Abstract
Hfq is a bacterial post-transcriptional regulator. It facilitates base-pairing between sRNA and target mRNA. Hfq mediates DsrA-dependent translational activation of rpoS mRNA at low temperatures. rpoS encodes the stationary-phase σ factor σS, which is the central regulator in general stress response. However, structural information on Hfq–DsrA interaction is not yet available. Although Hfq is reported to hydrolyze ATP, the ATP-binding site is still unknown. Here, we report a ternary crystal complex structure of Escherichia coli Hfq bound to a major Hfq recognition region on DsrA (AU6A) together with ADP, and a crystal complex structure of Hfq bound to ADP. AU6A binds to the proximal and distal sides of two Hfq hexamers. ADP binds to a purine-selective site on the distal side and contacts conserved arginine or glutamine residues on the proximal side of another hexamer. This binding mode is different from previously postulated. The cooperation of two different Hfq hexamers upon nucleic acid binding in solution is verified by fluorescence polarization and solution nuclear magnetic resonance (NMR) experiments using fragments of Hfq and DsrA. Fluorescence resonance energy transfer conducted with full-length Hfq and DsrA also supports cooperation of Hfq hexamers upon DsrA binding. The implications of Hfq hexamer cooperation have been discussed.
Keywords: Hfq RNA complex, crystal structure, NMR, DsrA, rpoS, Hfq cooperation, ADP
Hfq is a highly abundant bacterial post-transcriptional regulator found in around half of Gram-positive and Gram-negative bacteria, while its counterparts in eukaryotes, Sm/Lsm proteins, are involved in mRNA splicing and decay (Kambach et al. 1999; Tharun et al. 2000; Scofield and Lynch 2008). Hfq plays a central role in the small RNA (sRNA)-mediated regulation network and interacts with many bacterial sRNAs (Zhang et al. 2003). Polyadenylation-dependent (Hajnsdorf and Regnier 2000; Mohanty et al. 2004) and sRNA-mediated (Morita and Aiba 2011) mRNA decay in bacteria is also regulated by Hfq. Escherichia coli (Ec) Hfq consists of a conserved N-terminal Sm domain (Hfq65, amino acid residues 1∼65, an N-terminal α helix followed by five β strands) and a flexible C-terminal tail of 37 amino acid residues. The Sm domain forms a ring-like homohexameric structure (Sauter et al. 2003). One side of the ring is named the proximal side, which preferentially associates with U-rich ssRNA sequences, while the opposite side is called the distal side, which binds to A-rich sequences. It has been proposed that Hfq is capable of binding substrates simultaneously on the distal and proximal sides (Mikulecky et al. 2004; Brennan and Link 2007; Link et al. 2009), but direct structural evidence has not yet been reported.
The ability to sense and respond to environmental changes is crucial for the survival of bacteria. Ec sRNA mediates various stress responses, such as oxidative stress, UV irradiation, heat shock, hyperosmolarity, phosphosugar toxicity, and iron concentration (Gottesman et al. 2006). One particularly striking example of sRNA-mediated stress response is the regulation of rpoS mRNA translation by DsrA sRNA. The rpoS mRNA encodes the RNA polymerase subunit σS factor, which is the master regulator of the general stress response (Hengge-Aronis 2002). The sRNA DsrA is required for effective translation of rpoS at low temperatures (Lease et al. 1998; Majdalani et al. 1998). In the absence of activation factors, translation of rpoS mRNA is hindered by a stem–loop structure that sequesters the Shine-Dalgarno (SD) ribosome-binding site. DsrA base-pairs with one strand of this stem on rpoS and releases the occluded SD site for effective ribosome binding. The annealing of DsrA to rpoS requires the unwinding of stem–loops on both RNAs, and Hfq is demonstrated to facilitate this process (Muffler et al. 1996; Lease and Woodson 2004). RNase footprinting experiments suggest that Hfq preferentially binds the unpaired linker between DsrA stem–loop I (SLI) and SLII and parts of SLII (Brescia et al. 2003; Lease and Woodson 2004). The only three reported RNA–Hfq complex structures are A15 RNA bound to the distal side of Ec Hfq (Link et al. 2009); a canonical U-rich Sm recognition sequence, AU5G, bound to the proximal side of Staphylococcus aureus (Sa) Hfq (Schumacher et al. 2002); and Salmonella typhimurium (St) Hfq72 (C-terminal truncated after S72) in complex with U6 RNA (Sauer and Weichenrieder 2011). These structures provided important insights into U-rich and A-rich ssRNA-binding patterns on Hfq. However, these structures provide limited information on where and how Hfq binds to DsrA. Hfq has been reported to hydrolyze ATP with relatively low activity (Sukhodolets and Garges 2003; Brennan and Link 2007). Efforts have been made to predict the ATP-binding site on Hfq (Arluison et al. 2007; Lazar et al. 2010). However, structural information on ATP or ADP binding to Hfq is not available.
