Abstract
The response of cells to changes in their environment often requires coregulation of gene networks, but little is known about how this can occur at the post-transcriptional level. An important example of post-transcriptional coregulation is the selective translational regulation in response to growth conditions of mammalian mRNAs that encode protein biosynthesis factors and contain hallmark 5′-terminal oligopyrimidine tracts (5′TOP). However, the responsible trans-factors and the mechanism by which they coregulate 5′TOP mRNAs have remained elusive. Here we identify stress granule-associated TIA-1 and TIAR proteins as key factors in human 5′TOP mRNA regulation, which upon amino acid starvation assemble onto the 5′ end of 5′TOP mRNAs and arrest translation at the initiation step, as evidenced by TIA-1/TIAR-dependent 5′TOP mRNA translation repression, polysome release, and accumulation in stress granules. This requires starvation-mediated activation of the GCN2 (general control nonderepressible 2) kinase and inactivation of the mTOR (mammalian target of rapamycin) signaling pathway. Our findings provide a mechanistic explanation to the long-standing question of how the network of 5′TOP mRNAs are coregulated according to amino acid availability, thereby allowing redirection of limited resources to mount a nutrient deprivation response. This presents a fundamental example of how a group of mRNAs can be translationally coregulated in response to changes in the cellular environment.
Keywords: 5′TOP, TIA-1, TIAR, translation regulation
The activity of the eukaryotic translation machinery is highly regulated according to conditions of cell growth. This occurs in large part through regulation of the activity of two key general translation initiation factors: the mRNA cap-binding eukaryotic initiation factor 4E (eIF4E), and eIF2, which is responsible for recruiting the initiator tRNA (tRNAiMet) to the small ribosomal subunit (Hinnebusch 1997; Ma and Blenis 2009; Sonenberg and Hinnebusch 2009). eIF4E is negatively regulated by eIF4E-binding proteins (4E-BPs). Under favorable growth conditions, mTOR (mammalian target of rapamycin) inactivates 4E-BP1 by phosphorylation. In contrast, poor growth conditions can lead to mTOR inactivation and 4E-BP1 dephosphorylation, which increases 4E-BP1 binding to eIF4E and inhibits cap-dependent translation (Ma and Blenis 2009). Regulation of eIF2 occurs through phosphorylation of the eIF2 subunit eIF2α. This, in turn, leads to reduced accumulation of the cellular eIF2–GTP–tRNAiMet ternary complex and a general reduction in translation initiation. When mammalian cells experience starvation for certain amino acids, both eIF4E and eIF2 are repressed. eIF4E repression occurs through inactivation of mTOR, which is normally stimulated by amino acids through the small G-protein Rheb (Ras homolog enriched in brain), which in turn is regulated by the tuberous sclerosis complex (TSC1/2) (Ma and Blenis 2009). Phosphorylation of the eIF2α subunit of eIF2 is stimulated during amino acid starvation primarily by GCN2 (general control nonderepressible 2), which is activated by the accumulation of uncharged tRNAs (Dever et al. 1995; Dong et al. 2000; Qiu et al. 2001, 2002).
Despite this level of general translation repression, a network of mammalian mRNAs that contain hallmark 5′-terminal oligopyrimidine tracts (5′TOPs) become much more efficiently repressed during amino acid starvation than other cellular mRNAs (Avni et al. 1994, 1997; Iadevaia et al. 2008). 5′TOP elements are predominant in mammalian mRNAs encoding ribosomal proteins and translation factors (Meyuhas 2000; Hamilton et al. 2006; Iadevaia et al. 2008). Transcription of 5′TOP mRNAs invariantly initiates at a cytosine, which is followed by an uninterrupted 4- to 15-nucleotide (nt) pyrimidine tract (Hamilton et al. 2006), and many are produced from genes containing the recently described TCT promoter motif, which overlaps, at the DNA level, with the sequence encoding the 5′TOP (Parry et al. 2010). The 5′TOP element is, along with a short downstream sequence, necessary and sufficient for selective repression of translation during cell cycle arrest or nutrient starvation, and is thought to be important for inhibiting the energetically demanding process of ribosome biogenesis during poor growth conditions (Meyuhas 2000; Hamilton et al. 2006). Reactivation of 5′TOP mRNA translation after cell cycle arrest or nutrient starvation requires the activity of mTOR and PI3K/Akt signaling pathways (Tang et al. 2001; Stolovich et al. 2002; Patursky-Polischuk et al. 2009), but little is known about which pathways contribute to selective repression. Moreover, the specific factors involved in selectively repressing 5′TOP mRNA translation have remained elusive.
Early evidence suggested that regulation of 5′TOP mRNA translation involves an unknown titratable repressor (Biberman and Meyuhas 1999). Several candidates for such an activity have been proposed through their reported association with 5′TOP elements and immediate downstream regions. These include the abundant La antigen (La) (Pellizzoni et al. 1996; Crosio et al. 2000; Cardinali et al. 2003; Intine et al. 2003; Schwartz et al. 2004), ZNF9 (Pellizzoni et al. 1997, 1998), and AUF1 (Kakegawa et al. 2007). However, definitive evidence for regulatory roles of these proteins in 5′TOP mRNA translation is lacking. More recently, a microRNA (miR-10a) was reported to interact with a sequence immediately downstream from the 5′TOP sequence and activate translation of a number of 5′TOP mRNAs (Orom et al. 2008). Despite these recent advances, the mechanism by which translation of 5′TOP mRNAs becomes selectively inhibited during nutrient starvation has remained unknown.
Here we identify the stress granule (SG)-associated TIA-1 and TIAR proteins as key regulators of 5′TOP mRNA translation during amino acid starvation. TIA-1 and TIAR interact specifically with the 5′ end of 5′TOP mRNAs and show a significant increase in assembly into 5′TOP mRNPs during amino acid starvation. This stalls the 5′TOP mRNP at the translation initiation step, as evidenced by TIA-1/TIAR-dependent 5′TOP mRNA release from polysomes and accumulation in SGs, with a concomitant decrease in 5′TOP mRNA-directed de novo protein synthesis upon starvation. The position of TIA-1/TIAR binding within the mRNA is critical for this regulation, as mRNAs containing TIA-1/TIAR-binding sites in their 3′ untranslated regions (UTRs) are not released from polysomes during amino acid starvation. 5′TOP mRNA repression requires inactivation of mTOR and the activity of GCN2. This identifies TIA-1 and TIAR as key trans-factors in starvation-mediated 5′TOP mRNA repression regulated by mTOR and GCN2 signaling, and provides insights into the mechanism by which a network of mRNAs can be translationally coregulated according to cell growth conditions.
