends (Meek et al. 2008). In addition, DNA-PKcs autophosphorylation serves to control disassembly of the DNA-PK complex after DNA repair has been completed (Douglas et al. 2007). Importantly, however, relevant DNA-PK substrates other than DNA-PKcs have remained unidentified.
Here we describe experiments that demonstrate efficient interaction between NR4A2 and DNA-PKcs. The identification of DNA-PKcs as a NR4A2-interacting protein prompted us to investigate the potential role of NR4As in DNA repair. We analyzed NR4A localization in various cell types in response to DNA damage. Moreover, we used loss-of-function and gain-of-function experiments to assess the role of NR4As in the process of DNA repair. The results demonstrate that NR4A promotes DNA repair of DSBs via direct physical translocation to DNA repair foci and that NR4As are novel and relevant substrates of DNA-PK in the context of DNA repair.
Results
NR4A nuclear orphan receptors interact with DNA-PKcs and are recruited to DNA repair foci
NR4A2 harbors an unusual transactivation domain in its C terminus that fails to respond to typical nuclear receptor coactivators (Volakakis et al. 2006). We therefore searched for specific NR4A2 transcriptional coactivators via tandem affinity purification to isolate NR4A2-interacting proteins from human embryonic kidney (HEK) 293 cells in which NR4A2 is transcriptionally active (Supplemental Fig. 1A). By this approach, two major NR4A2-interacting proteins with approximate molecular weights of 70 and 450 kDa were identified (Fig. 1A). Mass spectrometry identified these proteins as heat-shock protein 70 (Hsp70) and the DNA-PKcs, respectively. While Hsp70 is known to interact relatively nonspecifically with many different proteins, we were intrigued by the interaction with DNA-PKcs. Further analysis by coimmunoprecipitation in HEK 293 cells transfected with expression vectors encoding Flag-tagged nuclear receptors revealed that other members of the NR4A subgroup were all interacting with DNA-PKcs, including the Drosophila homolog DHR38 (Fig. 1B). In contrast, related nuclear receptors (SF1 [NR5A1], ERR1 [NR3B1], and ERR2 [NR3B2]) did not interact with DNA-PKcs (Fig. 1B). Moreover, analysis by coimmunoprecipitation from human osteosarcoma U2OS cells showed interaction between endogenous DNA-PKcs and NR4A (Supplemental Fig. 1B). The interaction with DNA-PKcs was found to be unaffected by a mutation within the NR4A2 AF2 transactivation domain, suggesting that DNA-PKcs is not essential for NR4A2's transcriptional activity (Fig. 1A; Supplemental Fig. 1A). In line with this, NR4A2-induced reporter gene activity was only modestly affected by either overexpression or siRNA-induced down-regulation of DNA-PKcs in HEK 293 cells (Supplemental Fig. 1C). Similar results were obtained in DNA-PKcs-deficient (V3) and DNA-PKcs reconstituted (V3+DNA-PKcs) Chinese hamster ovary (CHO) cells (Supplemental Fig. 1C). We conclude that DNA-PKcs does not appear to function as a coactivator for NR4A receptors.
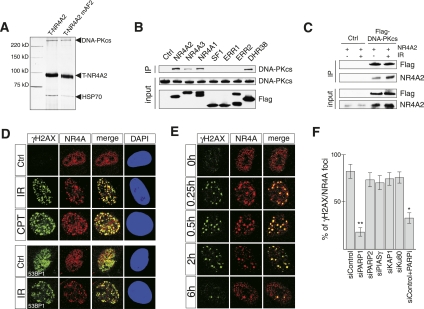
Figure 1.
NR4A receptors interact with DNA-PKcs and are recruited to DNA repair foci after DNA damage. (A) Silver staining of material copurifying with affinity-captured tagged NR4A2 (T-NR4A2) or mutant NR4A2 where the last 12 C-terminal amino acids were deleted (T-NR4A2 mAF2). (B) Immunoprecipitation of Flag-tagged nuclear receptors with endogenous DNA-PKcs from nuclear extracts from HEK 293 cells transfected with the indicated expression vectors. (C) Interaction of NR4A2 and Flag-tagged DNA-PKcs in HEK 293 cells transfected with expression vectors for NR4A2 and Flag-DNA-PKcs, as indicated. Cells were either untreated or exposed to IR (3 Gy) as indicated. (D, top panel) Immunofluorescence images of primary MEFs stained against NR4A (red) or γH2AX (green). Antibodies against 53BP1 were used in the bottom panel (green). Cells were untreated (control), exposed to IR (3 Gy), or treated with camptothecin for 30 min (CPT, 10 μm). Cells were fixed and imaged 30 min after IR treatment or camptothecin washout. (E) Immunofluorescence images of normal human fibroblasts (C5RO cells). Cells were exposed to IR (1 Gy), fixed, and stained for NR4A (red) and γH2AX (green) at the indicated time points. (F) Translocation of NR4A to DSB foci requires PARP-1. Quantification of NR4A translocation to DSB foci after 2 Gy of irradiation in U2OS cells transfected with siRNAs or treated with PARP inhibitor (PARPi) as indicated. The graph shows a percentage of γH2AX-positive foci that are also NR4A-positive at 30 min post-irradiation. Error bars indicate SD; n = 3; (*) significant at P < 0.05; (**) significant at P < 0.01.