We identified both A-rich and U-rich Hfq-binding regions on SLI and the linker between SLI and SLII of DsrA using florescence polarization (FP) experiments. Furthermore, we report the crystal structures of 1.7 Å resolution Hfq65–AU6A–ADP and 2.0 Å resolution Hfq65–ADP complexes. AU6A (nucleotides 28∼35) is the major U-rich Hfq-binding site on DsrA. Hfq65–AU6A–ADP is the first complex structure of Hfq with substrates bound simultaneously on the distal and proximal sides. It provides the first insight into how Hfq recognizes the internal U-rich region of a specific sRNA. Our crystal structures revealed that AU6A binds in a novel recognition pattern that is different from Sa Hfq recognition of the canonical U-rich recognition sequence AU5G (Schumacher et al. 2002) or St Hfq recognition of the U6 sequence (Sauer and Weichenrieder 2011). AU6A contacts the proximal side and the distal side of two different Hfq hexamers, suggesting that the cooperation of the Hfq distal and proximal sides from different hexamers is required in DsrA binding. This cooperation is also supported by our observations in solution nuclear magnetic resonance (NMR) paramagnetic relaxation enhancement (PRE) experiments, FP experiments conducted with fragments of DsrA sRNA and Hfq, and fluorescence resonance energy transfer (FRET) experiments conducted with full-length Hfq and DsrA. NMR chemical shift perturbation showed that both the distal and proximal sides, as well as residues on the outer rim of Hfq, are involved in binding to DsrA fragments. Multiple ADPs bind to the distal side of each Hfq hexamer. All ADPs position their phosphate groups above the distal side instead of burying them in a nearby cleft as previously postulated (Arluison et al. 2007; Lazar et al. 2010). The phosphate groups of some ADPs make polar contact to R16 and R17 or Q41 on the proximal side of another Hfq hexamer.
Results
Preferential Hfq-binding sites on DsrA
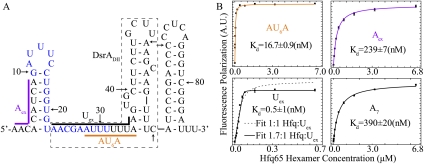
RNase footprinting experiments suggest that Hfq preferentially binds the unpaired linker between SLI and SLII and part of SLII (Lease and Belfort 2000; Lease and Woodson 2004), as well as an A-rich stretch at the 5′ end on DsrA (Brescia et al. 2003). To better understand the binding properties between Hfq and DsrA, multiple segments on DsrA were selected for binding investigations. Equilibrium dissociation constants (Kd) were determined by fitting the FP data of 5′-FAM-labeled oligonucleotides upon Hfq65 binding into a 1:1 model unless otherwise mentioned. These sequences are depicted on the DsrA secondary structure (Fig. 1A), and corresponding Kds are listed in Supplemental Table S2. Aex (nucleotides 1–9) binds Hfq65 with relatively low affinity (Kd = ∼240 nM), similar to that of short poly(A) stretch A7 (Kd = ∼390 nM) (Fig. 1B). AU6A (nucleotides 28∼35) is the major U-rich Hfq-binding site between SLI and SLII and was used in cocrystallization with Hfq65. It binds tightly to Hfq65 with a Kd of ∼17 nM. Uex (nucleotides 23∼36) consists of the linker between SLI and SLII, containing an A-rich segment extension to the 5′ end of the U-rich AU6A and one adenosine extension to the 3′ end. These extensions caused significant changes in the interaction properties between Uex and AU6A. The FP data of Uex fail to fit well to a simple 1:1 binding model, and a typical poorly fitted curve is shown as a dashed line in Figure 1B (bottom left panel). As Uex contains both A-rich and U-rich regions, which bind to distinct sides on Hfq with different affinities, interaction between Hfq and Uex could lead to very complicated behaviors. However, due to avid binding of Uex to Hfq65, an accurate apparent binding stoichiometry of ∼1.7:1 can be acquired. An approximately 1 nM apparent equilibrium dissociation constant at this stoichiometry can be obtained by the fitting of FP data (Fig. 1B). The near 2:1 Hfq:Uex binding stoichiometry and much higher affinity for Uex compared with AU6A indicate that A-rich and U-rich segments on Uex simultaneously bind to Hfq, and that more than one Hfq hexamer are involved in the interaction. This is supported by our crystallography, NMR, and FRET studies.