Results
TIA-1 and TIAR show starvation-stimulated association with 5′TOP mRNAs
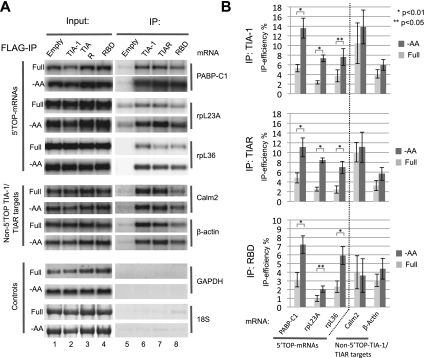
Based on previous studies implicating TIA-1 and TIAR in translational repression (Piecyk et al. 2000; Lopez de Silanes et al. 2005; Mazan-Mamczarz et al. 2006; Kim et al. 2007) and in binding to pyrimidine-rich sequences (Dember et al. 1996; Lopez de Silanes et al. 2005; Mazan-Mamczarz et al. 2006; Kim et al. 2007, 2011), we speculated that TIA-1 and TIAR may play a role in the control of 5′TOP mRNA translation. To first determine whether TIA-1 and TIAR exist in complex with 5′TOP mRNAs, we established human HEK293 cell lines stably expressing Flag-tagged TIA-1 or TIAR under control of tetracycline-inducible promoters. This allows control of Flag-tagged TIA-1 and TIAR expression to roughly match the levels of their corresponding endogenous proteins. As seen in the RNA immunoprecipitation (RIP) assays in Figure 1A, endogenous 5′TOP mRNAs encoding PABPC1, ribosomal protein L23a (rpL23a), and rpL36 all copurify with TIA-1 and TIAR (Fig. 1A, cf. lanes 6,7 and lane 5, input samples are shown in lanes 1–4). In addition, mRNAs for calmodulin 2 and β-actin, which lack 5′TOPs but have previously been described to bear TIA-1- and TIAR-binding sites in their 3′ UTRs (Lopez de Silanes et al. 2005; Mazan-Mamczarz et al. 2006; Kim et al. 2007), served as positive controls, and both copurify with TIA-1 and TIAR as expected (Fig. 1A, middle panels). In contrast, abundant negative control RNAs (GAPDH mRNA and 18S rRNA) did not copurify at levels above background (Fig. 1A, bottom panels). The interaction between TIA-1 and target mRNAs does not depend on the C-terminal glutamine/asparagine (Q/N)-rich prion-related domain (PRD) of TIA-1, a domain important for TIA-1-mediated formation of SGs (Gilks et al. 2004), as the mRNAs that copurify with full-length TIA-1 also copurify with a truncated version of TIA-1 containing only the RNA-binding domain (RBD), albeit at somewhat lower levels (Fig. 1A, cf. lanes 8 and 6). Thus, TIA-1 and TIAR exist in complex with 5′TOP mRNAs, as well as with a subset of other cellular mRNAs. This is consistent with previous observations from global analyses of TIA-1- and TIAR-associated mRNAs (Lopez de Silanes et al. 2005; Mazan-Mamczarz et al. 2006; Kim et al. 2007).
Figure 1.
Amino acid starvation stimulates assembly of TIA-1 and TIAR with 5′TOP mRNAs. (A) Northern blots comparing levels of mRNAs indicated on the right, in input fractions (lanes 1–4) versus pellet fractions (lanes 5–8) of immunopurified Flag-tagged TIA-1, TIAR, or C-terminally truncated TIA-1 (TIA-1 RBD) stably expressed in HEK293 cell lines, incubated for 2 h in either medium lacking amino acids (−AA) or with a full nutrient supplement (Full). (Lane 5) Immunopurification from cells expressing no Flag-tagged protein. Probed mRNAs are the 5′TOP-containing PABPC1, rpL23a, and rpL36 mRNAs (top three panels); calmodulin 2 and β-actin mRNAs, which lack 5′TOPs but contain TIA-1/TIAR-binding sites in their 3′ UTRs (middle two panels); and negative control GAPDH mRNA and 18S rRNA (bottom two panels). All mRNAs were detected with radiolabeled riboprobes, whereas 18S rRNA was detected using methylene blue staining. Input lanes represent 10% of the total RNA in immunoprecipitated samples. (B) Quantification of the data in A. Immunoprecipitation efficiencies were quantified as the percent of individual mRNAs found in pelleted material as compared with input fractions. Values and standard deviation were calculated from three independent experiments (n = 3). Dark-gray bars represent amino acid-starved cells (−AA), and light-gray bars represent cells in full medium (Full). P-values were calculated using Student's t-test (two tailed) and are indicated above the bars: (*) P < 0.01; (**) P < 0.05.
We next asked whether the interaction of TIA-1/TIAR with 5′TOP mRNAs is affected upon amino acid starvation. As seen in Figure 1A, the levels of 5′TOP mRNAs copurifying with TIA-1 and TIAR increased significantly (twofold to fourfold) after 2 h of amino acid starvation (Fig. 1A [cf. −AA and Full panels], quantified in B). In contrast, the levels of the non-5′TOP TIA-1/TIAR target mRNAs encoding calmodulin 2 and β-actin copurifying with TIA-1 and TIAR were not significantly elevated upon amino acid starvation (Fig. 1A [middle panels], quantified in B). Similar specificity was also observed for the C-terminally truncated TIA-1 containing only the RBD (Fig. 1A [lane 8], quantified in B [bottom panel]). GAPDH mRNA and 18S rRNA remained absent from TIA-1 and TIAR immunoprecipitates also after amino acid starvation (Fig. 1A). Thus, amino acid starvation stimulates the association of TIA-1 and TIAR proteins with 5′TOP mRNAs, but not with other target or nontarget mRNAs. Repeating the RIP assays with cell lysates subjected to DNA oligo-mediated RNase H cleavage revealed that TIA-1 and TIAR binding is restricted to the cap-proximal ∼40 nt of the 5′TOP rpL23a mRNA (Supplemental Fig. S1). This corresponds to the region of 5′TOP mRNAs critical for starvation-mediated translational control (Avni et al. 1994, 1997; Meyuhas 2000).
TIA-1 and TIAR repress 5′TOP mRNA translation during amino acid starvation
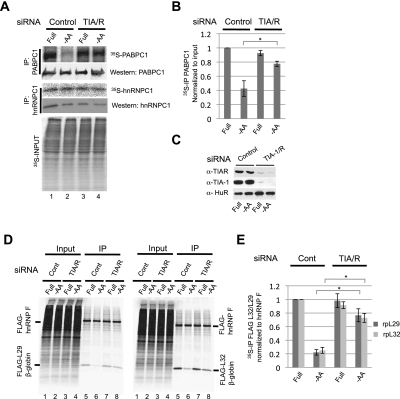
We next used metabolic 35S pulse-labeling assays to test whether the assembly of TIA-1 and TIAR with 5′TOP mRNAs controls 5′TOP mRNA translation during amino acid starvation. As seen in the immunoprecipitation assay in Figure 2A, incorporation of 35S-methionine into 5′TOP mRNA-encoded PABPC1 protein is significantly reduced after 2 h of amino acid starvation, consistent with translational repression (Fig. 2A [cf. lanes 1 and 2, top panel], quantified in B). However, siRNA-mediated codepletion of TIA-1 and TIAR proteins (knockdown efficiency shown in Fig. 2C) strongly impairs starvation-mediated repression of PABPC1 translation (Fig. 2A [cf. lanes 3 and 4, top panel], quantified in B). These effects are specific, as the synthesis of hnRNPC1, which is encoded by a non-5′TOP mRNA, and of bulk cellular protein is not detectably affected by TIA-1/TIAR depletion (Fig. 2A, middle and bottom panels).
Figure 2.
TIA-1/TIAR depletion derepresses 5′TOP mRNA translation during amino acid starvation. (A) Autoradiogram monitoring the incorporation of 35S-methionine over 1 h into PABPC1 (top panel), hnRNP C1 (middle panel), and total protein (bottom panel) in HeLa cells treated with siRNAs targeting either Luciferase (control siRNA) or TIA-1/TIAR and grown in full medium (Full) or medium lacking amino acids for 2 h (−AA) prior to 35S labeling. PABPC1 and hnRNP C1 proteins (top and middle panels) were isolated from the total protein (shown in the bottom panel) by immunoprecipitation, and the level of total immunopurified PABPC1 and hnRNP C1 was monitored by Western blotting as indicated. (B) Quantification of PABPC1-IP data in A. The levels of 35S-labeled PABPC1 protein were normalized to the 35S levels in the input samples. Values and error bars (standard deviation) were calculated from three independent experiments (n = 3). (C) Western blots showing knockdown efficiencies of TIA-1 and TIAR. HuR serves as a negative control. (D) Autoradiogram monitoring the incorporation of 35S-methionine over 1 h into immunoprecipitated Flag-tagged-hnRNP F (non-5′TOP control; top band, left and right panels) or either Flag-tagged rpL29-5′TOP-β-globin (bottom band, left panel) or Flag-tagged rpL32-5′TOP-β-globin (bottom band, right panel) in HeLa cells treated with siRNAs targeting either Luciferase (control siRNA) or TIA-1/TIAR and grown in full medium (Full) or medium lacking amino acids for 2 h (−AA), as indicated. (E) Values and error bars (standard deviation) were calculated from three independent experiments (n = 3). P-values were determined by Student's t-test (two tailed); (*) P < 0.01.