Given that NR4A activity was not drastically affected by the interaction with DNA-PKcs, we next considered the possibility that NR4A proteins are somehow involved in DSB repair. In support of this, an increased interaction between NR4A2 and DNA-PKcs was observed in cells exposed to ionizing radiation (IR), as revealed by increased NR4A2 coimmunoprecipitation in IR-exposed HEK 293 cells expressing Flag-DNA-PKcs (Fig. 1C; Supplemental Fig. 1B). The immunoblot experiment also indicated that overexpression of Flag-DNA-PKcs increased the steady-state levels of NR4A2 protein, most likely as a result of increased protein stability (input NR4A) (Fig. 1C; Supplemental Fig. 1D). Moreover, antibodies detecting NR4A proteins revealed a distinct redistribution of these factors into IR- or camptothecin-induced foci in treated mouse embryonic fibroblasts (MEFs) (Fig. 1D). The specificity of these antisera was confirmed by analyzing MEFs in which the main NR4A proteins expressed in these cells, NR4A1 and NR4A2, had been depleted by either gene targeting (NR4A2) or expression of shRNA (NR4A1) (Supplemental Fig. 2B,C). Loss of foci occurred when both NR4A1 and NR4A2 were down-regulated, showing that both proteins have the capacity to redistribute after IR exposure. Importantly, NR4A-rich foci were also labeled by antibodies detecting either phosphorylated histone variant 2AX (γH2AX) or 53-binding protein 1 (53BP1), two DSB markers, demonstrating that NR4A localizes with these proteins at DSBs (Fig. 1D). A similar localization of NR4A was seen in all other cells analyzed, including human osteosarcoma U2OS cells, HEK 293 cells, normal human fibroblasts, and mouse neurons (Supplemental Fig. 3). Similar localization of NR4A was also observed upon treatment of cells with other DSB-inducing agents, such as zeocin (bleomycin) and neocarzinostatin (data not shown). Thus, since NR4A-containing foci colocalizing with γH2AX were detected in different cell types with variable NR4A expression levels and in response to different DSB-inducing treatments, we conclude that its localization to DSBs appears to be a general phenomenon. Moreover, IR exposure did not increase NR4A mRNA, indicating that the NR4A protein expressed prior to IR exposure is redistributed to DSBs and sites of DNA repair (Supplemental Fig. 2A). Furthermore, in IR-exposed MEFs, NR4A and γH2AX accumulated at DNA repair foci with similar kinetics, indicating that NR4A is localized to these sites throughout the repair process (Fig. 1E). Taken together, our data clearly demonstrate that NR4A proteins are markers for DNA DSBs.
We wished to better understand how NR4A proteins are recruited to DSBs. IR exposure of DNA-PKcs-deficient MEFs did not disrupt the ability of NR4A to localize at γH2AX-positive foci, demonstrating that DNA-PKcs itself is not essential for NR4A translocation (Supplemental Fig. 4). NR4A proteins have previously been shown to interact with other proteins implicated in DNA repair (Galleguillos et al. 2004; Ohkura et al. 2008; Rambaud et al. 2009). Thus, the corresponding mRNAs (and the mRNA encoding the Ku80 subunit of DNA-PK) were down-regulated by siRNA transfection in U2OS cells, followed by exposure to IR and analysis of NR4A cellular distribution. Interestingly, down-regulation of poly(ADP-ribose) polymerase-1 (PARP1), but not any of the other tested candidates, severely disrupted the formation of NR4A at foci colocalizing with γH2AX (Fig. 1F; Supplemental Fig. 5). Moreover, a PARP small molecule inhibitor also abolished distribution of NR4A to DSB foci (Fig. 1F). Thus, the process of poly(ADP-ribosyl)ation is essential for the recruitment of NR4A to sites of DSB repair.
NR4A loss of function results in defective DSB repair
We next investigated whether NR4A proteins promote DSB repair. In support of such a role, γH2AX persisted longer in camptothecin-treated or IR-exposed NR4A2 knockout MEFs as compared with wild-type cells (Fig. 2A). Moreover, overexpression of NR4A2 using a NR4A2 lentivirus expression vector in NR4A2 knockout MEFs reversed the persisting γH2AX and instead resulted in drastically reduced levels of γH2AX expression already at 4 h after camptothecin treatment (Fig. 2B). Thus, both loss-of-function and gain-of-function experiments strongly suggest that NR4A proteins are important for normal DSB repair kinetics. Comet assays performed under neutral DSB-detecting conditions were used to further assess how NR4A influences DNA repair in MEFs and in human U2OS cells in which NR4A1 and NR4A2 are also the predominant NR4A isotypes (Supplemental Fig. 6). Consistent with delayed γH2AX resolution kinetics, NR4A2 knockout MEFs showed a modestly delayed decrease of DNA in comet tails after 3 h following IR (Fig. 2C, top panel). However, combined knockout of NR4A2 and knockdown (shRNA) of NR4A1 markedly increased the amount of DNA in tails, an effect that persisted even at 6 h after DNA damage induction (Fig. 2C, top panel; Supplemental Fig. 7). Interestingly, an even more dramatic effect on DNA repair was seen in human U2OS cells in which NR4A1 and NR4A2 had been down-regulated by siRNA transfection. Combined NR4A1 and NR4A2 deficiency in these cells resulted in almost unaltered amounts of DNA in comet tails after 6 h following IR exposure, indicating that the DNA repair process had been severely disrupted (Fig. 2C, bottom panel; Supplemental Fig. 8). A deficient repair process was also evident from quantification of the number of γH2AX foci that persisted in IR-exposed U2OS cells. Thus, when both NR4A1 and NR4A2 were down-regulated, γH2AX-positive foci persisted even at 24 h following IR exposure (Fig. 2D). Notably, the magnitude of the effect was comparable with that seen after siRNA-mediated down-regulation of DNA-PKcs, further pointing toward a critical role for NR4A in DSB repair.
Figure 2.
