Abstract
Transcriptional R loops are anomalous RNA:DNA hybrids that have been detected in organisms from bacteria to humans. These structures have been shown in eukaryotes to result in DNA damage and rearrangements; however, the mechanisms underlying these effects have remained largely unknown. To investigate this, we first show that R-loop formation induces chromosomal DNA rearrangements and recombination in Escherichia coli, just as it does in eukaryotes. More importantly, we then show that R-loop formation causes DNA replication fork stalling, and that this in fact underlies the effects of R loops on genomic stability. Strikingly, we found that attenuation of replication strongly suppresses R-loop-mediated DNA rearrangements in both E. coli and HeLa cells. Our findings thus provide a direct demonstration that R-loop formation impairs DNA replication and that this is responsible for the deleterious effects of R loops on genome stability from bacteria to humans.
Keywords: R loop, genome stability, transcription, replication
A transcriptional R loop is a structure in which a nascent transcript is hybridized with the template DNA strand, leaving the nontemplate strand unpaired. Such RNA:DNA hybrids have been detected in organisms from bacteria to humans under various physiological and/or pathological conditions (Li and Manley 2006). A growing body of evidence has revealed an inherent impact of R loops on the integrity of genomes in eukaryotes (Li and Manley 2006; Aguilera and Gomez-Gonzalez 2008). For example, R loops occur naturally in highly repetitive and GC-rich immunoglobulin gene switch (S) regions in mouse B cells, contributing to class switch recombination (CSR) (Yu et al. 2003; Huang et al. 2006). In Saccharomyces cerevisiae, formation of R loops underlies transcription-associated hyperrecombination phenotypes in mutants affecting components of the THO complex, which functions to link transcription and mRNP assembly in yeast (Huertas and Aguilera 2003). In chicken DT40 cells and human HeLa cells, R loops can accumulate in the absence of the SR splicing factor SRSF1 and lead to DNA double-strand breaks (DSBs) and DNA rearrangements (Li and Manley 2005). Furthermore, a genome-wide siRNA screen in HeLa cells identified a group of mRNA processing factors that function in preventing R-loop-mediated DNA breaks, providing evidence that R-loop formation constitutes a broad source of genomic instability in human cells (Paulsen et al. 2009).
R loops are known to form in organisms from bacteria to humans. Except in unusual situations such as CSR, they are deleterious, and cells employ a number of mechanisms to prevent their formation or eliminate them once formed (Li and Manley 2006). One is to prevent the nascent RNA from reannealing with template DNA, and this is accomplished in eukaryotes by rapid association with factors such as the THO complex in yeast and SR proteins in mammals (Li and Manley 2005), while translating ribosomes and Rho-mediated transcription termination serve this function in prokaryotes (Masse and Drolet 1999a; Broccoli et al. 2004). Second, topoisomerase I (Top I) has been identified as an evolutionarily conserved factor that suppresses R-loop formation (Drolet et al. 1995; Tuduri et al. 2009; El Hage et al. 2010). This is likely due to the ability of Top I to relieve negative supercoils, which can facilitate reannealing between the nascent transcript and template DNA strand by promoting opening of the DNA behind RNA polymerase (RNAP) (Drlica 1992; Drolet 2006). Additionally, Top I has been reported to prevent R-loop formation by enhancing the assembly of mRNPs in a SRSF1-dependent manner in mammalian cells (Tuduri et al. 2009). Finally, R loops can be actively removed once formed. For example, the suppressive effect of RNase HI overexpression on R-loop-mediated growth defects and genome instability suggest that R-loop formation can be removed by RNase H-mediated degradation (Drolet et al. 1995; Huertas and Aguilera 2003; Li and Manley 2005), and loss of RNase HI activity can provoke the SOS response in Escherichia coli (Kogoma et al. 1993). It is also of note that a recent study by Mischo et al. (2011) showed that Sen1, an RNA:DNA helicase thought to be involved in transcription termination, can prevent the occurrence of R loops in yeast, suggesting a possible role for helicases in R-loop resolution.
Despite the strong correlation between R-loop formation and loss of genome stability in eukaryotes, the underlying mechanisms are largely unclear. Studies on mechanisms for CSR targeting in Ig genes suggest that activation-induced deaminase (AID), a factor specifically expressed in active B cells, introduces DNA lesions in ssDNA regions resulting from transcriptional R loops in S regions (Petersen-Mahrt et al. 2002; Rada et al. 2002; Bransteitter et al. 2003; Chaudhuri et al. 2003; Imai et al. 2003; Yu et al. 2003). This likely initiates the process of CSR between the Sμ and acceptor S regions, although some debate remains (Stavnezer et al. 2008). Consistent with its proposed roles in R-loop-mediated genome instability, AID has the capacity to exacerbate transcription-dependent hypermutation and hyperrecombination phenotypes when expressed in yeast THO complex mutants (Gomez-Gonzalez and Aguilera 2007). However, AID is predominantly expressed in active B cells at the germinal center (Muramatsu et al. 1999). This raises the question of how transcriptional R-loop formation impairs genome integrity in AID-lacking cells, such as SRSF1-depleted HeLa cells.
It is also possible that R loops can induce genomic instability by interfering with DNA replication. Indeed, a growing body of evidence demonstrates that genomes are particularly vulnerable when undergoing replication (Labib and Hodgson 2007; Szilard et al. 2010). Aguilera and colleagues (Huertas and Aguilera 2003; Wellinger et al. 2006) have shown that R-loop-mediated hyperrecombination occurs concomitantly with a transcription-dependent slowdown of replication fork progression in plasmid-borne constructs in yeast hpr1Δ mutants. Tuduri et al. (2009) reported that depletion of Top I in mammalian cells resulted in transcription-dependent and RNase H-sensitive DNA breaks and replication defects. Notably, both the transcription-associated recombination phenotype in yeast hpr1Δ cells and DNA breaks resulting from Top I deficiency in mammalian cells occurred predominantly in S phase, suggesting a correlation between replication and R-loop-mediated genome instability. However, it remains unclear whether such R-loop-associated replication defects in fact account for the R-loop-mediated genomic instability that occurs in these cells.
Here we describe experiments establishing an evolutionarily conserved mechanism underlying R-loop-mediated genomic instability. We first present evidence that transcription through a fragment of the mouse Ig gene Sγ3 region in its physiological orientation results in R-loop-mediated DNA rearrangement and recombination in E. coli, indicating that transcriptional R-loop formation is a conserved threat to genome integrity throughout evolution. We further show, by analysis of the pattern of replication intermediates, that R-loop formation impedes replication fork progression and that the impact of R loops on genome stability is in fact dependent on the presence of active replication in both E. coli and SRSF1-depleted vertebrate cells. Our data establish that impairment of replication is an evolutionarily conserved mechanism that underlies the catastrophic effects of R loops on genome stability.
Results
Transcription through mouse Ig gene Sγ3 region in its physiological orientation results in plasmid loss and growth defects in E. coli
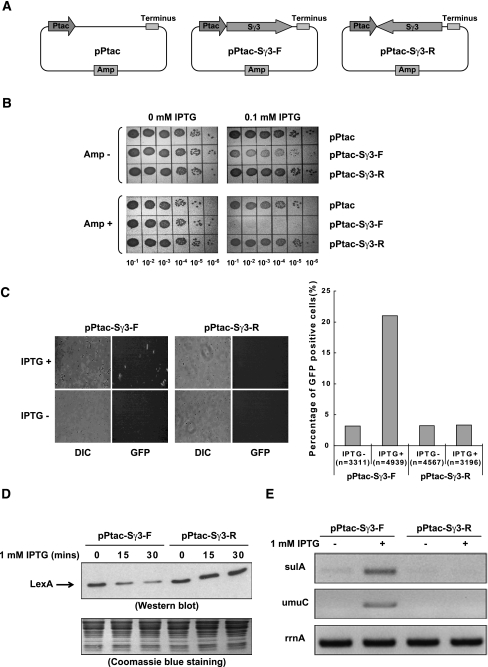
To continue our studies on the mechanisms of R-loop formation, we initially set out to clone a fragment of the mouse Ig gene Sγ3 region in an E. coli expression plasmid. However, we found that the Sγ3 region could only be inserted in the direction opposite to its physiological orientation relative to the lacZ promoter in the pBluescriptII KS vector. It has been documented that transcription through the S region in its physiological orientation results in formation of R loops both in vitro and in vivo (Reaban et al. 1994; Tian and Alt 2000; Yu et al. 2003; Duquette et al. 2004), and that R-loop formation is indeed a major problem linked to growth defects observed in E. coli cells deficient in Top I and in the dinG helicase mutant that carries a chromosomal inversion of the rRNA operons (Drolet et al. 1995; Boubakri et al. 2010). We therefore suspected that transcription of Sγ3 driven by the constitutively active lacZ promoter resulted in R-loop-associated growth defects, and that this ultimately eliminated cells transformed with plasmids containing the Sγ3 fragment in its physiological orientation relative to the lacZ promoter. To test this idea, we constructed plasmids carrying a fragment of the mouse Sγ3 region downstream from the IPTG-inducible Ptac promoter (Fig. 1A). In this context, the Sγ3 region was successfully inserted downstream from the Ptac promoter in both orientations at roughly equal efficiency (data not shown). Addition of IPTG had no observable effect on the growth of cells transformed with pPtac or pPtac-Sγ3-R (reverse) plasmids. However, induction of transcription with IPTG resulted in a significant growth arrest in cells transformed with the pPtac-Sγ3-F (forward) plasmid (Fig. 1B, top panel). Remarkably, the pPtac-Sγ3-F transformed cells were completely eliminated on plates bearing both IPTG and ampicillin (Amp) (Fig. 1B, bottom panel). As the amp resistance gene was carried by the pPtac-Sγ3-F plasmid, these results indicate that transcription through Sγ3 regions resulted in severe plasmid loss in a manner dependent on the orientation of transcription.