Of the commercially available antibodies against 5′TOP mRNA-encoded proteins tested, only anti-PABPC1 antibodies showed sufficient specificity for the assays in Figure 2A. We therefore additionally tested the effect of TIA-1/TIAR depletion on translation of exogenously expressed 5′TOP reporter mRNAs based on human β-globin mRNA containing at the 5′ end the 5′TOP element and the immediate downstream sequence of rpL29 or rpL32 mRNAs. Primer extension assays confirmed that these reporter genes correctly initiate transcription at the 5′ end of the 5′TOP (Supplemental Fig. S2). As seen in Figure 2D, protein synthesis from the 5′TOP reporter mRNAs was repressed fivefold during amino acid starvation (Fig. 2D [cf. lanes 6 and 5, bottom band], quantified in E). In contrast, this repression was strongly impaired upon knockdown of TIA-1/TIAR (Fig. 2D [cf. lanes 7 and 8], quantified in E). Exogenously expressed hnRNP F, which served as a negative control, remained unaffected. We conclude that TIA-1/TIAR proteins are critical for 5′TOP mRNA-specific translational repression. The small amount of residual translational repression that persists after TIA-1/TIAR knockdown (Fig. 2B,E) could be a result of either residual TIA-1/TIAR protein (Fig. 2C) or additional mechanisms contributing to translational repression during amino acid starvation.
5′TOP mRNAs accumulate with TIA-1 and TIAR in SGs upon amino acid starvation
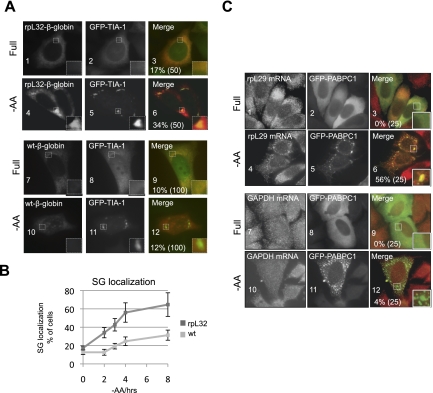
What is the mechanism of 5′TOP mRNA translational repression by TIA-1 and TIAR? The association of TIA-1 and TIAR with the 5′ end of 5′TOP mRNA indicated an effect on translation initiation. mRNAs that are stalled at the translation initiation step as a consequence of various cellular stress conditions often accumulate in cytoplasmic mRNP granules called stress granules (SGs) (Kimball et al. 2003; Mollet et al. 2008; Anderson and Kedersha 2009; Buchan and Parker 2009; Farny et al. 2009), whereas stalling mRNAs in the elongation step of translation inhibits SG formation (Kedersha et al. 2000). We therefore tested whether 5′TOP mRNAs accumulate in SGs upon amino acid starvation. As seen in the RNA fluorescence in situ hybridization (RNA-FISH) assays in Figure 3A, 2 h of amino acid starvation results in increased accumulation of the rpL32-β-globin 5′TOP reporter mRNA in SGs, as marked by coexpressed GFP-TIA-1 (Fig. 3A, cf. panels 4–6 and 1–3), but not of non-5′TOP wild-type β-globin mRNA (Fig. 3A, panels 7–12). The fraction of cells showing accumulation of the rpL32-β-globin 5′TOP mRNA in SGs increases steadily over time of starvation, reaching ≈65% of cells in 8 h (Fig. 3B, rpL32). In contrast, wild-type β-globin mRNA only slowly starts accumulating in SGs after extended times of starvation (Fig. 3B, wt). The accumulation of rpL32-β-globin mRNA in SGs is not an artifact of exogenous GFP-TIA-1 expression, as combined indirect immunofluorescence/RNA-FISH assays demonstrated enhanced colocalization of the 5′TOP mRNA also with endogenous TIAR after amino acid starvation (Supplemental Fig. S3A). 5′TOP sequences from PABPC1 and rpL29 mRNAs also directed SG accumulation of β-globin mRNA upon amino acid starvation (Supplemental Fig. S3B), and, in addition to TIA-1 and TIAR, the SGs forming during amino acid starvation also contain other SG components, including PABPC1 and eIF4G (Supplemental Fig. S3C).
Figure 3.
5′TOP mRNAs accumulate in SGs during amino acid starvation. (A) RNA-FISH for rpL32-β-globin 5′TOP reporter mRNA (panels 1,4) or wild-type (wt) β-globin mRNA (panels 7,10), transiently expressed in HeLa cells. (Panels 2,5,8,11) Transiently expressed GFP-tagged TIA-1 marks SGs. (Panels 3,6,9,12) Merged images (rpL32-β-globin, red; GFP-TIA-1, green). Panels 1–3 and 7–9 are from HeLa cells cultured in full DMEM medium (Full), whereas panels 4–6 and 10–12 are from HeLa cells under amino acid starvation conditions (−AA). Enlargements of boxed areas are shown at the bottom right of each picture, and the percent of cells with rpL32-β-globin mRNA colocalizing with GFP-TIA-1 in SGs are given in the merged panels, with the number of counted cells in parentheses. (B) Quantification of the fraction of cells in which the rpL32-β-globin 5′TOP reporter mRNA (rpL32) concentrates in SGs over time (0–8 h) of amino acid starvation, compared with wild-type β-globin mRNA (wt) lacking a 5′TOP. Error bars represent standard deviation calculated from three independent experiments (n = 3), in each of which 50 cells were counted. (C) RNA-FISH for endogenous rpL29 (panels 1,4) and GAPDH (panels 7,10) mRNAs in HeLa cells grown in full medium (Full) or under amino acid starvation conditions for 2 h (−AA), as indicated. (Panels 2,5,8,11) Transiently expressed GFP-PABPC1 marks SGs. Merged panels on the right show mRNAs in red and GFP-PABPC1 in green.
To test whether an endogenous 5′TOP mRNA can be observed in SGs upon amino acid starvation, the localization of endogenous rpL29 mRNA was monitored in HeLa cells transiently expressing GFP-PABPC1 to label SGs. As seen in Figure 3C, endogenous rpL29 mRNA localizes in a granular pattern throughout the cell under normal growth conditions (Fig. 3C, panels 1–3), but after 2 h of amino acid starvation, it can be observed to colocalize with PABPC1 in SGs in 56% of cells (Fig. 3C, panels 4–6). In contrast, endogenous GAPDH mRNA, which served as a negative control, was only rarely observed in SGs (Fig. 3C, panels 7–12). Thus, amino acid starvation stimulates the assembly of 5′TOP mRNAs into SGs in human HeLa cells, consistent with the accumulation of 5′TOP mRNPs repressed at the translation initiation step.
TIA-1 and TIAR promote release of 5′TOP mRNAs from polysomes upon amino acid starvation
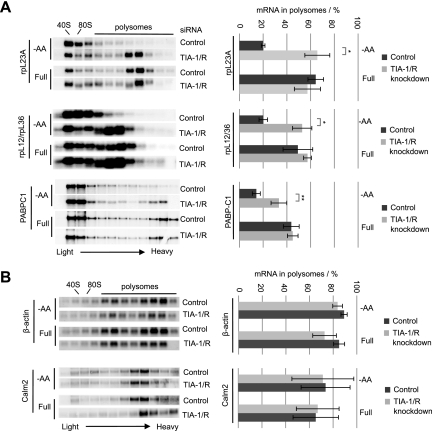
If TIA-1 and TIAR regulate 5′TOP mRNA translation at the initiation step, 5′TOP mRNAs should be released from polysomes during amino acid starvation in a TIA-1/TIAR-dependent manner. Consistent with this, the sucrose gradient polysome fractionation assays in Figure 4A show that, as expected, endogenous rpL23a, rpL12/rpL36, and PABPC1 5′TOP mRNAs all efficiently shift out of polysomes upon amino acid starvation (Fig. 4A, quantified in the right panels, and note that rpL36 mRNA migrates immediately below rpL12 mRNA). However, the ability of the 5′TOP mRNAs to shift out of polysomes during amino acid starvation is strongly diminished when TIA-1 and TIAR are depleted (Fig. 4A, quantified on the right). Thus, TIA-1 and TIAR are required for the release of 5′TOP mRNAs from polysomes upon amino acid starvation, consistent with the idea that they repress the translation initiation step. In the case of PABPC1 mRNA (Fig. 4A, bottom panel), a large fraction resides outside of polysomes under all tested conditions, likely reflecting its translational autorepression (Hornstein et al. 1999). General sucrose gradient A254 RNA profiles revealed that the overall shift of ribosomes toward subpolysomal fractions observed upon amino acid starvation is significantly reversed when TIA-1 and TIAR proteins are knocked down (Supplemental Fig. S4). This is consistent with TIA-1 and TIAR controlling the translational repression of the highly abundant 5′TOP mRNA network during amino acid starvation.