NR4A receptors regulate DSB repair in mammalian cells. (A) Antibodies against either γH2AX or H2AX were used in immunoblots detecting proteins from MEF cell extracts from cells treated as indicated. (B) Knockout NR4A2 MEFs (NR4A2−/−) were infected with GFP or NR4A2 lentiviruses. Cells were treated with camptothecin and used in immunoblotting as in A. (C) MEFs or U2OS cells were exposed to IR (15 Gy), and DNA repair was quantified by the neutral comet assays. Wild-type (wt) cells or NR4A2 knockout (NR4A2−/−) MEFs were infected with control shRNA (shControl) or NR4A1 shRNA (shNR4A1) lentiviruses. Error bars indicate SD; n = 4; (*) significant at P < 0.05 calculated against wild-type shControl sample with the corresponding time of DNA repair. U2OS cells were transfected with siRNAs (siControl; siNR4A1; siNR4A2) as indicated. The diagrams illustrate the percentage of DNA detected in comet tails. Error bars indicate SD; n = 4; (**) significant at P < 0.01 calculated against the siControl sample with the corresponding time of DNA repair. (D) DNA repair foci resolution kinetics at the indicated time points in U2OS cells exposed to IR (1 Gy). U2OS cells were transfected with the same siRNAs as those used in C. Error bars indicate SD; n = 3; (*) significant at P < 0.05, siControl versus siNR4A1/A2. (E) Immunofluorescence images of brain sections from wild-type (wt) and conditional NR4A2 knockout (cNR4A2−/−) mice stained against γH2AX. The small panels show individual cells in high magnification with or without DAPI staining as indicated. (F) Immunoblot images of γH2AX and H2AX levels in extracts derived from cerebral cortex of either wild-type (wt) or conditional NR4A2 knockout (cNR4A2−/−) mice; the hash mark (#) indicates different animals.
Although functional redundancy between coexpressed NR4A subtypes was observed in cell culture experiments, access to NR4A2 knockout mice prompted us to analyze whether defective DSB repair could be detected in vivo in such mutant mice. Since NR4A2-null mice are lethal at birth, we studied a strain of conditional NR4A2 knockout mice (Kadkhodaei et al. 2009) in which NR4A2 was ablated selectively within the forebrain. Although reduced cellularity, signs of degenerating neurons, or apoptosis was not observed in these animals (data not shown), a strongly increased γH2AX signal was detected by immunohistochemistry and immunoblot in the adult dorsal–lateral neocortex of 12- to 14-mo-old NR4A2 conditional knockout mice (Fig. 2E,F). Increased γH2AX immunostaining, distributed in a punctate pattern, was seen in all analyzed animals by both immunohistochemistry (n = 6) (Fig. 2E) and immunoblotting (n = 2) (Fig. 2F), suggesting that lack of NR4A2 impaired repair of endogenously formed DSBs. Although NR4A2 had been ablated throughout the forebrain, increased immunostaining was only detected in a relatively restricted area of the cortex, suggesting compensatory effects (presumably by other NR4A members) in other parts of the forebrain. Nonetheless, these results substantiate the findings in cultured cells and provide additional strong evidence for the involvement of NR4A proteins in DSB repair.
DNA-PK-mediated phosphorylation of NR4A is required for efficient DNA repair
In order to define which NR4A2 domain can interact with endogenous DNA-PKcs, bacterially expressed and purified GST-fused NR4A2 protein derivatives were used in pull-downs from HEK 293 cell nuclear extracts (Fig. 3A). As expected, the analysis demonstrated that DNA-PKcs interacted with full-length NR4A2 and, importantly, also with GST-fused peptides encompassing the NR4A DNA-binding domain (DBD), but not with the N-terminal or the C-terminal ligand-binding domains (Fig. 3A).
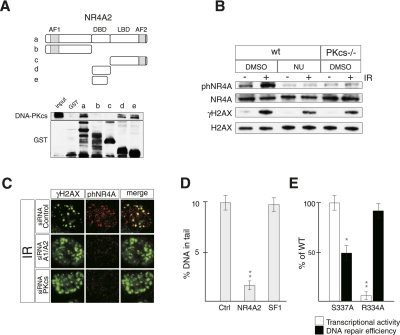
Figure 3.
DNA-PK-mediated S337 phosphorylation of NR4A2 is required for efficient DSB repair. (A) Indicated GST-NR4A2 fusion proteins (designated a–e) were incubated with HEK 293 cell nuclear extracts and used to pull down endogenous DNA-PKcs visualized by immunoblotting. (Top) The NR4A2 primary structure is illustrated. (AF1) N-terminal transactivation domain; (DBD) DNA-binding domain; (LBD) ligand-binding domain; (AF2) C-terminal transactivation domain. (B) Immunoblots showing S337 phosphorylation of NR4A (phNR4A) in cells exposed to IR (5 Gy). phNR4A, NR4A, γH2AX, and H2AX levels were detected with specific antibodies by immunoblotting of extracts from MEFs harvested at 15 min after IR exposure. Wild-type (wt) or DNA-PKcs knockout (PKcs−/−) MEFs were exposed to NU7026 (NU) or DMSO as indicated. (C) Immunofluorescence images of U20S cells exposed to IR (2 Gy) and stained against phNR4A (red) and γH2AX (green) 30 min after IR. Cells were transfected with siRNAs against DNA-PKcs (siRNA-PKcs), NR4A1 and NR4A2 (siRNA-A1/A2), or control siRNA (siRNA-Control), as indicated. (D) Quantification of comet assays in HEK 293 cells transfected with LacZ (Ctrl), NR4A2, or SF1 expression vectors as indicated. Error bars indicate SD; n = 3; (**) significant at P < 0.01. (E) Quantification of comet assays (for DNA repair efficiency) (black bars) and reporter gene assays (for transcriptional activity) (white bars) in HEK 293 cells transfected with NR4A2S337A (S337A) or NR4A2R334A (R334A) expression vectors. Cells were treated with camptothecin for 30 min, then allowed to repair DNA for 4 h. Error bars indicate SD; n = 3; (*) significant at P < 0.05; (**) significant at P < 0.01.