Figure 1.
Transcription through a plasmid-borne Sγ3 region in its physiological orientation induces cell growth defect, plasmid loss, and SOS damage response in E. coli. (A) Schematic diagram of pPtac, pPtac-Sγ3-F, and pPtac-Sγ3-R plasmids. Positions of the Ptac promoter, transcription terminus, Sγ3 region, and amp resistance gene are indicated. (B) Determination of the viability of transformants in the presence or absence of 0.1 mM IPTG. E. coli K-12 BW25113 cells were transformed by pPtac, pPtac-Sγ3-F, or pPtac-Sγ3-R plasmids, respectively. A single colony from each transformant was inoculated into LB with 100 μg/mL Amp. Tenfold dilutions of overnight cultures were grown on LB plates containing 0.1 mM IPTG and/or 100 μg/mL Amp. (C) Sγ3 transcription in its physiological orientation activates the SOS-inducible sulA promoter. pPtac-Sγ3-F or pPtac-Sγ3-R was transformed into an E. coli SMR8379 strain in which a chromosomally located gfp gene is under the control of a SOS-inducible sulA promoter. Transformants were cultured in the presence or absence of 1 mM IPTG for 30 min and allowed to recover for 2 h. Expression of GFP was monitored by fluorescent microscope. Representative images are presented. Quantification is graphically shown in the right panel. The number of cells counted for each sample is indicated. (DIC) Differential interference contrast image; (GFP) GFP fluorescence image. (D,E) E. coli K-12 BW25113 cells were transformed with pPtac-Sγ3-F or pPtac-Sγ3-R plasmids, respectively. Transformants were grown in the presence of 1 mM IPTG for the indicated times and allowed to recover for 30 min. (D, top panel) Cell lysates were assayed for LexA by Western blot. (Bottom panel) The amount of proteins loaded onto each lane of the SDS-PAGE gel was monitored by Coomassie blue staining. (E) The mRNA levels of the sulA (top panel), umuC (middle panel), and rrnA (bottom panel) genes were determined by RT–PCR.
Induction of the SOS-DNA damage response by transcription of the Sγ3 region
R-loop formation is known to impair DNA integrity in eukaryotes, but has not been documented in prokaryotes. The fact that transcription of Sγ3 resulted in plasmid loss in E. coli cells led us to suspect that transcription through the Sγ3 region in its physiological orientation might induce DNA damage. To test this, we first employed the sulAp-gfp reporter system to measure the DNA damage-induced SOS response in single living E. coli cells in the presence or absence of Sγ3 transcription (McCool et al. 2004; Pennington and Rosenberg 2007). For this, pPtac-Sγ3-F and pPtac-Sγ3-R plasmids were transformed into the E. coli strain SMR8379 (Supplemental Table S1), which contains a chromosomally located gfp reporter gene under control of the SOS-inducible sulA promoter (Pennington and Rosenberg 2007). If transcription through the Sγ3 region results in SOS response-inducing DNA damage, the cells will be fluorescent green. As shown in Figure 1C, 30-min IPTG exposure induced a significant increase in the expression of GFP in cells carrying the pPtac-Sγ3-F plasmid (from 3.2% to 21.0%, P < 0.0001), but not in cells carrying the pPtac-Sγ3-R plasmid (from 3.2% to 3.4%, with no statistical significance). Consistent with activation of the SOS response, IPTG caused filamentous growth of cells carrying the pPtac-Sγ3-F plasmid, but not of cells carrying pPtac-Sγ3-R (Fig. 1C; data not shown). Filamentous growth is one of the well-documented manifestations of the SOS response, resulting from induction of sulA expression (Walker 1985). These data suggest that transcription through the Sγ3 region in its physiological orientation in E. coli induces the SOS-DNA damage response.
We next wished to obtain direct evidence that an SOS response is indeed activated upon transcription of the Sγ3 region. The SOS response is initiated when RecA-ssDNA filaments stimulate autocleavage of the transcriptional repressor LexA, which in turn activates expression of >40 SOS response genes (Little and Mount 1982; Janion 2008). We therefore examined the integrity of the LexA repressor after induction of Sγ3 transcription. Consistent with the activation of the sulA promoter described above, addition of IPTG resulted in a significant decrease in the level of intact LexA repressor in whole-cell extracts from strains carrying pPtac-Sγ3-F, but not in extracts from cells carrying pPtac-Sγ3-R (Fig. 1D). In agreement with the degradation of LexA, the mRNA levels of two SOS genes, the sulA and umuC genes, were markedly increased in cells with Sγ3 transcription in its physiological orientation (Fig. 1E, bottom panel). Both the sulA and umuC genes contain strong SOS boxes with high affinity for LexA and are only transcriptionally activated when the LexA concentration is significantly reduced after high or persistent SOS induction (Michel 2005). The increase in the amount of transcripts of the two SOS genes was specific and significant because the mRNA levels of the rrnA gene, a constitutively expressed housekeeping gene, were comparable among samples (Fig. 1E). It is also noteworthy that the sulA gene encodes an inhibitor of cell division, and that the umuC gene encodes a subunit of the mutagenic DNA repair polymerase PolV. These data suggest that induction of SOS gene expression underlies the growth defect and filamentation phenotypes observed in E. coli cells with active Sγ3 transcription. Together, these results provide strong evidence that transcription through a plasmid-borne Sγ3 region in its physiological orientation activates the SOS-DNA damage response in E. coli.
Transcription through the Sγ3 region induces DNA rearrangements in the E. coli chromosome
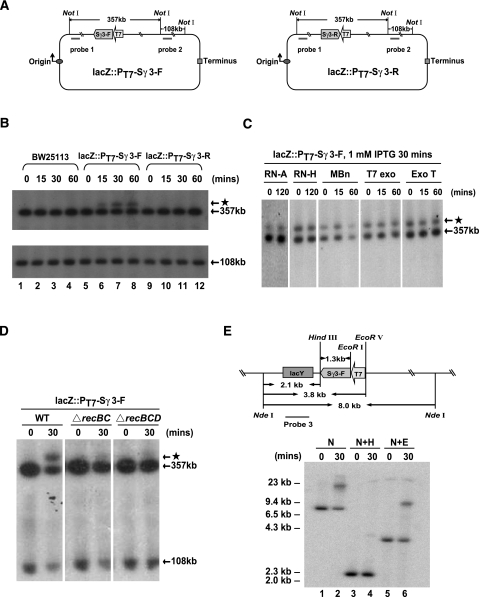
We next wished to determine whether transcription through the Sγ3 region in fact impairs the integrity of the E. coli genome. To this end, we constructed two derivatives of E. coli strain BW25113—lacZ::PT7-Sγ3-F and lacZ::PT7-Sγ3-R—by integrating a bacteriophage T7 promoter and a 1.2-kb fragment of the Sγ3 region into the E. coli chromosome in the lacZ locus in both orientations relative to the T7 promoter (Fig. 2A). An IPTG-inducible T7 RNAP expression construct was then introduced into these cells. After IPTG exposure for various times, chromosomal DNA was purified and digested with NotI endonuclease in agarose plugs. Digested DNAs were subjected to pulsed-field gel electrophoresis (PFGE), followed by Southern blot analysis using the probes shown in Figure 2A. Analogous to the parental strain BW25113, IPTG treatment had no effect on the integrity of the chromosomal DNA in the lacZ::PT7-Sγ3-R strain (Fig. 2B, lanes 1–4,9–12). However, the amount of a 357-kb DNA fragment, which contains the Sγ3 transcription unit, was modestly but repeatedly decreased in samples from the lacZ::PT7-Sγ3-F strain in a time-dependent manner after IPTG addition (Fig. 2B, lanes 5–8). Strikingly, a slowly migrating band, designated as the “starlet band,” was detected by probe 1 in the samples from the lacZ::PT7-Sγ3-F strain as early as 15 min after IPTG addition (Fig. 2B, lanes 6–8). The starlet band was specifically induced by transcription of the Sγ3 region in its physiological orientation, as it was absent in samples from the parental strain and the lacZ::PT7-Sγ3-R strain at all time points tested after IPTG addition (Fig. 2B, lanes 1–4,9–12).
Figure 2.