Figure 4.
5′TOP mRNAs are released from polysomes by TIA-1/TIAR proteins upon amino acid starvation. (A,B) Northern blots of sucrose gradient polysome fractions, monitoring endogenous 5′TOP mRNAs encoding rpL23a, rpL36/rpL12, and PABPC1 (A), or control mRNAs lacking a 5′TOP but containing 3′ UTR TIA-1/R-binding sites, encoding β-actin and calmodulin 2 (B). Cells were either starved for amino acids (−AA, top panels) or left in full medium (Full, bottom panels). Polysomal fractions are denoted on the basis of A254 profiles recorded during fractionation and by staining gels for ribosomal RNAs using methylene blue (data not shown). Quantifications of the fraction (in percent of the total amount) of 5′TOP mRNA found in the polysomal fractions (indicated by bars above each gel) were performed using a PhosphorImager and are shown on the right. The data presented are based on three independent knockdown experiments (n = 3), and error bars indicate standard deviation from the mean. P-values were determined by Student's t-test (two tailed); (*) P < 0.01; (**) P < 0.05.
In contrast to 5′TOP mRNAs, β-actin and calmodulin 2 mRNAs, which contain TIA-1/TIAR-binding sites in their 3′ UTRs (Fig. 1; Lopez de Silanes et al. 2005; Mazan-Mamczarz et al. 2006; Kim et al. 2007), do not shift toward smaller polysomal fractions during amino acid starvation and are unaffected by the depletion of TIA-1 and TIAR (Fig. 4B). Similarly, exogenously expressed β-globin mRNAs containing the rpL32 5′TOP sequence or high-affinity binding sites for TIA-1 or TIAR in the 3′ UTR fail to shift out of polysomes upon amino acid starvation (Supplemental Fig. S5). Thus, the position within the mRNA of TIA-1/TIAR binding is critical for TIA-1/TIAR-dependent translational control in response to starvation.
TIA-1 and TIAR repress 5′TOP mRNA translation in a redundant manner, and the PRD of TIA-1 is not required
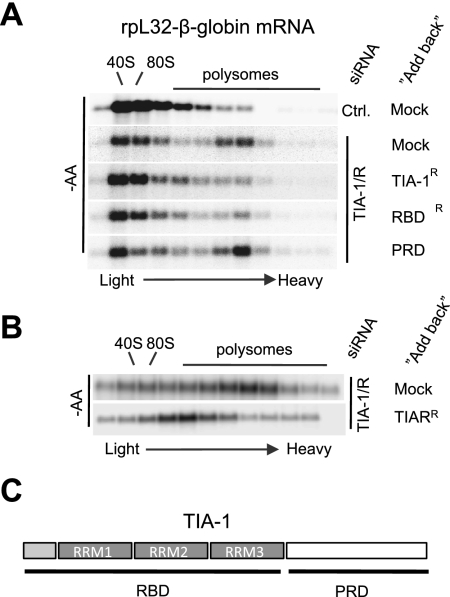
All of the above-mentioned experiments addressing TIA-1/TIAR function were performed under conditions of codepletion of TIA-1 and TIAR. It was therefore unclear which of the two proteins is responsible for 5′TOP mRNA repression. To address this question, the ability of exogenous TIA-1 or TIAR to complement the depletion of endogenous TIA-1/TIAR proteins was tested using plasmids expressing siRNA-resistant mRNAs encoding for TIA-1 or TIAR. The sucrose gradient polysome assays in Figure 5 show that expression of TIA-1 (Fig. 5A) or TIAR (Fig. 5B) alone is sufficient to repress the association of a rpL32-β-globin 5′TOP reporter mRNA with polysomes during amino acid starvation when endogenous TIA-1/TIAR proteins are knocked down. However, TIAR appears to be less efficient in doing so than is TIA-1 (Fig. 5, cf. A and B). Thus, TIA-1 and TIAR appear, at least in part, to function redundantly in the repression of 5′TOP mRNA translation upon amino acid starvation. This also rules out the formal possibility that the 5′TOP regulation observed in the above-mentioned experiments was due to off-target effects of the used siRNA.
Figure 5.
TIA-1 and TIAR act in a redundant manner. (A) Northern blots showing sucrose gradient polysome profiles of the rpL32-β-globin 5′TOP mRNA reporter in HeLa cells treated with the siRNA targeting TIA-1 and TIAR (TIA-1/R) or a control siRNA targeting Luciferase (Ctrl). Cells were subjected to 2 h of amino acid starvation prior to polysome fractionation. “Add-back” indicates cotransfected “siRNA-resistant” expression plasmid encoding TIA-1, TIA-1 RBD, or TIA-1 PRD. (B) Same as A, but adding back exogenous TIAR instead of TIA-1. (C) Schematic of TIA-1 showing the RNA-binding RRM domains (RBDs) and the PRD (white box).
The complementation assay also enabled us to test the importance for 5′TOP mRNA translational repression of the C-terminal PRD of TIA-1. This domain is critical for TIA-1-mediated mRNP assembly into SGs (Gilks et al. 2004). As seen in Figure 5A, the TIA-1 RBD alone (Fig. 5A, RBD lacking the PRD) is able to rescue the function of endogenous TIA-1/TIAR in releasing the 5′TOP rpL32-β-globin reporter mRNA from polysomes. In contrast, the TIA-1 PRD lacking the TIA-1 RBD fails to rescue TIA-1/TIAR function as expected (Fig. 5A, PRD). Thus, although a stimulatory role cannot be ruled out, the PRD of TIA-1 is not required for translational repression of 5′TOP mRNA. This suggests that the assembly of TIA-1 into microscopically visible SGs, which is mediated by this domain (Gilks et al. 2004), is not essential for keeping 5′TOP mRNA dissociated from polysomes during amino acid starvation.
Starvation-mediated repression of 5′TOP mRNA translation requires inactivation of mTOR and activation of GCN2
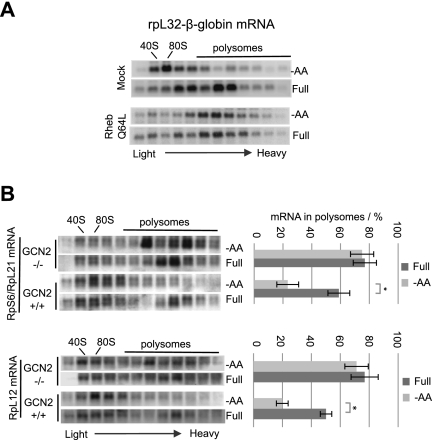
Which signaling pathways are important for the translational repression of 5′TOP mRNA during amino acid starvation? Previous studies showed an important role of mTOR kinase in reactivating repressed 5′TOP mRNA after the readdition of nutrients, including amino acids, which specifically relay their signaling to mTOR via the GTPase Rheb (Meyuhas 2000; Tang et al. 2001; Ma and Blenis 2009; Patursky-Polischuk et al. 2009). We therefore asked whether mTOR inactivation is critical for 5′TOP mRNA translational repression during amino acid starvation. As seen in the polysome fractionation assays in Figure 6A, expression of mutant Rheb Q64L, which constitutively activates mTOR (Li et al. 2004), renders the rpL32-β-globin 5′TOP mRNA insensitive to amino acid starvation (Fig. 6A, cf. bottom and top panels). Thus, starvation-mediated mTOR inactivation is critical for 5′TOP mRNA repression.