We next concluded that NR4A2 can be phosphorylated by purified DNA-PK, since DNA-dependent phosphorylation of bacterially expressed and purified GST-fused NR4A2 was detected in vitro (Supplemental Fig. 9B). The possibility that NR4A may be phosphorylated within the DNA-PKcs-interacting DBD was next evaluated. The amino acid sequence of NR4A proteins, including the Drosophila homolog DHR38, contains a highly conserved putative DNA-PK phosphorylation site localized at the C-terminal end of their DBDs (S337 in NR4A2). This site (TDSLKG) is conserved in all NR4A members (Supplemental Fig. 9A) and resembles the “hydrophobic” DNA-PK consensus phosphorylation site with a serine or threonine followed by a hydrophobic amino acid residue rather than a glutamine (Q), which is the more typical site for all related DNA damage-responsive kinases (DNA-PK, ATM, and ATR) (Traven and Heierhorst 2005). A phosphopeptide composed of NR4A2 sequences, including the phosphorylated S337, was synthesized and used to generate antibodies recognizing phosphorylated NR4A proteins (phNRA4). Several experiments strongly suggest that NR4A is a relevant substrate of DNA-PK in vivo. First, the specificity of phNR4A antibodies was verified in immunoblots showing that in vitro phosphorylated GST-NR4A2, but not GST-NR4A2S337A harboring a mutation substituting S337 for an alanine, could be detected by phNR4A antibodies (Supplemental Fig. 9C). Second, phNR4A antibodies detected increased levels of endogenous phNR4A in immunoblots from IR-treated wild-type MEFs, but not if cells had been pretreated with NU7026, a selective DNA-PKcs inhibitor (Veuger et al. 2003), or in genetically ablated DNA-PKcs-deficient MEFs (Fig. 3B). Third, phNR4A antibodies detected distinct foci that colocalized with γH2AX foci in IR-exposed U2OS cells (Fig. 3C) and MEFs (Supplemental Fig. 9D). Importantly, these foci were not detected in U2OS cells in which NR4A1 and NR4A2 or DNA-PKcs had been down-regulated by siRNAs (Fig. 3C). Peptide-blocking experiments also indicated that phNR4A antibody specifically recognizes the S337 phosphorylated form of NR4A in DNA repair foci (Supplemental Fig. 9E). Of note, phosphorylation at S337 was not required for NR4A distribution to DSB repair foci, as endogenous NR4A localized to foci also in DNA-PK-deficient MEFs (Supplemental Fig. 4), a conclusion that was corroborated by experiments showing that the NR4A2S337A mutant localized normally to DSB foci after IR exposure in U2OS cells (Supplemental Fig. 10). Taken together, NR4A proteins are associated with DSB recognition and repair and are phosphorylated by DNA-PK at DSBs.
We next aimed to assess whether NR4A phosphorylation influences its ability to promote DSB repair. For this purpose, HEK 293 cells, which express very low levels of endogenous NR4A proteins (Supplemental Fig. 3; data not shown), were transfected with different expression vectors encoding mutated NR4A2 derivatives, exposed to camptothecin, and then analyzed for DNA repair capacity by the comet assay (Fig. 3D; Supplemental Fig. 11B,C). The transcriptional activity of these NR4A2 derivatives was also analyzed in parallel in HEK 293 cells (Fig. 3E). Overexpression of wild-type NR4A2 drastically reduced the amount of DNA in comet tails as compared with controls at 4 h following camptothecin exposure (Fig. 3D; Supplemental Fig. 11B). In contrast, expression of NR4A2S337A resulted in a much more modest reduction in comet formation, indicating that deficient phosphorylation at S337 interferes with the ability of NR4A2 to promote DNA repair (Fig. 3E; Supplemental Fig. 11C). In contrast, this mutation did not interfere with NR4A2's ability to activate transcription or promote cell cycle arrest of a neural cell line, an activity of NR4A2 that previously has been shown to depend on its gene regulatory function (Supplemental Fig. 12; Castro et al. 2001), further supporting the hypothesis that the DSB repair function is a separate activity of NR4A. Since our preliminary analysis suggests that other sites outside of the NR4A DBD are also phosphorylated by DNA-PK (data not shown), it seems likely that the reduced DNA repair activity of NR4A2S337A underestimates the importance of DNA-PK-mediated NR4A phosphorylation in the repair process.
Interestingly, an additional mutant, NR4A2R334A, which harbors a mutation within its DBD that renders it unable to bind cognate DNA-binding sites (NBREs), was as efficient as wild-type NR4A2 in promoting DNA repair (Fig. 3E; Supplemental Fig. 11C). This result was also corroborated by an in vivo plasmid recircularization assay for DSB repair (Supplemental Fig. 13A) and in a NHEJ reporter cell line (Supplemental Fig. 13B). Moreover, a point mutation in the AF2 transcriptional activation domain that severely affects the transcriptional activity of NR4A2 was as efficient as wild-type NR4A2 in promoting DNA repair (Supplemental Fig. 14). To further assess whether gene regulatory functions are important, the mRNA expression levels of genes involved in NHEJ were analyzed by quantitative PCR (qPCR) after NR4A2 or NR4A2S337A overexpression in HEK 293 cells. None of the analyzed genes were significantly affected by wild-type or mutant NR4A2, further substantiating the hypothesis that effects in DSB repair are distinct from NR4A's gene regulatory activity (Supplemental Fig. 15). Thus, phosphorylation is important, while, surprisingly, sequence-specific DNA-binding and transcriptional activity seem dispensable for NR4A's ability to promote DSB repair.