Transcription through the Sγ3 region in its physiological orientation induces DNA rearrangement in the chromosome of E. coli. (A) Schematic of chromosomes of E. coli strains lacZ::PT7-Sγ3-F (left panel) and lacZ::PT7-Sγ3-R (right panel). Relative positions of the probe-binding sites, NotI cleavage sites, T7 promoter (T7), and Sγ3 region (Sγ3-F or Sγ3-R) are indicated. (B) Southern blot analysis of the chromosomal regions surrounding the Sγ3 transcription unit. BW25113, lacZ::PT7-Sγ3-F, and lacZ::PT7-Sγ3-R strains were grown in the presence of 1 mM IPTG for the indicated times. Genomic DNAs from these strains were purified in agarose plugs and digested with NotI. Digested DNAs were subjected to PFGE and Southern blot analysis with probe 1 (top panel) and probe 2 (bottom panel), respectively. Sizes of targeted DNA fragments are indicated. The starlet band, a slowly migrating band detected by probe 1, is indicated by an asterisk. (C) RNA- and ssDNA-specific nuclease treatments have no effect on the migration of the starlet band. The lacZ::PT7-Sγ3-F strain was grown in the presence of 1 mM IPTG for 30 min. NotI-digested DNAs were subsequently treated with RNase A (RN-A), RNase H (RN-H), mung bean nuclease (MBn), T7 exonuclease (T7 exo), or exonuclease T (Exo T), respectively, for the indicated times prior to PFGE. Southern blot analysis was performed as described above. (D) The RecBCD recombination pathway is required for the appearance of the starlet band. NotI-digested DNAs from lacZ::PT7-Sγ3-F, lacZ::PT7-Sγ3-FΔrecBC, and lacZ::PT7-Sγ3-FΔrecBCD strains were subjected to PFGE and Southern blot analysis as described above. (E) A DNA rearrangement event appears in the Sγ3 region upon induction of Sγ3 transcription in its physiological orientation. The lacZ::PT7-Sγ3-F strain was grown in the presence of 1 mM IPTG for the indicated times. DNAs were digested with NdeI, NdeI plus HindIII, or NdeI plus EcoRV, respectively, and hybridized with probe 3 specific to the lacY gene located immediately upstream of the Sγ3 transcription unit in the chromosome of the lacZ::PT7-Sγ3-F strain.
The starlet band could reflect a DNA rearrangement event induced by Sγ3 transcription or a mobility shift of the 357-kb DNA fragment due to either the presence of associated transcripts or a DNA conformation change. To distinguish between these possibilities, we first treated the NotI-digested DNA agarose plugs with RNase A-, RNase H-, or ssDNA-specific nucleases prior to PFGE. The results showed that the starlet band was resistant to all of the nuclease treatments except for a 60-min incubation with mung bean nuclease (MBn). While MBn is a nuclease that prefers ssDNA over dsDNA, MBn has the capacity to degrade dsDNA upon longer incubations (http://www.neb.com/nebecomm/products/faqproduct M0250.asp). Because the signals in both the starlet band and the 357-kb band were decreased after the 60-min MBn treatment, and because both bands were resistant to a 15-min MBn treatment, we conclude that the degradation observed upon the 60-min MBn treatment was not due to the appearance of a ssDNA region in the starlet band. Taken together, these data indicate that neither transcripts of the Sγ3 region nor DNA conformation affect the mobility of the starlet band (Fig. 2C). In contrast, consistent with the model that the appearance of the starlet band reflects a DNA rearrangement, the amount of the starlet band markedly decreased in recBC- and recBCD-null mutant derivatives of the lacZ::PT7-Sγ3-F strain upon IPTG induction (Fig. 2D), indicative of the involvement of the RecBCD recombination pathway in its formation.
The above data indicate that activation of Sγ3 transcription induces a DNA rearrangement, reflected in the appearance of the starlet band. To investigate this further, we next asked where in the E. coli genome the rearrangement occurs. To this end, genomic DNAs from cells with or without 30-min IPTG exposure were digested with NdeI, NdeI plus HindIII, or NdeI plus EcoRV (see Fig. 2E, top panel). Digested DNAs were subjected to Southern blot analysis using a probe specific for the lacY gene, which is located immediately upstream of the Sγ3 transcription unit in the chromosome of the lacZ::PT7-Sγ3-F strain (Fig. 2E, top panel). Consistent with the data presented above, Sγ3 transcription resulted in a significant decrease in the amount of the DNA fragment carrying the lacY gene and in the appearance of a slowly migrating band in both the Nde I-digested and the NdeI plus EcoRV-digested samples (Fig. 2E, lanes 1,2, and 5,6, respectively). However, neither the decrease of the lacY-containing fragment nor the appearance of the slowly migrating band was observed in the NdeI plus HindIII-digested samples (Fig. 2E, lanes 3,4). As there is only one HindIII site, located immediately upstream of the Sγ3 region, in the targeted NdeI–EcoRV fragments from the chromosome of the lacZ::PT7-Sγ3-F strain, these data indicate that a DNA rearrangement event occurred in the Sγ3 region upon transcription of Sγ3 in its physiological orientation.
Transcription of the Sγ3 region stimulates homologous recombination in an orientation-dependent manner
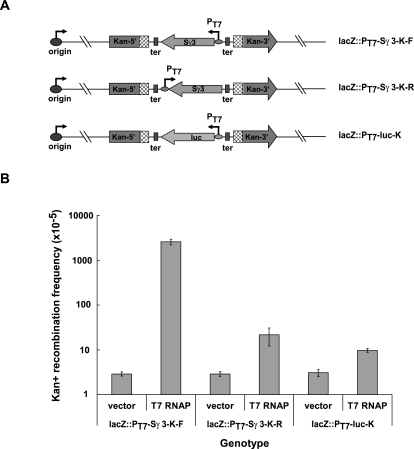
We next wished to determine whether transcription of the Sγ3 region provokes DNA recombination in the E. coli chromosome. To this end, two incomplete copies of the kanamycin resistance (kan) gene with 240-base-pair (bp) direct repeats were inserted into the genome of the lacZ::PT7-Sγ3-K-F and lacZ::PT7-Sγ3-K-R strains, respectively (Fig. 3A, top and middle panels). Given that a homologous recombination event between the two nonfunctional copies restores Kan resistance, recombination rates can be measured by the number of Kan-resistant colonies. To determine whether transcription of the Sγ3 region induces Kan resistance, vector alone or an IPTG-inducible T7 RNAP expression construct was first introduced into these strains. Transformants were subcultured in the presence of IPTG for 30 min, allowed to recover for 1 h, and then plated on Kan-containing LB plates to select cells with a functional kan gene. As shown in Figure 3B, Kan-positive recombination rates were similar in both strains in the absence of T7 RNAP. Strikingly, compared with the lacZ::PT7-Sγ3-K-R strain, the recombination rate was significantly increased in the lacZ::PT7-Sγ3-K-F strain in the presence of T7 RNAP (Fig. 3B). Although transcription of Sγ3 is codirectional with replication in the lacZ::PT7-Sγ3-K-R strains, the difference in recombination rates was not due to the fact that transcription of Sγ3 is head-on with replication in the lacZ::PT7-Sγ3-K-F strains, as the recombination rate in the lacZ::PT7-luc-K strain, which contains a T7 promoter-driven luciferase (luc) gene head-on with replication, was actually slightly lower than that of the lacZ::PT7-Sγ3-K-R strain upon induction of T7 RNAP expression (Fig. 3A [bottom panel], B). In agreement with the involvement of homologous recombination events in reconstruction of the kan gene, the increase of Sγ3 transcription-induced Kan-positive recombination rates in a recA-null mutant derivative of the lacZ::PT7-Sγ3-K-F strain was ∼10-fold lower than that in the parental lacZ::PT7-Sγ3-K-F strain (Supplemental Fig. S1). Together, these data indicate that transcription through the Sγ3 region in its physiological orientation significantly increased the homologous recombination rate in the surrounding chromosomal regions.
Figure 3.
Sγ3 transcription in its physiological orientation increases homologous recombination rates in its surrounding regions. (A) Schematic of chromosomes of E. coli strains lacZ::PT7-Sγ3-K-F, lacZ::PT7-Sγ3-K-R, and lacZ::PT7-luc-K. Relative positions and orientations of the replication origin (origin), two nonfunctional kan resistence gene expression cassettes (Kan-5′ and Kan-3′), directly repeated regions (boxes filled with rhombuses), the Sγ3 region (Sγ3), and luciferase (luc) transcription units are indicated. (B) Rates of recombination were measured using the reconstitution of Kan resistance as an indicator. Relative rates of recombination were calculated and are shown in the graph. Error bars represent the average deviation from three independent experiments.