Figure 6.
mTOR and GCN2 kinase pathways are required for 5′TOP-mRNA regulation. (A) Northern blots showing sucrose gradient polysome profiles of the rpL32-β-globin 5′TOP mRNA reporter in HeLa cells transfected with either empty plasmid (Mock, top panel) or plasmid encoding Rheb Q64L (bottom panel) while being grown in the absence (−AA) or presence (Full) of amino acids as indicated. (B) Northern blots of sucrose gradient polysome fractions, monitoring endogenous mouse 5′TOP mRNAs encoding rpS6/rpL21 (top panel) and rpL12 (bottom panel). Quantification of three independent experiments, showing polysomal association as described in Figure 4, are shown on the right. Error bars indicate standard deviation. P-values were determined by Student's t-test (two tailed); (*) P < 0.01.
Amino acid starvation is, via elevated levels of uncharged tRNAs, known to activate the kinase GCN2, which phosphorylates eIF2α, thereby reducing cellular levels of the eIF2–GTP–tRNAiMet ternary complex and causing general reduction in translation initiation (Dong et al. 2000; Bruhat et al. 2009). As seen in Figure 6B, 5′TOP mRNAs encoding rpS6, rpL21, and rpL12 all remain associated with polysomal fractions in mouse embryonic fibroblast cells (MEFs) knocked out for GCN2 (GCN2−/−), whereas they efficiently shifted into the subpolysomal fractions in wild-type (GCN2+/+) MEFs during amino acid starvation (Fig. 6B, quantified in the right panels, and note that rpL21 mRNA migrates immediately below rpS6 mRNA). GCN2-dependent regulation is specific for 5′TOP mRNAs, since non-5′TOP β-actin mRNA remains associated with polysomes regardless of the presence of GCN2 or amino acids (Supplemental Fig. S6). Furthermore, the general A254 RNA profiles revealed loss of overall translation repression in GCN2−/− MEFs upon amino acid starvation (Supplemental Fig. S6). Collectively, these observations show that inactivation of the mTOR pathway and activation of the GCN2 kinase is critical for 5′TOP mRNA repression upon amino acid starvation.
Discussion
Our studies identify the paralogous RNA-binding proteins TIA-1 and TIAR as key factors in the translational regulation of the 5′TOP mRNA network (Fig. 7). Upon amino acid starvation, TIA-1 and TIAR assemble with the 5′ end of 5′TOP mRNAs (Fig. 1; Supplemental Fig. S1) and thereby repress the production of 5′TOP mRNA-encoded protein biosynthesis factors (Figs. 2–5). The position of TIA-1/TIAR binding at the 5′ end of the target mRNA is critical for this regulation, as mRNAs with TIA-1/TIAR-binding sites in the 3′ UTR are not regulated in a similar manner (Figs. 1, 4; Supplemental Fig. S5). Starvation-mediated repression of 5′TOP mRNAs requires activation of the GCN2 kinase and inactivation of mTOR (Fig. 6), which is consistent with evidence suggesting that mTOR and GCN2 are functionally linked (Cherkasova and Hinnebusch 2003). These observations explain how the large network of cellular mRNAs encoding protein biosynthesis factors can be coregulated in response to nutrient availability.
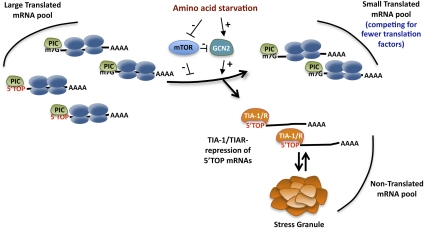
Figure 7.
TIA-1 and TIAR sequesters 5′TOP mRNAs from the translational pool during amino acid starvation. Model illustrating how TIA-1 and TIAR remove 5′TOP mRNAs from the translational pool during amino acid starvation, resulting in a smaller pool of mRNAs competing for a limiting translation machinery (see the text for details). The importance of GCN2 and mTOR signaling is indicated.
While the detailed mechanism by which TIA-1 and TIAR control 5′TOP mRNA translation remains to be investigated, our findings suggest that repression occurs at the translation initiation step. First, amino acid starvation causes accumulation of 5′TOP mRNAs in SGs (Fig. 3), which is characteristic of mRNPs stalled during the translation initiation step (Anderson and Kedersha 2009; Buchan and Parker 2009). Second, 5′TOP mRNAs are released from polysomes in a TIA-1/TIAR-dependent manner upon amino acid starvation (Fig. 4), consistent with ribosome runoff after repression. Third, TIA-1 and TIAR associate specifically with the 5′ end of 5′TOP mRNA (Supplemental Fig. S1), and association of TIA-1 and TIAR with the mRNA 3′ UTR fails to promote starvation-mediated repression (Fig. 4B; Supplemental Fig. S5). Collectively, these observations suggest that TIA-1/TIAR interfere with a step in translation initiation from the mRNA 5′end. The ability of TIA-1 to promote accumulation of 5′TOP mRNAs within SGs does not appear to be critical for translational repression, as mutant TIA-1 lacking the PRD retains the ability regulate 5′TOP mRNAs in response to starvation (Fig. 5). This suggests that although SGs are known to harbor mRNPs stalled at the translation initiation step, these may form as a consequence of translational inhibition rather than being the cause of translational repression. This is consistent with recent observations regarding other repressed mRNPs accumulating in mRNP granules such as SGs and processing bodies (PBs) (Eulalio et al. 2007; Franks and Lykke-Andersen 2007, 2008; Ohn et al. 2008). Understanding the detailed mechanism by which TIA-1 and TIAR repress 5′TOP translation is an important topic for future studies.
How is the activity of TIA-1 and TIAR on 5′TOP mRNAs controlled by amino acid starvation? Our observations show that amino acid starvation enhances association of TIA-1/TIAR proteins with the 5′ end of 5′TOP mRNAs (Fig. 1; Supplemental Fig. S1). It seems unlikely that this is due to an alteration in the intrinsic affinity of TIA-1/TIAR for the pyrimidine tract, since no significant enhancement of the association of TIA-1/TIAR with mRNAs containing TIA-1/TIAR-binding sites in their 3′ UTRs was observed upon starvation (Fig. 1). Enhanced 5′TOP mRNA association could instead be mediated by starvation-stimulated cooperative binding with other factors associated with the 5′ end of 5′TOP mRNAs, such as, for example, La, ZNF9, or AUF1 (Pellizzoni et al. 1996, 1997, 1998; Crosio et al. 2000; Cardinali et al. 2003; Intine et al. 2003; Schwartz et al. 2004; Kakegawa et al. 2007). Alternatively, starvation could trigger the release of factors from the 5′TOP, such as translation initiation complexes, which under conditions stimulatory to cell growth may prevent the binding of TIA-1/TIAR to the 5′TOP. What could be the role of mTOR and GCN2 signaling in this? One possibility is that the general repression of translation initiation factors that occurs upon starvation-mediated mTOR repression and GCN2 activation allows TIA-1 and TIAR to effectively compete with initiation factors for binding to the 5′ end of 5′TOP mRNAs, leading to selective repression of the 5′TOP mRNA network under these conditions. Alternatively, mTOR and/or GCN2 could more directly modify TIA-1/TIAR proteins, or potential associated factors, to modulate their activities.
Our observations suggest a mechanism by which starved cells redirect their limited resources to the production of proteins essential for the starvation response and cell survival. Given that 5′TOPs are found in a large number of highly expressed mRNAs (Hamilton et al. 2006; Iadevaia et al. 2008), TIA-1/TIAR-mediated release of 5′TOP mRNAs from the translational pool during amino acid starvation should result in substantial reduction in the overall pool of mRNAs competing for the translation machinery (Fig. 7). Consistent with this idea, TIA-1/TIAR knockdown results in significant recovery of the overall polysomal pool under starvation conditions (Supplemental Fig. S4). This is potentially critical for the starved cell, which in addition to selectively repressing 5′TOP mRNAs also dampens the activity of general translation factors. Thus, the dampening of the general translation machinery, together with selective sequestration of the abundant 5′TOP mRNA network by TIA-1 and TIAR, could help ensure lower demands on cellular resources, and the continued production of proteins essential for cell survival and for mounting a nutrient deprivation response even under conditions of starvation and limited translational capacity.