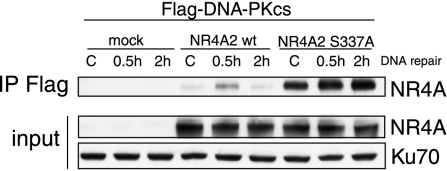
It remains unclear why the phosphorylation of NR4A is important. However, in coimmunoprecipitation experiments, we noted that antibodies against Flag-DNA-PKcs immunoprecipitated much more NR4A2S337A compared with wild-type NR4A2. Moreover, in contrast to wild-type NR4A2, NR4A2S337A was not decreased in precipitates even 2 h after irradiation (Fig. 4). These observations may suggest that NR4A is important in a transient interaction with DNA-PKcs and that phosphorylation may facilitate NR4A release from the DNA-PK complex.
Figure 4.
NR4A2 S337A mutant shows increased basal and DNA damage-induced interaction with DNA-PKcs. 293 cells were transfected with Flag-DNA-PKcs expression vector and either NR4A2 wild-type (wt) or NR4A2 S337A expression plasmids. Twenty-four hours after transfection, cells were irradiated (10 Gy) and then harvested at the indicated time points. Whole-cell extracts were used for immunoprecipitations with anti-Flag antibodies. Precipitated material was immunoblotted and probed with anti-NR4A antibodies (NR4A-IP Flag). Expression levels of NR4A are also shown (NR4A input).
A human severe combined immunodeficiency (SCID)-causing mutation in DNA-PKcs disrupts DNA-PKcs-mediated phosphorylation of NR4A
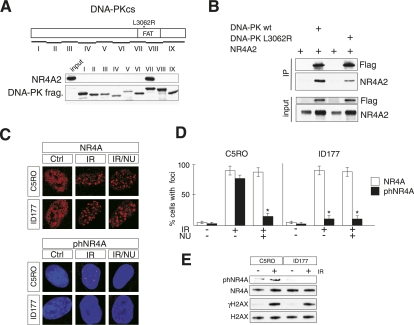
To gain further insight into the relationship between NR4A and DNA-PK, the region in DNA-PKcs that interacts with NR4A was identified. DNA-PKcs fragments fused to GST were purified and used in pull-down experiments of in vitro translated 35S-labeled NR4A2. Interaction was only seen with fragment VII, which includes a portion of the so-called FAT domain (Fig. 5A; Dip and Naegeli 2005). A mutation (L3062R) in the DNA-PKcs FAT domain was recently identified in a human patient with SCID (DNA-PKcsL3062R) (van der Burg et al. 2009). Primary fibroblasts (ID177) from this patient are highly radiosensitive due to inefficient DSB repair even though kinase activity is intact, autophosphorylation of DNA-PKcsL3062R appears normal, and DNA-PKcsL3062R is recruited to DSBs in IR-exposed ID177 cells. Thus, it remains unclear how the L3062R mutation influences DNA repair. We therefore investigated the ability of mutated human DNA-PKcs to interact with NR4A. Flag-tagged derivatives of human DNA-PKcs or DNA-PKcsL3062R were immunoprecipitated from extracts derived from transfected HEK 293 cells overexpressing either of these two proteins together with NR4A2. Strikingly, although not completely abolished, coimmunoprecipitation of NR4A2 with DNA-PKcsL3062R was severely reduced (Fig. 5B). Moreover, although NR4A foci, which were detected with NR4A antibodies, formed normally in IR-exposed ID177 cells, foci containing phNRA4, which were detected with phNR4A antibodies, were almost completely absent (Fig. 5C; Supplemental Fig. 16). Quantification demonstrated that the reduction in phNRA4-positive foci formation was as severe as when IR-exposed control cells were treated with NU7026 inhibitor (Fig. 5D). These findings were also corroborated in immunoblots derived from IR-exposed control or immortalized ID177 fibroblasts in which phNR4A was essentially undetectable (Fig. 5E). Thus, these data provide additional strong evidence for a functional link between NR4A and DNA-PK in DNA repair.
Figure 5.
A human SCID-causing L3062R DNA-PKcs mutation shows diminished interaction with NR4A and drastically reduced S337 NR4A phosphorylation. (A) Mapping the NR4A interaction domain in DNA-PKcs. GST-DNA-PKcs fusion proteins were incubated with in vitro translated NR4A2. Pulled-down NR4A2 was visualized by immunoblotting. The position of the L3062R mutation within the FAT domain in DNA-PKcs fragment VII is indicated. (B) Immunoprecipitation experiment showing defective interaction of DNA-PKcsL3062R with NR4A2. HEK 293 cells were transfected with Flag-tagged DNA-PKcs and NR4A2 expression vectors. Immunoprecipitates from whole-cell extracts were used for immunoblotting using antibodies against either Flag or NR4A, as indicated. (C) Immunofluorescence of normal human fibroblasts (C5RO) and human SCID primary cells (ID177) stained against NR4A (top panel) or phNR4A (red) and DAPI (blue) (bottom panel) in control cells (Ctrl), cells exposed to IR (1 Gy), or cells pretreated with NU7026 (NU) before exposure to IR (IR/NU). (D) Quantification of immunofluorescence staining from C as the percentage of cells containing foci. Error bars indicate SD; n = 4; (*) significant at P < 0.05. (E) Immunoblotting of extracts from control cells (C5RO) and TERT-immortalized SCID cells (ID177) was used with specific antibodies to detect phNR4A, NR4A, γH2AX, and H2AX, as indicated. Cells were either untreated or harvested 30 min after exposure to IR (5 Gy).
Discussion
The experiments presented in this study revealed that NR4A proteins localize at DSB foci and regulate DSB repair. NR4As localizing at DSB foci were found in all analyzed cell types. Some of these cells express low levels of NR4A mRNA, but it is notable that protein expression was nevertheless sufficient for clearly detectable accumulation of NR4A at DSBs. Thus, it seems likely that NR4As contribute to DSB repair in most, if not all, types of tissues, including cells in which NR4As are expressed only at low or moderate levels. It should also be noted that NR4A loss-of-function experiments probably underestimate the role played by NR4A proteins in DNA repair, since only one or two out of three NR4A genes were down-regulated/knocked out in these experiments.