RNase H overexpression suppresses chromosomal DNA rearrangement and recombination resulting from Sγ3 transcription
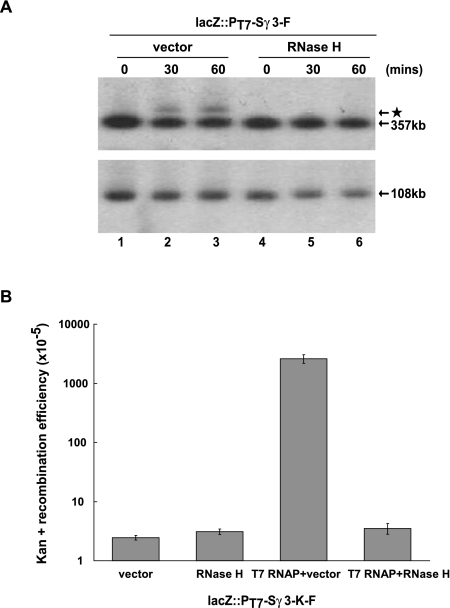
The above data reveal that transcription through a chromosomally located Sγ3 fragment impairs the integrity of the E. coli genome, reflected in the rapid appearance of DNA rearrangement and recombination events in chromosomal regions surrounding the Sγ3 transcription unit. As transcription through the S region results in the formation of R loops, which have been demonstrated to have deleterious effects on genomic integrity in eukaryotes (Huertas and Aguilera 2003; Li and Manley 2005; Paulsen et al. 2009), we asked whether the DNA rearrangement and recombination events induced by Sγ3 transcription in E. coli are indeed attributable to R-loop formation. If this were the case, destroying R loops by overexpression of RNase H would be expected to suppress these phenotypes. To test this, we introduced vector alone or an E. coli RNase HI expression construct into the lacZ::PT7-Sγ3-F strain. Southern blot analysis was performed to measure the amount of the starlet band resulting from a DNA rearrangement event induced by Sγ3 transcription. As shown in Figure 4A, while the starlet band was induced by Sγ3 transcription in vector-alone transformants (lanes 1–3), it disappeared completely in cells overexpressing RNase H (lanes 4–6).
Figure 4.
RNase H overexpression suppresses DNA rearrangement and recombination resulting from Sγ3 transcription. (A) Southern blot analysis of chromosomal regions surrounding the Sγ3 transcription unit. The lacZ::PT7-Sγ3-F strain was transformed with vector or an RNase H expression construct, respectively. Transformants were grown in the presence of 1 mM IPTG for the indicated times and subjected to Southern blot analysis as described above. (B) The lacZ::PT7-Sγ3-K-F strain was transformed with vector, an RNase H expression construct, vector plus a T7 RNAP expression construct, or RNase H plus T7 RNAP expression constructs, respectively. Transformants were grown in the presence of 1 mM IPTG for 30 min, allowed to recover for 1 h, and then plated on Kan-containing LB plates to select for cells carrying the functional kan resistance gene. Relative rates of recombination were calculated and are shown in the graph. Error bars represent the average deviation from three independent experiments.
We next examined whether RNase H overexpression suppressed Sγ3 transcription-induced DNA recombination by using reconstitution of Kan resistance as an indicator. To this end, T7 RNAP expression constructs, together with empty vector or the RNase HI expression plasmid, were introduced into the lacZ::PT7-Sγ3-K-F strains. Compared with the vector-alone transformants, overexpression of RNase H resulted in an ∼750-fold decrease in the rate of recombination between direct repeats in the two incomplete copies of the kan gene that was induced by Sγ3 transcription (Fig. 4B). This correlates well with the decrease in the amount of the starlet band detected in the Southern blot analysis (Fig. 4A). Together, these results demonstrate that R-loop formation accounts for the negative effect of Sγ3 transcription on the integrity of the E. coli genome.
Transcription through a plasmid-borne Sγ3 region provokes R-loop-mediated replication fork stalling
The above data, together with previous studies, indicate that R-loop formation constitutes a threat to genome stability conserved from E. coli to humans. We next wished to investigate the mechanism of this evolutionarily conserved phenomenon. One possibility is suggested by the fact that R-loop formation has the capacity to impair transcription elongation (see the Discussion). The halted or slowed transcription machinery might in turn block the progression of DNA replication (Tehranchi et al. 2010). A possible outcome of this could be replication fork stalling and the resultant impairment of DNA integrity (Mirkin and Mirkin 2007). We therefore asked whether transcription through the Sγ3 region could lead to R-loop-induced replication fork stalling.
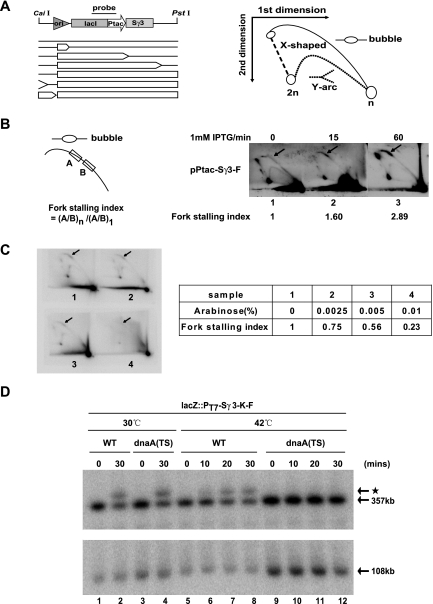
We first examined whether Sγ3 transcription could impede replication fork progression in plasmid-borne constructs. To this end, we used two-dimensional (2D) gel electrophoresis to analyze the pattern of replication intermediates on the pPtac-Sγ3-F and pPtac-Sγ3-R plasmids in the presence or absence of IPTG. Transcription of the Sγ3 region in both plasmids is orientated codirectionally with replication from the ColE1 origin to eliminate the inhibitory effect of head-on transcription on replication fork progression (French 1992; Liu and Alberts 1995; Pomerantz and O'Donnell 2010; Srivatsan et al. 2010). As ColE1 is a unidirectional origin, the replicating plasmid, cleaved upstream of the origin and downstream from the Sγ3 region, should be a bubble-shaped, Y-shaped, or X-shaped structure. If a replication fork stalls around the Sγ3 transcription unit, the bubble-shaped intermediates will accumulate, and this should lead to the appearance of a bulge on the bubble arc. To test this, the transformants were grown in the presence or absence of IPTG for 30 min. Plasmid DNAs were purified and subjected to CaiI and PstI digestion (Fig. 5A). Replication intermediates were analyzed by 2D gel electrophoresis followed by Southern blot analysis using a probe specific for the lacI gene, which is located immediately upstream of the Sγ3 insertion. As shown in Figure 5B, induction of Sγ3 transcription resulted in a sharp increase of bubble-shaped replication intermediates in the pPtac-Sγ3-F plasmid, reflected in the accumulation of a bulge on its bubble arc in a time-dependent manner. In contrast, no visible bulge appeared in the bubble arc when Sγ3 transcription was induced in the pPtac-Sγ3-R plasmid (Supplemental Fig. S2A,B, bottom right panel). These data indicate that transcription through the Sγ3 region in its physiological orientation blocks replication fork progression.
Figure 5.
R-loop-mediated chromosomal DNA rearrangement is dependent on the presence of active replication in E. coli. (A) Schematic representation of 2D gel electrophoresis for plasmids containing the Sγ3 transcription unit. (Top left panel) Locations of the replication origin (ori), the Sγ3 region (Sγ3), the Ptac promoter (Ptac), the lacI gene (lacI), and the CaiI and PstI endonuclease cleavage sites are indicated. Plasmids were linearized by CaiI and PstI double digestion prior to 2D gel electrophoresis. Structures of linear replication intermediates are graphically presented in the left panel. The size of a bubble reflects the extent of replication. The right panel diagrams the separation of linear replication intermediates by 2D gel electrophoresis. (B) Transcription through a plasmid-borne Sγ3 region in its physiological orientation induces replication fork stalling. Cells were transformed with pPtac-Sγ3-F plasmids. Transformants were grown in the presence or absence of IPTG for the indicated times before harvest. Replicating plasmids were purified and analyzed as described in the Materials and Methods. The definition of the fork stalling index is schematically depicted in the left panel. Representative photographs of the corresponding autoradiographs are shown in the right panel. The arrow indicates replication fork stalling sites. The fork stalling index for each panel is indicated below the graphs. (C) RNase H overexpression alleviates Sγ3 transcription-induced replication fork blockage. An arabinose-inducible RNase H expression construct was introduced into cells containing the pPtac-Sγ3-F plasmid. Replication intermediates of the pPtac-Sγ3-F plasmid were analyzed as described above. Representative photographs of the corresponding autoradiographs are shown in the left panel. The arrow indicates replication fork stalling sites. The fork stalling index for each sample is shown in the right panel. (D) Wild-type lacZ::PT7-Sγ3-F strain (WT) and lacZ::PT7-Sγ3-F dnaA46(TS) tnaA::Tn10 strain [dnaA(TS)] were grown for 2.5 h at 30°C or 42°C prior to IPTG treatment for the indicated times. Genomic DNAs from these strains were then purified in agarose plugs and digested with NotI. Digested DNAs were subjected to PFGE and Southern blot analysis with probe 1 (top panel) and probe 2 (bottom panel) as described in Figure 2A. Sizes of targeted bands are indicated. The starlet band is indicated by an asterisk.
We next wished to determine whether transcription-induced replication fork blockage resulted from R-loop formation. To test this, we introduced an arabinose-inducible RNase H expression construct into cells carrying the pPtac-Sγ3-F plasmid. The intensity of hybridization signals in the bulge was normalized among samples by comparison with that of its nearby region on the same bubble arc, and this generates an index of fork stalling. As shown in Figure 5C, in contrast to the cells that were grown in the presence of glucose (without arabinose), RNase H overexpression significantly alleviated transcription-induced replication fork stalling, reflected in the appearance of a 77% decrease in the fork stalling index in cells that were grown in the presence of 0.01% arabinose (Fig. 5C). These data indicate that transcription of a plasmid-borne Sγ3 induces R-loop-mediated replication fork stalling.