Materials and methods
Plasmids and stable cell lines
Plasmids expressing rpL32-β-globin, PABPC1-β-globin, and rpL29-β-globin 5′TOP reporter mRNAs are based on pcTET2Bwt and pcTET2Bwt-3MS2 constructs expressing, under the control of tetracycline-regulatable promoters, wild-type β-globin mRNA (β-wt) or β-globin mRNA containing six MS2 coat protein-binding sites in the 3′ UTR (β-6bs), described previously (Lykke-Andersen and Wagner 2005). To ensure transcription initiation at the start of the 5′TOP sequence in these constructs, the entire promoter region of the pcTET2Bwt plasmid was amplified by PCR using a reverse primer containing the respective 5′TOP sequences, a downstream HindIII restriction site, and a forward primer complementary to a region upstream of the plasmid promoter, which includes an MluI restriction site. This allowed ligation into the MluI–HindIII sites of pcTET2Bwt and pcTET2Bwt-3MS2. To create the reporter constructs containing TIA-1- or TIAR-binding sites in the 3′ UTR of β-globin mRNA, the following sequences for TIA-1 (5′-TTCTTCTTTACTTAAGTTATGTAATAGTTGAAAGGAA-3′) (Lopez de Silanes et al. 2005; Mazan-Mamczarz et al. 2006) and TIAR (5′-TTGCCACCTCCTTGCTCCTGCCCAGACAG-3′) (Kim et al. 2007) were inserted into the NotI site of pcTET2-Bwt. To create the reporter construct containing the 5′TOP element, including downstream sequences, from human rpL32 mRNA in the 3′ UTR of β-globin mRNA, the sequence 5′-CCCTTCTCTCTCGGCGGCTGCCTACGGAGGTGGCAGCCATCTCCTTCTCGGCT-3′ was inserted between NotI and XbaI sites of pcTET2-Bwt. A plasmid encoding a tetracycline-responsive activator was used to activate transcription of reporter mRNAs (pTet-tTA; Clontech). Plasmids expressing human TIA-1 or TIAR proteins pcNEGFP-TIA-1/TIAR, pcDNA3-Flag-TIA-1/TIAR, or pcDNA5-Flag-TIA-1/TIAR were created by inserting PCR products encompassing the entire ORFs of TIA-1 or TIAR, amplified from a random hexamer-primed HeLa cDNA library, into BamHI and NotI sites of the vectors pcNEGFP (Franks and Lykke-Andersen 2007), pcDNA3-Flag (Lykke-Andersen 2002), and pcDNA5-frt-TO (Invitrogen), respectively, the latter of which was modified to encode an N-terminal Flag peptide. Truncated versions of TIA-1 were generated in the same manner, using primers amplifying specific parts of the TIA-1 ORF (TIA-1 RBD: amino acids 1–278; TIA-1 PRD: amino acids 279–375). siRNA-resistant clones were generated by site-specific mutagenesis using standard protocols (QuickChange, Stratagene).
The plasmid pcNEGFP-hDcp1a, which was used as a PB marker, has been described previously (Lykke-Andersen 2002). pcNEGFP-PABPC1 was generated by insertion of the ORF of human PABPC1 between BamHI and NotI sites of pcNEGFP. Stable HEK293S cell lines expressing TIA-1, TIAR, or truncated versions of TIA-1 were generated using the Flp-In T-Rex system (Invitrogen) by site-specific integration using the pcDNA5-frt-TO-based TIA-1/TIAR plasmids described above. Plasmids used to generate radiolabeled riboprobes for Northern blotting were prepared by inserting RT–PCR-generated fragments of human rpL23a, rpS6, rpL21, rpL12, rpL36, PABPC1, β-actin, calmodulin 2, and GAPDH cDNAs into pcDNA3. Plasmid sequences are available on request.
RNAi
HeLa Tet-Off cells (Clontech) were seeded at low density (20%–25% confluency) in 10-cm plates in full medium (DMEM, 10% fetal bovine serum [FBS], 1% penicillin/streptomycin solution) for 24 h prior to transfection. Cells were transfected twice with either control siRNA targeting Luciferase/GFP or siRNA targeting both TIA-1 and TIAR (UUCCAGAGAUGUGACAGAA) at a final concentration of 20 nM. First transfection was conducted using siLentFect Lipid (Bio-Rad) according to the manufacturer's protocol. Twenty-four hours later, cells were washed in phosphate-buffered saline (PBS) and supplemented with full medium (DMEM, 10% FBS). Another 24 h later, cells were transfected with siRNA at a final concentration of 20 nM and, where appropriate, 6 μg of reporter and/or expression plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. When using tetracycline-regulated plasmids, 50 ng/mL tetracycline was included in transfections to inhibit reporter expression. Twenty-four hours later, cells were washed in PBS and supplemented with DMEM, 10% FBS, and 1% penicillin/streptomycin solution, including 50 ng/mL tetracycline. Eight hours to 12 h prior to harvest, cells were washed twice and incubated in medium lacking tetracycline to induce transcription of the reporter gene.
Indirect immunofluorescence and in situ hybridization assays
HeLa cells in DMEM/10% FBS at ∼50% confluency in six-well plates were transfected using Mirus TransIT HeLaMonster reagent according to the manufacturer's protocols, with a total of 2 μg of plasmid. Cells were split to chamber slides 24 h later. For indirect immunofluorescence experiments, cells were fixed in 4% paraformaldehyde for 15 min, and permeabilized and blocked with PBS/1% goat serum (or horse serum)/0.1% Triton X-100 for 20 min. Cells were then incubated for 1 h with goat anti-TIA-1 (Santa Cruz Biotechnologies, SC-1751), goat anti-TIAR (Santa Cruz Biotechnologies, SC-1749), rabbit anti-PABPC1 (Abcam, ab21060-100), or mouse anti-eIF4G (a generous gift from Dr. Nahum Sonenberg) antibodies at 1:500, 1:500, 1:2000, and 1:2000 dilutions, respectively. Following removal of the primary antibody, cells were incubated for 1 h with 4 μg/mL secondary anti-IgG antibodies labeled with either Alexa 594, Alexa 488, or Texas-Red fluorophore (Molecular Probes).
For in situ hybridization experiments, HeLa cells in six-well plates were transfected using Mirus TransIT HeLaMonster reagent in the presence of 50 ng/mL tetracycline with 300 ng of reporter mRNA expression plasmid, 300 ng of pTet-tTA, and 100 ng of pcNEGFP-hDcp1a, pcNEGFP-TIA-1, pcNEGFP-TIAR, or pcNEGFP-PABPC1. pcDNA3 was added to a total of 2 μg of plasmid in each experiment. After 24 h, cells were trypsinized and reseeded onto eight-well chamber slides (Permanox). Approximately 36 h after transfection, transcription of reporter mRNAs was initiated by washing cells in PBS and placing them in DMEM/10% FBS containing no tetracycline. After a transcriptional pulse of 8–12 h, cells were fixed in 4% paraformaldehyde for 15 min and permeabilized overnight in 70% ethanol. Cells were then rehydrated for 10 min in 50% formamide and 2× SSC (300 mM NaCl, 30 mM Na-citrate at pH 7.0). Next, cells were incubated overnight at 37°C in a solution containing 50% formamide, 2× SSC, 0.02% bovine serum albumin (BSA) (Invitrogen), 2 mM vanadyl–ribonucleoside complexes (New England Biolabs), 1 μg/mL total yeast RNA (Roche), and 0.1 mg/mL dextran sulfate (Invitrogen). In order to detect the localization of the β-globin mRNA, four Texas-Red-labeled 50-nt DNA probes (Invitrogen) complementary to sequences in exons 1, 2, and 3 of β-globin mRNA were added to the mixture at a concentration of 20 ng/mL each (Franks and Lykke-Andersen 2007). Cells were washed twice for 30 min at 37°C in 50% formamide and 2× SSC, followed by four 5-min washes in PBS and an additional brief wash in nuclease-free water prior to visualization. Endogenous rpL29 and GAPDH mRNAs were visualized in HeLa cells transfected with pNEGFP-PABPC1 as a marker for SGs. For each mRNA, three 50-mer DNA probes, each containing five Cy5 fluorophores conjugated to amino-allyl-modified thymidines (DNA Technology A/S), were used for in situ hybridization. Probe sequences were as follows (with “5” representing amino-allyl-T): for GAPDH mRNA, #1: 5GGTGATGGG5TTCCATTGA5GACAAGCTTCCCG5TCTCAGCCT5GAC, #2: G5GGTGCAGGAGGCA5TGCTGATGATC5TGAGGCTGT5GTCATACTTC5C, and #3: ACGA5ACCAAAGTTG5CATGGATGACC5TGGCCAGGGG5GCTAAGCAG5T; and for rpL29 mRNA, #1: 5TCTGTGCCAT5TTCGGGACTGG5TGTGTGTGGTG5GGTTCTTGGACT5G, #2: 5GGGGTCCACCCCC5TAAGAGATTCG5ATCTTTGTGA5CGGGGTTTCT5G, and #3: C5TTGGGATCT5GGGCTTAACC5CCTTGGGCTT5ACGAGGGCCTTGA5AG. Cells were seeded on glass coverslips in six-well plates, transfected 24 h later with 300 ng of pNEGFP-PABPC1 using HeLaMONSTER reagent (Mirus) according to the manufacturer's recommendations, and incubated for another 24 h. Treatment, fixation, and permeabilization steps were done as described above. Cells were then washed twice in PBS-M (PBS + 5 mM MgCl2) for 5 min at room temperature, and rehydrated for 10 min in 50% formamide and 2× SSC (300 mM NaCl, 30 mM Na-citrate at pH 7.0). Next, cells were incubated for 3 h at 37°C in a solution containing 50% formamide, 2× SSC, 0.25 mg/mL Escherichia coli tRNA, 0.25 mg/mL salmon sperm DNA (Invitrogen), 2.5 mg/mL BSA (Roche), and 0.5 ng/μL fluorescently labeled probes. Post-hybridization washing steps were done as described above.