NR4A proteins have recently been shown to interact with PARP1 via the NR4A DBD (Ohkura et al. 2008). DNA-PK and PARP1, which is known to interact with and become activated at DSB DNA ends, may function to promote DSB repair via the same DSB repair pathway (Mitchell et al. 2009), and it is therefore tempting to hypothesize that at least some DSB repair defects observed in PARP1 knockout cells are related to deficient recruitment of NR4As to DNA repair foci. We still do not know why PARP1 is important for NR4A assembly at DSBs. Since a PARP inhibitor also blocked NR4A translocation, PARP enzymatic activity is essential. Via direct interaction with NR4As, PARP1 could function as a platform for NR4A assembly, and, although direct ribosylation of NR4As by PARP1 has not been demonstrated, such modifications could potentially stabilize NR4As at DSBs. Alternatively, PARP1 may influence NR4A translocation indirectly in various ways. For example, it has been shown that histones are poly(ADP-ribosyl)ated in vivo (Ball and Yokomori 2011), and it is therefore possible that PARP1 is inducing a modified chromatin state that can facilitate the binding of NR4As and other components of the DSB repair machinery. Future studies should provide further insight into the exact relationship between these two proteins in the DNA repair process.
Although the detailed mechanism whereby NR4A proteins promote DSB repair remains to be elucidated, the process appears to be associated with DNA-PK-mediated phosphorylation and localization of S337 phosphorylated NR4A at DSBs. This conclusion is substantiated by the results showing that the human SCID-causing mutant of DNA-PKcs is impaired in its ability to phosphorylate NR4A proteins, although its catalytic activity and autophosphorylation at S2056 are intact. Although some putative DNA-PKcs substrates have been identified, substrates linked to DSB repair at DSB foci have remained elusive (van Gent and van der Burg 2007; Meek et al. 2008; Jackson 2009). Intriguingly, our data identify such a phosphorylated target and emphasize the important role of DNA-PKcs substrate phosphorylation at DSBs. It is possible that the human SCID mutation, which does not impair DNA-PK kinase activity, may affect a domain of DNA-PKcs that is important for interaction with not only NR4A2, but also other as-yet-unidentified DNA-PK substrates.
One role of DNA-PK autophosphorylation appears to be to control the access of DNA repair factors to DSBs, but also to promote dissociation of DNA-PKcs from the Ku heterodimer (Douglas et al. 2007). Since our analysis suggests that at least the S337 NR4A phosphorylation is important for the dissociation of NR4A from the DNA-PKcs (Fig. 4), it is tempting to speculate that DNA-PK-mediated autophosphorylation and phosphorylation of NR4A may play related roles in controlling the assembly/disassembly of DNA repair complexes. It is also interesting to note that, in contrast to NR4A, the DNA-PK complex fails to form stable foci at DSBs (Bekker-Jensen et al. 2006). This may be explained by a transient nature of the interaction of DNA-PK components with broken DNA ends. Given that NR4As form stable foci at DSBs, it is conceivable that they provide a mechanism to increase association of DNA-PKcs and chromatin surrounding the DNA break. Such chromatin association has previously been found necessary for efficient activation of related DNA damage-responsive ATM kinases (Kim et al. 2009).
Finally, it is notable that numerous studies have implicated NR4A transcription factors in gene regulatory responses that counteract stressful insults, including oxidative stress, which often leads to induction of DSBs via the generation of reactive oxygen species (Rassool et al. 2007; Volakakis et al. 2010). Thus, the results presented here show that NR4A proteins use dual mechanisms that are both dependent on and independent of gene regulatory functions to provide protection against such stressful insults.
Materials and methods
Cell culture and transfection
Cell lines (SV40-transformed MEFs, 293FT [Invitrogen], 293H [Invitrogen], U2OS, TERT-immortalized normal human fibroblasts [C5RO], and primary or TERT-immortalized human SCID fibroblasts [ID177]) were cultured in DMEM medium with 10% FCS (fetal calf serum). Neurons were derived from mouse embryonic stem cells differentiated as described previously (Volakakis et al. 2010). Unless stated otherwise, cells were transfected with Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions.
Derivation and culture of MEFs
MEFs were derived from embryonic day 14 (E14) wild-type or NR4A2 knockout mouse littermate embryos after crossing heterozygous NR4A2 knockout mice (Zetterström et al. 1997). Embryos were dissected, organs were removed, and the tissue was cut and digested in 10 mL of 0.05% trypsin-EDTA for 30 min at 37°C. Trypsin was inactivated by the addition of 2 mL of FCS. The cell suspension was plated on a 15-cm dish in DMEM medium containing 10% FCS. Cells were used for experiments after reaching confluence at 48 h after plating (passage 1). All experiments were done with cells in passages 2–4.
Chemicals
Cycloheximide (CHX), polybrene, ionomycin, and NU7026 were purchased from Sigma. NU7026 was used at 10 μM final concentration, and cells were pretreated with NU for 1 h before the irradiation. Camptothecin was purchased from Sigma and was used at 10 μM final concentration. For induction of DNA damage, cells were treated with camptothecin for 30 min; after that, camptothecin was washed out, and cells were incubated for the indicated times to follow DNA repair. PARP inhibitor PJ-34 was purchased from Santa Cruz Biotechnologies and used at 1 μM final concentration.
Affinity purification of tagged NR4A (T-NR4A) protein complexes
Protein A moiety followed by streptavidin-binding peptide was fused in-frame to the N terminus of NR4A2 full-length cDNA (T-NR4A2) or AF2-deleted NR4A2 (T-NR4A mAF2, which harbors a deletion of the last 12 amino acids of NR4A2). Resulting T-NR4A2 constructs were transiently transfected into 293 cells with the use of Lipofectamine Plus (Invitrogen) reagent according to the manufacturer's instructions. Nuclear extracts from transfected 293 cells were incubated with streptavidin beads (Pierce). Bound proteins were eluted with biotin, and elutes were incubated with IgG beads (GE Healthcare). Beads were washed, and bound proteins were eluted with IgG elution buffer (glycine, pH 2.5). Eluted proteins were concentrated by phenol/ether extraction, run on SDS-PAGE, and used for silver staining followed by mass spectrometry analysis of individual silver-stained bands.