Active replication is required for Sγ3 transcription-induced chromosomal rearrangements
The fact that transcription through a plasmid-borne Sγ3 fragment blocks replication fork progression raises the possibility that the effect of R-loop formation on the integrity of the E. coli genome might be replication-dependent. To test this possibility, we constructed a derivative of the lacZ::PT7-Sγ3-F strain, lacZ::PT7-Sγ3-F dnaA46(TS) tnaA::Tn10, bearing a temperature-sensitive dnaA gene (Pennington and Rosenberg 2007). As initiation of chromosomal DNA replication is mediated by dnaA, new rounds of chromosomal DNA replication from oriC cannot be initiated in the dnaA(TS) mutant at the nonpermissive temperature (42°C) (Hirota et al. 1970). We then asked whether inactivation of dnaA could in fact suppress R-loop-mediated chromosomal DNA rearrangement upon induction of Sγ3 transcription. For this, cells were grown for 2.5 h at 30°C or 42°C before addition of IPTG. When replication was permitted in dnaA(TS) mutants at 30°C, IPTG treatment induced the appearance of the starlet band at a level comparable with that in the strain carrying the wild-type dnaA gene (Fig. 5D, lanes 1–4). However, while temperature shift had no effect on formation of the starlet band in the wild-type strains, the starlet band disappeared in the dnaA(TS) mutants when the cells were grown at 42°C (Fig. 5D, lanes 5–12). These results indicate that the effect of Sγ3 transcription on E. coli genomic integrity is dependent on the presence of active replication.
Active replication is required for SRSF1 depletion-induced DNA damage in vertebrate cells
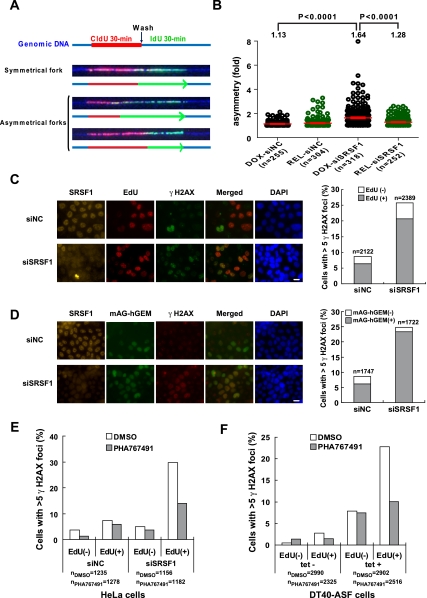
The data presented above has revealed that R-loop formation impairs genomic integrity in a replication-dependent manner in E. coli. Given that R loops pose a conserved threat to genomic stability throughout evolution, an important question is whether replication impairment is a general mechanism underlying R-loop-mediated genomic instability in other species. As our previous studies have established that depletion of splicing factor SRSF1 results in R-loop-mediated genomic instability, reflected by the appearance of DSBs, in both chicken DT40 cells and human HeLa cells (Li and Manley 2005), we first wished to determine whether SRSF1 depletion results in R-loop-mediated replication fork stalling in HeLa cells. To this end, we took advantage of a stable HeLa cell derivative, HeLa-HRH, which contains a tetracycline (tet) transactivator (tTA) expression cassette and a copy of the human RNase HI cDNA driven by a tet-repressible promoter (F Yang, Y Wei, J Tu, J Liu, Y Zhai, and X Li, unpubl.). Thus, expression of exogenous RNase H could be simply activated in these cells by withdrawal of doxycycline (dox) from the medium. After growth in the presence or absence of dox for 72 h, HeLa-HRH cells were transfected with either an SRSF1 (siSRSF1) or control (siNC) siRNA duplex for 60 h and subjected to two sequential rounds of 30-min pulse-labeling with different halogenated nucleotides before harvest. Genomic DNA was then purified in agarose plugs and stretched onto silanized glass slides by molecular combing (Michalet et al. 1997). Replicating DNA molecules were detected by immunofluorescence with antibodies against CldU (Fig. 6A, red) and IdU (Fig. 6A, green). As shown in Figure 6A, an ongoing replication fork is represented as a double-halogenated nucleotide-labeled track. The genomic DNAs were visualized with a third fluorescently labeled antibody against ssDNA (Fig. 6A, blue).
Fork stalling leads to asymmetrical forks that contain unequal IdU and CldI tracks (Lebofsky et al. 2006). We therefore used the asymmetry factor (AF), which is calculated as the ratio of the longest halogenated nucleotide track to the shortest one in each ongoing fork, to quantify replication fork stalling in cells transfected with siSRSF1 or siNC (Letessier et al. 2011). As shown in Figure 6B, in the presence of dox, the mean value of the AF significantly increased in the siSRSF1 transfected cells as compared with that in the cells transfected with control siRNA (from 1.13 to 1.64, P < 0.0001), indicating that SRSF1 depletion significantly increases replication fork stalling. Strikingly, withdrawal of dox resulted in a 70.6% reduction in the AF in siSRSF1-treated cells (from 1.64 to 1.28, P < 0.0001) (Fig. 6B), indicating that RNase H overexpression markedly suppressed SRSF1 depletion-induced fork stalling in HeLa cells.
Figure 6.
Depletion of SRSF1 induces replication-dependent DSBs in HeLa cells. (A) Representative images of DNA fibers carrying ongoing replication forks. A schematic representation of the DNA fibers (blue) and replication tracks (CldU in red and IdU in green) is shown below each fiber. Arrowheads indicate the direction of fork progression. (B) Distributions of fork asymmetry are shown graphically. Only nontruncated forks, as determined by DNA counterstaining (blue), were recorded. Forks in dox-treated cells are represented by black circles and those in dox-released cells are represented by green circles. Horizontal red lines represent the mean values of fork AFs with the standard error of the mean (SEM). Means, P-values, and the number of fibers for each sample are indicated. (C) γ-H2AX foci induced by SRSF1 depletion form predominantly in S phase. HeLa cells were transfected with control siRNA (siNC) or SRSF1 siRNA (siSRSF1), respectively. At 48 h after siRNA transfections, cells were pulse-labeled with 10 μM EdU for 30 min and analyzed by indirect immunofluorescence analysis with antibodies against SRSF1 (orange), EdU (red), and γ-H2AX (green). Representative images are shown in the left panel. Bar, 10 μm. The percentage of γ-H2AX foci-positive cells (whole bar) and the percentage of γ-H2AX foci-positive cells in the EdU-labeled population (hatched section) are shown in the right panel. The number of cells for each sample is indicated. (D) SRSF1 depletion did not exacerbate γ-H2AX foci formation in the cells in G1 phase. mAG-hGEM cells were transfected with siNC or siSRSF1. At 48 h after siRNA transfection, cells were fixed and subjected to indirect immunofluorescence analysis with antibodies against SRSF1 (orange) and γ-H2AX (red). Cells containing mAG-hGEM protein showed fluorescent green. Representative images are presented in the left panel. Bar, 10 μm. The percentage of γ-H2AX foci-positive cells (whole bar) and the percentage of γ-H2AX foci-positive cells in the mAG-hGEM-positive population (hatched section) are shown in the right panel. The number of cells for each sample is indicated. (E) HeLa cells were transfected with siNC or siSRSF1, respectively. At 48 h after transfection, cells were treated with DMSO or 5 μM PHA767491 for 6 h before harvest. Cells were then pulse-labeled with 10 μM EdU for 30 min and analyzed by indirect immunofluorescence analysis with antibodies against SRSF1, EdU, and γ-H2AX. The percentage of γ-H2AX foci-positive cells is shown graphically. The number of cells for each sample is indicated. (F) DT40-ASF cells were grown in the presence or absence of 1 μg/mL tet for 40 h and subjected to a 4-h treatment with DMSO or 5 μM PHA767491. Cells were analyzed as described in E. The percentage of γ-H2AX foci-positive cells is shown graphically. The number of cells for each sample is indicated.
We next wished to determine whether replication impairment accounts for the formation of DSBs in HeLa cells upon SRSF1 depletion. To this end, HeLa cells were first treated with siSRSF1 or siNC for 2 d and then subjected to a 30-min 10 μM EdU pulse-labeling before harvest (Salic and Mitchison 2008). Consistent with a striking increase in levels of phosphorylated histone H2AX (γ-H2AX) in SRSF1-depleted cells, as described previously (Li and Manley 2005), we observed a sharp increase in the number of cells with more than five nuclear γ-H2AX foci (Fig. 6C). γ-H2AX foci are one of the earliest DNA damage repair foci to occur at DSBs (Rogakou et al. 1998). Notably, SRSF1 depletion-induced γ-H2AX foci appeared predominantly in the EdU-positive cells, suggesting that loss of SRSF1 induced an S-phase-dependent DNA damage response.