Polysome profiling by sucrose gradient fractionation
HeLa Tet-Off cells (Clontech) were grown in 10-cm plates in full medium (DMEM, 10% FBS, 1% penicillin/streptomycin solution) for 24 h prior to transfection at ∼80% confluency. Four micrograms of β-globin reporter plasmid and, in the experiments in Supplemental Figure S4, 2 μg of plasmids encoding siRNA-resistant TIA-1, TIAR, or TIA-1 mutants, and pcDNA3 plasmid up to a total amount of 12 μg of plasmid were transfected using Lipofectamine 2000 (Invitrogen) or TransIT HeLa-MONSTER reagent (Mirus) according to the manufacturer's protocols. Cells were incubated overnight, and transcription of reporter genes was pulsed 8–12 h prior to harvest by removing tetracycline as described above. Cells were washed twice in PBS and either supplemented with fresh full medium (DMEM, 10% FBS) or starved for amino acids by adding Krebs-Ringer medium (KRM) (1.8 g/L glucose, 0.5 mM MgCl2, 4.5 mM KCl, 120 mM NaCl, 0.9 mM Na2HPO4, 2 mM NaH2PO4 at pH 7.4, supplemented with 10% dialyzed FBS [Invitrogen]) for 2 h. Cells were then treated with 100 μg/mL cycloheximide for 5 min followed by two washes in ice-cold PBS containing 100 μg/mL cycloheximide. Cells were subsequently lysed directly in the cell culture plate in 900 μL of 1× polysome extraction buffer (1× PEB; 50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 7.5 mM MgCl2, 0.5% Triton X-100, 500 μg/mL Heparin, 100 μg/mL cycloheximide), scraped off, and placed for 5 min on ice. Lysates were cleared by centrifugation at 15,300g for 15 min at 4°C. Complexes were separated in 11 mL of linear 1× PEB/10%–50% sucrose gradients by ultracentrifugation at 35,000 rpm in a Beckman SW41 rotor for 2.5 h at 4°C. Twelve 1-mL fractions were collected using a Teledyne/ISCO fractionation system. Four-hundred microliters from each fraction was vortexed for 20 sec after addition of 600 μL of 8 M guanidine-HCl and 600 μL of 2-propanol and precipitated overnight at −20°C. Precipitates were isolated by centrifugation at 17,900g for 30 min at 4°C, and pellets were subsequently dissolved in Trizol solution (Invitrogen). RNA was isolated according to the manufacturer's protocol. Total RNA from each fraction was run on a 1.2% formaldehyde-agarose gel for Northern blotting and probed with internally 32P-labeled riboprobes generated by SP6 polymerase-directed in vitro transcription of linearized plasmid templates.
Immunoprecipitation of TIA-1/TIAR mRNP complexes
For immunoprecipitation of Flag-tagged TIA-1 and TIAR, HEK293S stable cell lines were grown in 10-cm plates to ∼50% confluency, and the expression of Flag-tagged TIA-1 and TIAR was induced by addition of 60 ng/mL tetracycline 36 h prior to harvest. Two hours prior to harvest, cells were supplemented with full DMEM medium or KRM medium lacking amino acids as described above. Cells were washed in cold PBS and lysed for 5 min on ice in 900 μL of hypotonic lysis buffer (10 mM Tris-HCl at pH 7.4, 10 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, and either 0.5 mM PMSF, 2 μg/mL Aprotinin, 2 μg/mL Leupeptin, or Complete Mini EDTA-free [Roche]). The NaCl concentration was subsequently readjusted to 150 mM. Lysates were cleared by centrifugation at 20,000g and 800 μL of supernatant was loaded onto pre-equilibrated anti-Flag M2 agarose (20-μL bead volume; Sigma) and nutated for 4 h at 4°C. After a total of eight 1-mL washes in NET-2 buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 0.1% Triton X-100) containing 0.5 μg/mL Flag-peptide (Sigma), complexes were eluted twice into 60 μL of NET-2 containing 200 μg/mL Flag peptide by moderate shaking for 30 min at 10°C or eluted by direct addition of 1 mL of Trizol to the Flag-M2 agarose. Flag-eluates were combined and supplemented with 1 mL of Trizol (Invitrogen), and RNA was isolated according to the manufacturer's protocol. Input and immunoprecipitated RNA was detected by Northern blotting. For RNase H cleavage prior to immunoprecipitation, 1.5 mL of cleared cell lysates from 15-cm plates were split into three aliquots with either none or 1 μM final concentration targeting DNA-oligos (12-mers), supplemented with 40 mM KCl and 7 mM MgCl2, and incubated for 10 min at 37°C. Forty units of RNase H (Invitrogen) was added, and the reaction was incubated for an additional 20 min at 30°C. The following Flag immunoprecipitation was performed as described above.
Primer extension
Immunoprecipitated purified RNA was dissolved in 4.8 μL of nuclease-free H2O and used in two primer extension reactions. Ten picomoles of RT-primer was 5′ end-labeled using T4 polynucleotide kinase (Fermentas) and 32P-ATP (γ) according to the manufacturer's protocol. Primers were purified in a 10% denaturing acrylamide gel, and gel slices were soaked in 200 μL of TE for overnight elution. Three microliters of primer solution was mixed with 0.6 μL of 10× anneal buffer (100 mM Tris at pH 6.9, 400 mM KCl, 5 mM EDTA) and 2.4 μL of purified RNA. Samples were denatured for 1 min at 90°C and allowed to anneal for 20 min at 50°C and for 2 min at 37°C. Four microliters of extension mix (2 μL of 5× RT buffer, 1.5 μL of 2.5 mM dNTP, 0.5 μL of SuperScript II [Invitrogen]) was added, and the reaction was incubated for 30 min at 37°C. The reaction was stopped by addition of 0.3 M NaAc (pH 6.0) and 1 mM EDTA, and cDNA was precipitated with 125 μL of 96% ethanol. Pellet was dissolved in 6 μL of formamide load buffer and denatured for 2–3 min at 90°C prior to loading on a 6% denaturing PAGE.