Immunofluorescence stainings
After treatment, cells were washed with PBS and fixed directly on the slide with 4% PFA for 10 min at room temperature. Subsequently, slides were recovered and blocked with 5% goat serum containing 0.2% Triton X-100 in PBS for at least 1 h at 4°C. Cells were then stained with primary antibodies in blocking buffer overnight at 4°C. Cells were washed with PBS 0.2% Tween and stained with goat FITC- or Cy3-labeled secondary antibodies (Jackson Immunoresearch) in blocking buffer containing DAPI for 1 h at room temperature. Cells were mounted in Vectashield mounting medium (Vector Laboratories) and imaged on a confocal microscope (Zeiss).
siRNA-mediated knockdown in cultured cells
siRNAs were purchased from Santa Cruz Biotechnologies (siControl, sc-37007; siNR4A1, sc-156146; siNR4A2, sc-36111; si-DNA-PKcs, sc-35200; siPARP1, sc-29437; siPARP2, sc-106356; siPIASγ, sc-40851; siKAP1, sc-38550). siRNAs were transfected into U2OS and 293H cells at 50 nM final concentration with the use of Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. Cells were assayed 48–72 h after RNAi transfections.
shRNA-mediated knockdown of NR4A1
Generation of lentiviruses encoding GFP (Lenti-GFP) or NR4A2 (Lenti-NR4A2) has been described elsewhere (Volakakis et al. 2010). MEFs were infected with lentiviruses at 5 MOI (multiplicity of infection) in six-well plates. Cells were used for experiments 48 h after addition of the virus. In the initial experiments, knockdown of NR4A1 mRNA in MEFs was carried out with ready-made lentiviruses purchased from Santa Cruz Biotechnologies (control, sc-108080; NR4A1 targeting, sc-36110-V). MEFs were plated in six-well plates, and virus (5 MOI) was applied together with polybrene (5 μg/mL). Cells were analyzed 48 h after virus application. Similar results were obtained with viruses made in-house. For in-house virus production, 293FT cells were transfected with packaging plasmids as described in Volakakis et al. (2010), including shRNA-encoding plasmid shControl (SHC002) and shmNR4A1 (TRCN0000218931 and TRCN0000234021, Mission System, Sigma). MEFs were infected with in-house-made lentivirus at 10 MOI as described above but without polybrene.
In-gel digestion and mass spectrometry
The chosen silver-stained gel bands were treated for in-gel digestion. Briefly, the bands were destained using farmer's reagent, and trypsin (porcine, modified, sequence grade; Promega) was introduced to the dried gel pieces. After overnight tryptic digestion, the peptides were bound to a C18 μZipTip and, after washing, were eluted with acetonitrile-containing matrix (alfa-cyano 4-hydroxy cinnamic acid) directly onto the target plate. The mass lists were generated by MALDI-TOF mass spectrometry on an Ultraflex I TOF/TOF from Bruker Daltonics. The search for identity was performed by scanning the NCBInr sequence database with the tryptic peptides using the current version of the search engine ProFound (http://prowl.rockefeller.edu/prowl-cgi/ProFound). The spectrum was internally calibrated using autolytic tryptic peptides, and the error was set at ±0.07 Da. One missed cleavage was allowed, and methionine could be oxidized. The significance of the identity was judged from the search engine's scoring system and other parameters such as the similarity between the empiric and calculated peptides' masses. The DNA-PKcs-containing silver-stained band revealed 52 peptides covering 13% of DNA-PKcs protein. The Hsp70-containing silver-stained band revealed 16 peptides covering 29% of the Hsp70 protein.
Measurement of DNA repair by counting DSB foci
U2OS cells were transfected with siRNA on eight-well culture clusters (BD Biosciences) and, 72 h later, were irradiated with 1 Gy. Cells were fixed at indicated time points (30 min was chosen as the 0-h repair time; i.e., peak of foci count) and stained with γH2AX antibodies to visualize DNA repair foci. At least 50 cells were counted for foci content at each data point. For calculations of statistical significance, a Student's paired t-test was used.
Acknowledgments
We thank Eliza Joodmardi for breeding NR4A2 knockout mice; Hugo Fernandes for RT–PCR analysis; and Birgitta Mörk, Dali Zong, and Hogir Salim for help with cell irradiations. Pierre Chambon and Daniel Metzger are acknowledged for providing conditional NR4A2 knockout mice. We also thank Drs. Robert Haché and Sébastien Soubeyrand for generous help with reagents, Hanna IJspeert for sending human cell lines, and Dr. Kazi Fattah for help with the NHEJ reporter cell line. This work was supported by the Swedish Research Council via a Linnaeus grant (to T.P.), the Swedish Strategic Research Foundation (to T.P.), the Swedish Cancer Society (CAN 2009/1115) (to R.L.), the Stockholm Cancer Society (Projects 10:1291 and 10:1493) (to R.L.), and the National Institutes of Health (CA50519 and PO1-CA92584) (to D.C.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.16872411.