To confirm that SRSF1 depletion-induced DNA damage indeed occurred in S phase, we constructed a stable HeLa cell derivative, HeLa-mAG-hGEM, which carries an expression cassette of the mAG fluorescent protein-tagged truncated form of human Geminin (mAG-hGEM). Analogous to endogenous Geminin, levels of mAG-hGEM are tightly regulated by ubiquitin-mediated proteolysis in a cell cycle-dependent manner (Sakaue-Sawano et al. 2008). Thus, mAG-hGEM is highly expressed during S, G2, and M phases and is completely degraded at the end of mitosis. The amount of mAG-hGEM thus functions as an indicator for the cell cycle transition from G1 to S phase (Sakaue-Sawano et al. 2008). Similar to what was observed in the EdU pulse-labeled cells, the number of cells with γ-H2AX foci markedly increased in the population expressing mAG-hGEM upon SRSF1 depletion (Fig. 6D). Strikingly, compared with cells treated with the control siRNA, SRSF1 siRNA treatment did not exacerbate γ-H2AX foci formation in the mAG-hGEM-negative cells, which were at the G1 phase of the cell cycle.
We next wished to obtain evidence that active replication is indeed required for the generation of DNA damage in SRSF1-depleted cells. For this, HeLa cells transfected with control or SRSF1 siRNA for 48 h were grown in the presence or absence of 5 μM PHA767491, a CDC7 kinase inhibitor (Montagnoli et al. 2008), for 6 h and then subjected to a 30-min EdU pulse label prior to harvest. PHA767491 treatment sharply decreased the number of EdU-positive cells in both control and SRSF1 siRNA transfected cells (Supplemental Table S2), consistent with the essential role of Cdc7 in replication origin firing and some previous reports (Bell and Dutta 2002; O'Donnell et al. 2010). Remarkably, compared with control DMSO-treated cells, PHA767491 treatment significantly decreased the percentage of γ-H2AX foci-positive cells (from 34.7% to 17.6%, P < 0.0001) upon SRSF1 depletion, indicating that active replication is required for the appearance of DSBs in SRSF1-depleted cells (Fig. 6E).
We previously constructed a derivative of chicken DT40 cells, DT40-ASF, in which the only copy of the SRSF1 gene is a human cDNA under the control of a tet-repressible promoter (Wang et al. 1996). Addition of tet results in in vivo depletion of SRSF1. Analogous to what was observed in siSRSF1-treated HeLa cells, tet treatment induced a striking increase in the number of γ-H2AX foci in EdU-positive DT40-ASF cells (from 2.86% to 23.4%, P < 0.0001) (Figure 6F). The appearance of γ-H2AX foci was also sensitive to PHA767491 treatment in DT40-ASF cells (31.4% in DMSO-treated cells and 17.9% in PHA767491-treated cells, P < 0.0001) (Fig. 6F). Notably, a significant fraction of γ-H2AX-positive cells (25.7%) appeared in the EdU-negative population and was resistant to PHA767491 treatment in SRSF1-depleted DT40-ASF cells (Fig. 6F), indicating that there is at least one other mechanism underlying SRSF1 depletion-induced DNA damage in DT40 cells.
Taken together, these data strongly support the notion that SRSF1 depletion induced the replication-dependent DNA damage in both chicken DT40 and human HeLa cells.
Discussion
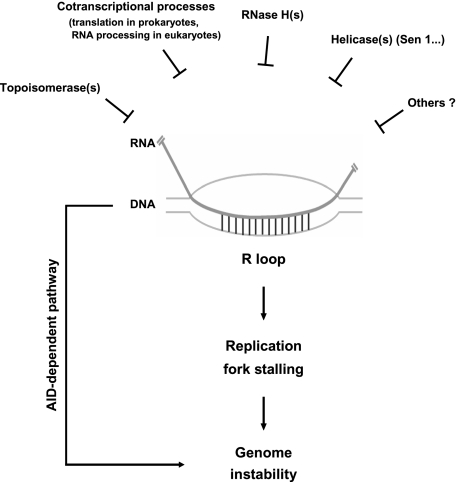
Our experiments provide significant new insights into the evolutionarily conserved phenomenon of transcription-induced R loops and the mechanism by which they can lead to genomic instability. The data presented here indicate that R-loop formation can have a striking negative impact on genome integrity in E. coli. We also found in both E. coli and HeLa cells that the impact of R-loop formation on genome stability is dependent on active DNA replication, indicative of a cause–effect relationship between replication impairment and R-loop-mediated genomic instability. From these experiments, we conclude that creation of an impediment to replication fork progression is an evolutionarily conserved mechanism underlying R-loop-induced genomic instability (Fig. 7). Below, we discuss how R-loop formation blocks replication fork progression and how this in turn leads to genome instability in an evolutionarily conserved manner.
Figure 7.
Mechanisms employed by cells to resolve transcription-induced R loops, and pathways by which R loops can lead to genomic instability. (Top) Cells have developed multiple protection systems to avoid the accumulation of transcription-induced R loops. For example, Top I suppresses R-loop formation by relieving negative supercoils behind RNAP, which can facilitate reannealing between the nascent transcript and template DNA strand. The tight coupling between transcription and various cotranscriptional processes keeps nascent RNA away from DNA as soon as it emerges from the polymerase. The activities of RNase H and perhaps RNA helicases, such as Sen1 in yeast, can disrupt R loops that form despite the above defense mechanisms. If R loops nonetheless persist, they can lead to DNA DSBs and rearrangements by two known mechanisms. One is an AID-dependent pathway that functions in active B cells. The other, as we showed here, is the creation of an impediment to replication fork progression, which constitutes a general and evolutionarily conserved mechanism underlying R-loop-induced genomic instability.
Early evidence for in vivo transcriptional R-loop formation was obtained with E. coli strains containing mutations affecting the TopA gene (Drolet et al. 1995; Masse et al. 1997; Masse and Drolet 1999b). Consistent with our observations, formation of cotranscriptional R loops was demonstrated to be a major problem linked to growth defects in topA mutants (Drolet et al. 1995; Drolet 2006). A recent study by Boubakri et al. (2010) has also shown that formation of R loops impairs viability of the dinG helicase mutant strain that contains a chromosomal inversion of the rRNA operons. However, it remained unclear whether R-loop-mediated genomic instability occurs in E. coli. Our data thus provide the first evidence showing that transcriptional R-loop formation can impair genome integrity in prokaryotes. Together with previous studies in yeast and vertebrate cells, our results establish that R-loop formation presents an obstacle to the maintenance of genome stability throughout evolution.
A key aspect of our findings is that active replication is in fact required for R-loop-mediated genomic instability in both E. coli and SRSF1-depleted vertebrate cells. Previous studies in yeast (Huertas and Aguilera 2003; Wellinger et al. 2006) and mammals (Tuduri et al. 2009) detected a correlation between replication and R-loop-mediated genomic instability (see above). Gomez-Gonzalez et al. (2009) have reported that R-loop-forming yeast THO-null mutants require functional S-phase checkpoint functions for survival under replicative stress. Our data show that, even in the absence of exogenous replication stress, persistent R-loop formation leads to DNA damage and rearrangements in a replication-dependent manner. Furthermore, our 2D analysis of replication intermediates in E. coli and DNA combing analysis of replication fork progression in HeLa cells provide direct evidence indicating that R loops are intrinsic impediments to replication fork progression. As a stalled replication fork, if not properly processed, can result in loss of genome integrity (Aguilera and Gomez-Gonzalez 2008; Branzei and Foiani 2009), we conclude that blockage of replication fork progression is a general, evolutionarily conserved mechanism that explains the deleterious effects of transcription-coupled R-loop formation on genome stability.
A significant question is: How mechanistically do R loops impede replication? One possibility is raised by observations that R-loop formation impairs transcription elongation both in vitro and in vivo. For example, Hraiky et al. (2000) observed that rRNA synthesis was significantly decreased in E. coli topA-null cells due to transcriptional blocks, and overexpression of RNase H effectively reduced this phenotype. Likewise, Huertas and Aguilera (2003) demonstrated that cotranscriptional R-loop formation impairs transcription elongation in yeast hpr1 mutant cells. Finally, a preformed RNA:DNA hybrid in a plasmid-borne transcription template reduces RNAP progression in vitro (Tous and Aguilera 2007). Although the impact of R loops on transcription in mammalian cells remains to be determined, it is of note that SR proteins, which play crucial roles in preventing R-loop formation in mammalian cells, have been shown to be important for efficient transcription elongation (Das et al. 2007; Lin et al. 2008). As the stalled transcription machinery could be an obstacle to replication (French 1992; Prado and Aguilera 2005; Mirkin and Mirkin 2007; Azvolinsky et al. 2009; Tehranchi et al. 2010; Merrikh et al. 2011), we propose that R-loop formation interferes with replication fork progression by provoking conflicts between the ongoing replication fork and the stalled transcription apparatus.