35S pulse-chase labeling and immunoprecipitation analysis
HeLa Tet-Off cells (Clontech) grown in 10-cm plates were used for double siRNA transfection targeting TIA-1/TIAR or Luciferase as described above. Forty-eight hours after the last siRNA transfection, cells were grown in DMEM lacking cysteine and methionine (Invitrogen) for 2 h. Subsequently, cells were supplemented with either DMEM/10% dialyzed FBS including 0.2 mCi 35S-cysteine/methionine (Perkin Elmer, Easy Tag Express Labeling Mix) or KRM/10% dialyzed FBS including 0.2 mCi 35S-cysteine/methionine for 1 h. Subsequently, cells were lysed in 1 mL of RIPA buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Igepal 630, 1% sodium deoxycholate, protease inhibitors [Complete Mini EDTA-free, Roche]) and homogenized by passing through a 20-guage syringe five times. Lysates were cleared by centrifugation at 20,000g, for 10 min at 4°C. Lysates were preincubated with 20 μL of RIPA-equilibrated protein-A sepharose for 30 min at 4°C with slow nutation. Supernatants were incubated with 20 μL (bead volume) of protein-A sepharose already conjugated to polyclonal anti-PABPC1 or mouse monoclonal anti-hnRNP C1 and incubated overnight at 4°C with slow nutation. Beads were washed six times in 1 mL of RIPA buffer at 4°C, and bound protein was eluted by boiling in SDS load buffer (10% mercaptoethanol, 100 mM Tris-HCl at pH 6.8, 2% sodium dodecyl sulphate [SDS], 0.02% bromphenol blue). Lysates were separated in a 10% PAGE/SDS gel and either exposed to PhosphorImager screen or blotted for Western analysis.
Acknowledgments
Drs. Amy Pasquinelli (UCSD) and Jim Kadonaga (UCSD) are thanked for comments on the manuscript. We thank Drs. Nancy Kedersha and Paul Anderson (Brigham and Women's Hospital) for anti-TIA-1 and -TIAR antibodies, Dr. Nahum Sonenberg (McGill University) for anti-eIF4G, and Dr. Seraphin Pinol-Roma for anti-hnRNPC1/C2 antibodies. Claire Egan and Line Kristensen are thanked for technical assistance. This study was supported by grants from the National Institutes of Health (R01 GM077243) and the American Cancer Society (RSG GMC111896) to J.L.-A., and from the Alfred Benzon Foundation (Copenhagen, Denmark), the Danish National Research Council (FNU), The Lundbeck Foundation (Denmark), and the Novo Nordisk Foundation (Denmark) to C.K.D.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17355911.
References
- Anderson P, Kedersha N 2009. Stress granules. Curr Biol 19: R397–R398 doi: 10.1016/j.cub.2009.03.013 [DOI] [PubMed] [Google Scholar]
- Avni D, Shama S, Loreni F, Meyuhas O 1994. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol Cell Biol 14: 3822–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni D, Biberman Y, Meyuhas O 1997. The 5′ terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res 25: 995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biberman Y, Meyuhas O 1999. TOP mRNAs are translationally inhibited by a titratable repressor in both wheat germ extract and reticulocyte lysate. FEBS Lett 456: 357–360 [DOI] [PubMed] [Google Scholar]
- Bruhat A, Cherasse Y, Chaveroux C, Maurin AC, Jousse C, Fafournoux P 2009. Amino acids as regulators of gene expression in mammals: molecular mechanisms. Biofactors 35: 249–257 [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R 2009. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinali B, Carissimi C, Gravina P, Pierandrei-Amaldi P 2003. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J Biol Chem 278: 35145–35151 [DOI] [PubMed] [Google Scholar]
- Cherkasova VA, Hinnebusch AG 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev 17: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Boyl PP, Loreni F, Pierandrei-Amaldi P, Amaldi F 2000. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res 28: 2927–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember LM, Kim ND, Liu KQ, Anderson P 1996. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem 271: 2783–2788 [DOI] [PubMed] [Google Scholar]
- Dever TE, Yang W, Astrom S, Bystrom AS, Hinnebusch AG 1995. Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol Cell Biol 15: 6351–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 6: 269–279 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27: 3970–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farny NG, Kedersha NL, Silver PA 2009. Metazoan stress granule assembly is mediated by P-eIF2α-dependent and -independent mechanisms. RNA 15: 1814–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2007. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev 21: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2008. The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TL, Stoneley M, Spriggs KA, Bushell M 2006. TOPs and their regulation. Biochem Soc Trans 34: 12–16 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J Biol Chem 272: 21661–21664 [DOI] [PubMed] [Google Scholar]
- Hornstein E, Harel H, Levy G, Meyuhas O 1999. Overexpression of poly(A)-binding protein down-regulates the translation or the abundance of its own mRNA. FEBS Lett 457: 209–213 [DOI] [PubMed] [Google Scholar]
- Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F 2008. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA 14: 1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intine RV, Tenenbaum SA, Sakulich AL, Keene JD, Maraia RJ 2003. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol Cell 12: 1301–1307 [DOI] [PubMed] [Google Scholar]
- Kakegawa T, Ohuchi N, Hayakawa A, Hirata S, Matsuda M, Kogure K, Kobayashi H, Inoue A, Kaspar RL 2007. Identification of AUF1 as a rapamycin-responsive binding protein to the 5′-terminal oligopyrimidine element of mRNAs. Arch Biochem Biophys 465: 274–281 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kuwano Y, Zhan M, Pullmann R Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M, et al. 2007. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol Cell Biol 27: 6806–6817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Wilce MC, Yoga YM, Pendini NR, Gunzburg MJ, Cowieson NP, Wilson GM, Williams BR, Gorospe M, Wilce JA 2011. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res 39: 1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol 284: C273–C284 doi: 10.1152/ajpcell.00314.2002 [DOI] [PubMed] [Google Scholar]
- Li Y, Inoki K, Guan KL 2004. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol Cell Biol 24: 7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M 2005. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol 25: 9520–9531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & Dev 19: 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J 2009. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M 2006. Translational repression by RNA-binding protein TIAR. Mol Cell Biol 26: 2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O 2000. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem 267: 6321–6330 [DOI] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D 2008. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell 19: 4469–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 10: 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- Parry TJ, Theisen JW, Hsu JY, Wang YL, Corcoran DL, Eustice M, Ohler U, Kadonaga JT 2010. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev 24: 2013–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patursky-Polischuk I, Stolovich-Rain M, Hausner-Hanochi M, Kasir J, Cybulski N, Avruch J, Ruegg MA, Hall MN, Meyuhas O 2009. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol 29: 640–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Cardinali B, Lin-Marq N, Mercanti D, Pierandrei-Amaldi P 1996. A Xenopus laevis homologue of the La autoantigen binds the pyrimidine tract of the 5′ UTR of ribosomal protein mRNAs in vitro: implication of a protein factor in complex formation. J Mol Biol 259: 904–915 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Lotti F, Maras B, Pierandrei-Amaldi P 1997. Cellular nucleic acid binding protein binds a conserved region of the 5′ UTR of Xenopus laevis ribosomal protein mRNAs. J Mol Biol 267: 264–275 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Lotti F, Rutjes SA, Pierandrei-Amaldi P 1998. Involvement of the Xenopus laevis Ro60 autoantigen in the alternative interaction of La and CNBP proteins with the 5′UTR of L4 ribosomal protein mRNA. J Mol Biol 281: 593–608 [DOI] [PubMed] [Google Scholar]
- Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, et al. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J 19: 4154–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Dong J, Hu C, Francklyn CS, Hinnebusch AG 2001. The tRNA-binding moiety in GCN2 contains a dimerization domain that interacts with the kinase domain and is required for tRNA binding and kinase activation. EMBO J 20: 1425–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Dong J, Hinnebusch AG 2002. Mutations that bypass tRNA binding activate the intrinsically defective kinase domain in GCN2. Genes Dev 16: 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EI, Intine RV, Maraia RJ 2004. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5′-terminal oligopyrimidine mRNA metabolism. Mol Cell Biol 24: 9580–9591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolovich M, Tang H, Hornstein E, Levy G, Cohen R, Bae SS, Birnbaum MJ, Meyuhas O 2002. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol Cell Biol 22: 8101–8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O 2001. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol 21: 8671–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]