References
- Baker KD, Shewchuk LM, Kozlova T, Makishima M, Hassell A, Wisely B, Caravella JA, Lambert MH, Reinking JL, Krause H, et al. 2003. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell 113: 731–742 [DOI] [PubMed] [Google Scholar]
- Ball AR Jr, Yokomori K 2011. Damage site chromatin: open or closed? Curr Opin Cell Biol 23: 277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo SO, Baffi JS, Palkovits M, Goldstein DS, Kopin IJ, Witta J, Magnuson MA, Nikodem VM 1998. Dopamine biosynthesis is selectively abolished in substantia nigra/ventral tegmental area but not in hypothalamic neurons in mice with targeted disruption of the Nurr1 gene. Mol Cell Neurosci 11: 36–46 [DOI] [PubMed] [Google Scholar]
- Castro DS, Hermanson E, Joseph B, Wallén A, Aarnisalo P, Heller A, Perlmann T 2001. Induction of cell cycle arrest and morphological differentiation by Nurr1 and retinoids in dopamine MN9D cells. J Biol Chem 276: 43277–43284 [DOI] [PubMed] [Google Scholar]
- Dip R, Naegeli H 2005. More than just strand breaks: the recognition of structural DNA discontinuities by DNA-dependent protein kinase catalytic subunit. FASEB J 19: 704–715 [DOI] [PubMed] [Google Scholar]
- Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, Ye R, Morrice N, Lees-Miller SP, Meek K 2007. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol Cell Biol 27: 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galleguillos D, Vecchiola A, Fuentealba JA, Ojeda V, Alvarez K, Gomez A, Andres ME 2004. PIASγ represses the transcriptional activation induced by the nuclear receptor Nurr1. J Biol Chem 279: 2005–2011 [DOI] [PubMed] [Google Scholar]
- Jackson SP 2009. The DNA-damage response: new molecular insights and new approaches to cancer therapy. Biochem Soc Trans 37: 483–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadkhodaei B, Ito T, Joodmardi E, Mattsson B, Rouillard C, Carta M, Muramatsu S-I, Sumi-Ichinose C, Nomura T, Metzger D, et al. 2009. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci 29: 15923–15932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, Kraemer KH, Shiloh Y, Bustin M 2009. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol 11: 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Dang V, Lees-Miller SP 2008. DNA-PK: the means to justify the ends? Adv Immunol 99: 33–58 [DOI] [PubMed] [Google Scholar]
- Mitchell J, Smith GC, Curtin NJ 2009. Poly(ADP-Ribose) polymerase-1 and DNA-dependent protein kinase have equivalent roles in double strand break repair following ionizing radiation. Int J Radiat Oncol Biol Phys 75: 1520–1527 [DOI] [PubMed] [Google Scholar]
- Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, Conneely OM 2007. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med 13: 730–735 [DOI] [PubMed] [Google Scholar]
- Ohkura N, Nagamura Y, Tsukada T 2008. Differential transactivation by orphan nuclear receptor NOR1 and its fusion gene product EWS/NOR1: possible involvement of poly(ADP-ribose) polymerase I, PARP-1. J Cell Biochem 105: 785–800 [DOI] [PubMed] [Google Scholar]
- Pearen MA, Muscat GEO 2010. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol 24: 1891–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnio T, Burton Q, Pereira FA, Wu DK, Conneely OM 2002. The nuclear receptor Nor-1 is essential for proliferation of the semicircular canals of the mouse inner ear. Mol Cell Biol 22: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaud J, Desroches J, Balsalobre A, Drouin J 2009. TIF1β/KAP-1 is a coactivator of the orphan nuclear receptor NGFI-B/Nur77. J Biol Chem 284: 14147–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Herrick AM, Mullican SE, Sheehan AM, Conneely OM 2011. Reduced NR4A gene dosage leads to mixed myelodysplastic/myeloproliferative neoplasms (MDS/MPN) in mice. Blood 117: 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassool FV, Gaymes TJ, Omidvar N, Brady N, Beurlet S, Pla M, Reboul M, Lea N, Chomienne C, Thomas NS, et al. 2007. Reactive oxygen species, DNA damage, and error-prone repair: a model for genomic instability with progression in myeloid leukemia? Cancer Res 67: 8762–8771 [DOI] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM 1998. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci 95: 4013–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirin O, Lukov GL, Mao R, Conneely OM, Goodell MA 2010. The orphan nuclear receptor Nurr1 restricts the proliferation of haematopoietic stem cells. Nat Cell Biol 12: 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Luk N, Newton RA, Roberts DW, Sturm RA, Muscat GE 2008. Melanocortin-1 receptor signaling markedly induces the expression of the NR4A nuclear receptor subgroup in melanocytic cells. J Biol Chem 283: 12564–12570 [DOI] [PubMed] [Google Scholar]
- Traven A, Heierhorst J 2005. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. BioEssays 27: 397–407 [DOI] [PubMed] [Google Scholar]
- van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, Mari PO, Tezcan I, Chen DJ, Zdzienicka MZ, et al. 2009. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest 119: 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, van der Burg M 2007. Non-homologous end-joining, a sticky affair. Oncogene 26: 7731–7740 [DOI] [PubMed] [Google Scholar]
- Veuger SJ, Curtin NJ, Richardson CJ, Smith GCM, Durkacz BW 2003. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res 63: 6008–6015 [PubMed] [Google Scholar]
- Volakakis N, Malewicz M, Kadkhodai B, Perlmann T, Benoit G 2006. Characterization of the Nurr1 ligand-binding domain co-activator interaction surface. J Mol Endocrinol 37: 317–326 [DOI] [PubMed] [Google Scholar]
- Volakakis N, Kadkhodaei B, Joodmardi E, Wallis K, Panman L, Silvaggi J, Spiegelman BM, Perlmann T 2010. NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc Natl Acad Sci 107: 12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NP, Perlmann T 2003. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423: 555–560 [DOI] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ 2008. Ku recruits XLF to DNA double-strand breaks. EMBO Rep 9: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterström RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T 1997. Dopamine neuron agenesis in Nurr1-deficient mice. Science 276: 248–250 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Bruemmer D 2010. NR4A orphan nuclear receptors: transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler Thromb Vasc Biol 30: 1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]