Our data showed that transcriptional R loops lead to genomic instability by interfering with DNA replication fork progression. However, stable and extensive R loops occur naturally in S regions of the Ig gene in mouse and likely other mammalian B cells undergoing CSR (Yu et al. 2003; Huang et al. 2006). S regions are normal components of mammalian genomes, and CSR is a natural process in the generation of antibody diversification in B cells (Maizels 2005). A considerable body of evidence demonstrates that formation of R loops by activating the promoter upstream of an acceptor S region is indeed required for a specific CSR targeting event (Jung et al. 1993; Xu et al. 1993; Bottaro et al. 1994). An important question is how the deleterious effects of R-loop formation on genome stability are restricted strictly to the active B cells that are stimulated by pathogens. Previous studies revealed that cells have evolved at least two safeguard systems to address this challenge. First, the promoters that exist upstream of S regions are only activated when B lymphocytes receive a specific extracellular cytokine signal (Stavnezer et al. 2008). Thus, formation of R loops in S regions is confined to a subset of B cells in the germinal center. Second, it has been well documented that transcription through the S region is necessary but not sufficient to confer genetic alteration to the IgH constant region (CH). AID, which is specifically expressed in active B cells, is also essential for the initiation of CSR in IgH (Muramatsu et al. 2000; Okazaki et al. 2002; Longerich et al. 2006). Thus, mechanisms for restricting AID expression to active B cells provide an important layer to the control of the risk of genome instability that is associated with R-loop formation in S regions. However, our data presented here suggest that there should be additional mechanisms for preventing the impediment posed by R loops on DNA replication at the CH region in active B cells. Consistent with this possibility, Petersen et al. (2001) observed that DNA damage associated with CSR, reflected in the appearance of DSBs at the CH region, occurs predominantly in the G1 phase of the cell cycle in cells undergoing CSR. It will be of great interest in the future to determine whether germline transcription and/or formation of R loops in S regions are in fact temporally separated from active replication in cells undergoing CSR.
The data presented in this study indicate that cotranscriptional R-loop formation poses a significant barrier to DNA replication and genome stability in cells from bacteria to humans. It is likely that R loops have a propensity to form at numerous genomic sites. For example, it has been known for some time that G-rich regions in nascent transcripts favor R-loop formation due to the exceptional stability of rG:dC base pairs (Sugimoto et al. 1995). While stable R loops have only been detected under physiological conditions in S regions of Ig genes, which indeed produce G-rich transcripts, there are, of course, numerous G-rich motifs with various densities and lengths spread throughout all genomes. It is thus likely that R-loop formation and interference with replication fork progression, and thus DNA DSBs and rearrangements, can occur at numerous transcribed regions. This highlights the importance of understanding in more detail the molecular determinants and pathological consequences of transcriptional R-loop formation, including its possible role in human disease.
Materials and methods
Strains, plasmids, and media
Bacterial strains used in this study are summarized in Supplemental Table S1. Mutations were introduced by P1 transduction (Miller 1992) or by using the λ red recombinase-mediated gene targeting method (Datsenko and Wanner 2000) essentially as described.
To construct the pPtac vector, a fragment containing the lacI gene and Ptac promoter replaced the araC gene and the PBAD promoter in the pBAD18 vector (Guzman et al. 1995). A 1.2-kb fragment of the mouse Sγ3 region was inserted into the pPtac vector in both orientations downstream from the Ptac promoter, giving rise to the constructs pPtac-Sγ3-F and pPtac-Sγ3-R. Details of plasmid constructions are available on request.
E. coli cells were routinely grown in LB medium containing 1% Bacto Tryptone, 0.5% yeast extract (Difco), and 1% NaCl with appropriate antibiotics. The following antibiotics were used at the concentrations indicated: 100 μg/mL Amp, 34 μg/mL chloramphenicol, 30 μg/mL Kan, and 10 μg/mL Tet.
Measurements of SOS response by using the sulAp-gfp reporter system
E. coli strain SMR8379 was kindly provided by Susan M. Rosenberg (Pennington and Rosenberg 2007), which carries a chromosomally located gfp gene encoding GFP controlled by a SOS-inducible sulA promoter (Pennington and Rosenberg 2007). Cells were transformed with pPtac-Sγ3-F or pPtac-Sγ3-R plasmid, respectively. Single colonies from each transformant was grown in LB medium until mid-log phase (OD600 = 0.5). Cultures were continuously grown in the presence or absence of 1 mM IPTG for 30 min and recovered in LB medium without IPTG for 2 h. Filamentous growth and expression of GFP were monitored by Nikon ECLIPSE 90i fluorescence microscope essentially as described (McCool et al. 2004). A Nikon DS-Qi1Mc CCD camera and NIS-Elements D3.1 software were used for all image acquisition and processing. The image acquisition parameters for fluorescence were identical for all samples examined.
RT–PCR and LexA protein analysis
Cells were grown in LB medium until mid-log phase (OD600 = 0.5) and treated with 1 mM IPTG for various times (indicated in the text). Cultures were transferred to LB medium without IPTG for 30 min and harvested for Western blot analysis and RT–PCR analysis as described (Li and Manley 2005). Antibody against LexA protein was purchased from Thermo Scientific.
PFGE
Cells were grown in LB medium until mid-log phase (OD600 = 0.5) and treated with 1 mM IPTG for 30 min at 37°C. For the dnaA(TS) mutant, a culture grown overnight at 30°C was used to inoculate two experimental cultures. One was grown until mid-log phase at 30°C and the other was first grown until early-log phase at 30°C (OD600 = 0.2) and then shifted for 2.5 h to 42°C.
Samples for PFGE were prepared essentially as described, with several modifications (Herschleb et al. 2007). Briefly, cells were washed and resuspended with TNE buffer (10 mM Tris, 200 mM NaCl, 100 mM EDTA at pH 7.2). After cells were incubated for 5 min at 37°C, an equal volume of 1.2% LMP agarose was added to each cell suspension, and the mixture was pipetted into CHEF plug molds (Bio-Rad) and allowed to solidify at 4°C. Plugs were incubated in EC lysis buffer (6 mM Tris–HCl at pH 7.6, 1 M NaCl [w/v], 100 mM EDTA, 0.5% Brij-58 [w/v], 0.2% deoxycholate [w/v], 0.5% N-laurylsarcosine [w/v], 1 mg/mL lysozyme, 20 mg/mL RNase A) overnight at 37°C with gentle shaking. Plugs were then washed and incubated in NDSK buffer (0.5 M EDTA, 1% N-laurylsarcosine [w/v], 1 mg/mL Proteinase K) overnight at 50°C with gentle shaking. Plugs were washed three times with TE and subjected to digestion with proper restriction enzymes. Digested DNA fragments were separated by PFGE on a 1% agarose gel. PFGE was carried out on a CHEF-DRIIPFGE system (Bio-Rad) with the following parameters: The gradient voltage was at 6 V/cm. The included angle was 120°. The initial and final switch times were 2.2 sec and 54.2 sec, respectively. The total run time was 22 h at 14°C.
Southern blot analysis
Southern blot analysis was performed as described (Sambrook et al. 1989). 32P-labeled RNA probes were prepared in an in vitro transcription reaction with T7 RNAP. Locations of probes are described in the text.
Recombination assay
Cells were grown in 2 mL of LB medium to an OD600 of 0.5. After continuously growing in the presence of 1 mM IPTG for 30 min, cells were washed and grown in LB medium for 1 h at 37°C. Serially diluted cultures were plated on LB plates with or without 30 μg/mL kanamycin to determine the number of cells plated and the number of Kan-resistant cells. Surviving colonies were scored, and the relative rate of recombination was calculated as Kan + recombination frequency = (kanamycin-resistant colonies observed)/(number of cells plated).
Isolation of replication intermediates
Plasmid replication intermediates were isolated essentially as described (Martin-Parras et al. 1991). Briefly, cells from overnight cultures were inoculated in LB medium and grown to an OD600 of 0.6 at 37°C. After treatment with or without IPTG for 30 min, the culture was cooled rapidly with ice-cold 0.9% NaCl (w/v) solution and collected by centrifugation. The cell pellet was lysed in 4 mL of lysis buffer (1% Brij-58 [w/v], 0.4% sodium deoxycholate [w/v], 63 mM EDTA, 50 mM Tris–HCl at pH 8.0). After incubating on ice for 10 min, the lysate was centrifuged at 30,000g for 1 h at 4°C. Plasmid DNA was precipitated from the supernatant by 8 mL of precipitation solution (1.5 M NaCl, 25% PEG8000). The precipitated DNA pellet was dissolved in 300 μL of TE and treated with RNase A for 20 min at 37°C. Coprecipitated proteins were hydrolyzed in a deproteinization solution (1 M NaCl, 10 mM Tris-HCl at pH 9.0, 1 mM EDTA, 0.1% SDS [w/v], 100 mg/mL proteinase K) for 20 min at 65°C, followed by a phenol/chloroform extraction. Plasmid DNA was then precipitated with ethanol.
2D gel electrophoresis
Replication intermediates were doubly digested by CaiI and PstI (Fermentas) and precipitated with ethanol. DNA was first separated in a 0.4% agarose gel in 1× TBE buffer at 1 V/cm for 34 h at room temperature. The slice of the gel containing all of the linear replication intermediates was excised and implanted into a 1% agarose gel with 0.3 μg/mL ethidium bromide. The second direction of electrophoresis was run at a 90° angle with respect to the first direction in 1× TBE buffer at 6 V/cm for 12 h at 4°C with continuous buffer recirculation.
DNA combing analysis
Cells were sequentially labeled with 100 μM CldU (30 min) and 100 μM IdU (30 min). Genomic DNA was extracted, stained, and combed as previously described (Michalet et al. 1997). After stretching, the DNA was denatured in alkali-denaturing buffer (0.1 N NaOH in 70% ethanol and 0.1% β-mercaptoethanol) for 15 min and fixed by addition of 0.5% glutaraldehyde for 5 min. Halogenated nucleotides were detected with a mouse anti-IdU monoclonal antibody (Becton Dickinson) or a rat anti-CldU monoclonal antibody (Accurate). DNA were detected with a mouse anti-human ssDNA monoclonal antibody (Chemicon). Alexa Fluor 488-conjugated goat anti-mouse, Alexa Fluor 568-conjugated goat anti-rat, and Alexa Fluor 647-conjugated goat anti-mouse antibodies were all purchased from Invitrogen.
Indirect immunofluorescence analysis
HeLa or HeLa mAG-hGEM cells were grown on glass coverslips and transfected with siNC or siSRSF1. At 48 h after transfection, cells were pulse-labeled with 10 μM EdU for 30 min. DT40-ASF were grown in the presence or absence of tet for 44 h and pulse-labeled with 10 μM EdU for 30 min.
Cells were harvested and then fixed for 15 min with 3.7% formaldehyde at room temperature followed by two PBS washes. Fixed cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min and blocked for 30 min in blocking solution (5% normal goat serum, 5% BSA in PBS). Coverslips were then incubated with primary antibodies for 60 min at room temperature. After PBS washes, the coverslips were incubated with proper fluorescence-labeled secondary antibody for 60 min at room temperature and washed three times with PBS. The coverslips were mounted with the prolong gold anti-fade reagent (Invitrogen). The presence of EdU was monitored by Click-It Chemistry (Invitrogen). Antibodies were purchased from Millipore (anti-γ-H2AX) and Invitrogen (anti-SRSF1).
Acknowledgments
We thank Mr. C. Zhou for assistance with analysis of replication intermediates. We are grateful to Dr. S. Rosenberg, Dr. D. Chattoraj, Dr. A. Miyawaki, and the Coli Genetic Stock at Yale University and the National BioResource Project: E. coli (NIG, Japan) for strains and reagents. We thank Mr. M. Li and Y. Wei for their assistance in DNA combing analysis, and Mr. J. Tu for his assistance in data analysis. This work was supported by grant 2007AA02Z1A8 from the Ministry of Science and Technology, China, to X.L. J.L.M. was supported by a grant from the NIH (R01 GM048259).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17010011.
References
- Aguilera A, Gomez-Gonzalez B 2008. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bottaro A, Lansford R, Xu L, Zhang J, Rothman P, Alt FW 1994. S region transcription per se promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. EMBO J 13: 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubakri H, de Septenville AL, Viguera E, Michel B 2010. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci 100: 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2009. The checkpoint response to replication stress. DNA Repair (Amst) 8: 1038–1046 [DOI] [PubMed] [Google Scholar]
- Broccoli S, Rallu F, Sanscartier P, Cerritelli SM, Crouch RJ, Drolet M 2004. Effects of RNA polymerase modifications on transcription-induced negative supercoiling and associated R-loop formation. Mol Microbiol 52: 1769–1779 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422: 726–730 [DOI] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R 2007. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell 26: 867–881 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K 1992. Control of bacterial DNA supercoiling. Mol Microbiol 6: 425–433 [DOI] [PubMed] [Google Scholar]
- Drolet M 2006. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 59: 723–730 [DOI] [PubMed] [Google Scholar]
- Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ 1995. Overexpression of RNase H partially complements the growth defect of an Escherichia coli ΔtopA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci 92: 3526–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N 2004. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes & Dev 18: 1618–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, Tollervey D 2010. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes & Dev 24: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S 1992. Consequences of replication fork movement through transcription units in vivo. Science 258: 1362–1365 [DOI] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Aguilera A 2007. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci 104: 8409–8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Felipe-Abrio I, Aguilera A 2009. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol 29: 5203–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschleb J, Ananiev G, Schwartz DC 2007. Pulsed-field gel electrophoresis. Nat Protoc 2: 677–684 [DOI] [PubMed] [Google Scholar]
- Hirota Y, Mordoh J, Jacob F 1970. On the process of cellular division in Escherichia coli. 3. Thermosensitive mutants of Escherichia coli altered in the process of DNA initiation. J Mol Biol 53: 369–387 [DOI] [PubMed] [Google Scholar]
- Hraiky C, Raymond MA, Drolet M 2000. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J Biol Chem 275: 11257–11263 [DOI] [PubMed] [Google Scholar]
- Huang FT, Yu K, Hsieh CL, Lieber MR 2006. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci 103: 5030–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol 4: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Janion C 2008. Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int J Biol Sci 4: 338–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Rajewsky K, Radbruch A 1993. Shutdown of class switch recombination by deletion of a switch region control element. Science 259: 984–987 [DOI] [PubMed] [Google Scholar]
- Kogoma T, Hong X, Cadwell GW, Barnard KG, Asai T 1993. Requirement of homologous recombination functions for viability of the Escherichia coli cell that lacks RNase HI and exonuclease V activities. Biochimie 75: 89–99 [DOI] [PubMed] [Google Scholar]
- Labib K, Hodgson B 2007. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep 8: 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A 2006. DNA replication origin interference increases the spacing between initiation events in human cells. Mol Biol Cell 17: 5337–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M 2011. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123 [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378 [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL 2006. Cotranscriptional processes and their influence on genome stability. Genes & Dev 20: 1838–1847 [DOI] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15: 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW, Mount DW 1982. The SOS regulatory system of Escherichia coli. Cell 29: 11–22 [DOI] [PubMed] [Google Scholar]
- Liu B, Alberts BM 1995. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 267: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Longerich S, Basu U, Alt F, Storb U 2006. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol 18: 164–174 [DOI] [PubMed] [Google Scholar]
- Maizels N 2005. Immunoglobulin gene diversification. Annu Rev Genet 39: 23–46 [DOI] [PubMed] [Google Scholar]
- Martin-Parras L, Hernandez P, Martinez-Robles ML, Schvartzman JB 1991. Unidirectional replication as visualized by two-dimensional agarose gel electrophoresis. J Mol Biol 220: 843–853 [DOI] [PubMed] [Google Scholar]
- Masse E, Drolet M 1999a. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J Biol Chem 274: 16659–16664 [DOI] [PubMed] [Google Scholar]
- Masse E, Drolet M 1999b. R-loop-dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J Mol Biol 294: 321–332 [DOI] [PubMed] [Google Scholar]
- Masse E, Phoenix P, Drolet M 1997. DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in Escherichia coli. J Biol Chem 272: 12816–12823 [DOI] [PubMed] [Google Scholar]
- McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ 2004. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol Microbiol 53: 1343–1357 [DOI] [PubMed] [Google Scholar]
- Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P 2011. Co-directional replication-transcription conflicts lead to replication restart. Nature 470: 554–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. 1997. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science 277: 1518–1523 [DOI] [PubMed] [Google Scholar]
- Michel B 2005. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol 3: e255 doi: 10.1371/journal.pbio.0030255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH 1992. A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Mirkin EV, Mirkin SM 2007. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71: 13–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C, et al. 2008. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol 4: 357–365 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem 274: 18470–18476 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563 [DOI] [PubMed] [Google Scholar]
- O'Donnell L, Panier S, Wildenhain J, Tkach JM, Al-Hakim A, Landry MC, Escribano-Diaz C, Szilard RK, Young JT, Munro M, et al. 2010. The MMS22L–TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell 40: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T 2002. The AID enzyme induces class switch recombination in fibroblasts. Nature 416: 340–345 [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington JM, Rosenberg SM 2007. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet 39: 797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. 2001. AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature 414: 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418: 99–103 [DOI] [PubMed] [Google Scholar]
- Pomerantz RT, O'Donnell M 2010. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science 327: 590–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 24: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol 12: 1748–1755 [DOI] [PubMed] [Google Scholar]
- Reaban ME, Lebowitz J, Griffin JA 1994. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin α switch region. J Biol Chem 269: 21850–21857 [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132: 487–498 [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci 105: 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: a laboratory manual. Cold Sprng Harbor Laboratory Press, Cold Spring Harbor [Google Scholar]
- Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD 2010. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet 6: e1000810 doi: 10.1371/journal/pgen.1000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE 2008. Mechanism and regulation of class switch recombination. Annu Rev Immunol 26: 261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M 1995. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34: 11211–11216 [DOI] [PubMed] [Google Scholar]
- Szilard RK, Jacques PE, Laramee L, Cheng B, Galicia S, Bataille AR, Yeung M, Mendez M, Bergeron M, Robert F, et al. 2010. Systematic identification of fragile sites via genome-wide location analysis of γ-H2AX. Nat Struct Mol Biol 17: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD 2010. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Alt FW 2000. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem 275: 24163–24172 [DOI] [PubMed] [Google Scholar]
- Tous C, Aguilera A 2007. Impairment of transcription elongation by R-loops in vitro. Biochem Biophys Res Commun 360: 428–432 [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11: 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GC 1985. Inducible DNA repair systems. Annu Rev Biochem 54: 425–457 [DOI] [PubMed] [Google Scholar]
- Wang J, Takagaki Y, Manley JL 1996. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev 10: 2588–2599 [DOI] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A 2006. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Gorham B, Li SC, Bottaro A, Alt FW, Rothman P 1993. Replacement of germ-line ε promoter by gene targeting alters control of immunoglobulin heavy chain class switching. Proc Natl Acad Sci 90: 3705–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4: 442–451 [DOI] [PubMed] [Google Scholar]