Figure 1.
Different Hfq-binding properties to selected segments on DsrA. (A) Schematic of the secondary structure of DsrA. Colored lines ([purple] Aex; [black] Uex; [orange] AU6A) along the structure show the fragments selected for investigation. The names of these fragments are colored accordingly. The nucleotides for rpoS base-pairing are colored in blue. DsrA domain II (DsrADII) consists of nucleotides in the dashed box. (B) FP of selected RNA fragments upon Hfq titration. A7 is a single-stranded poly(A) RNA segment. Equilibrium dissociation constants (Kd) of AU6A, Aex, and A7 were obtained by fitting to a 1:1 binding model. (Bottom left panel) Fitting of Uex in a 1:1 model (dashed lines) failed and ∼1.7:1 Hfq:Uex stoichiometry was required for good fitting of data points. Fitted curves are colored as in A for fragments from DsrA. Equilibrium dissociation constants are indicated in the graphs.
AU6A and ADP contact proximal and distal sides from different Hfq65 hexamers in the crystal structure
ssRNA 5′-AUUUUUUA-3′ (AU6A, nucleotides 28–35 of DsrA) was chosen for cocrystallization with the Sm domain of Ec Hfq (Hfq65) and ADP. AU6A is a major U-rich Hfq-binding site on the linker between DsrA SLI and SLII that was previously reported to associate with the Hfq proximal side (Lease and Woodson 2004; Mikulecky et al. 2004; Sun and Wartell 2006; Updegrove et al. 2011). AU6A is different from the canonical U-rich sequence AU5G recognized by Sm proteins. Actually, the AU5G sequence is quite different from many U-rich Hfq recognition sites on sRNAs (Brennan and Link 2007; Sauer and Weichenrieder 2011). Structural information of Hfq bound to specific RNA substrates is essential for understanding Hfq–RNA interactions. We demonstrated the strong binding of AU6A to Hfq65 using FP (Fig. 1B). The phase of the Hfq65–AU6A–ADP and Hfq65–ADP complex crystal structures was determined by molecular replacement using the apo Ec Hfq structure (1HK9) as the search model. The structure of the Hfq65–AU6A–ADP complex was refined to 1.7 Å resolution with Rwork and Rfree values of 16.5% and 20.5%, respectively. Each asymmetric unit contained clear density for one Hfq65 hexamer, one AU6A molecule, four ADP molecules, and one purine of ADP (Fig. 2A–C). The structure of the Hfq65–ADP complex was refined to 2.0 Å resolution with Rwork and Rfree values of 19.6% and 23.8%, respectively. Each asymmetric unit contained a clear density for two Hfq65 hexamers and four ADP molecules (Supplemental Fig. S1A,B). The structural and refinement statistics of these two structures are listed in Supplemental Table S1.
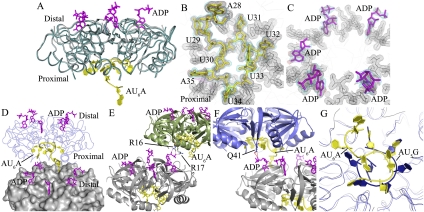
Figure 2.
Global structure of the Hfq–AU6A–ADP complex. (A) Each asymmetric unit contains one Hfq hexamer (dark-cyan ribbon), five ADPs (purple stick), and one AU6A molecule (yellow cartoon). The apo Hfq (gray ribbon) structure was not significantly affected by the binding of AU6A RNA and ADP. The apo Ec Hfq was fitted to bond Hfq in the Hfq65–AU6A–ADP complex using PyMol. (B,C) Electron density maps of AU6A and ADP, respectively. AU6A RNA is in yellow, and ADP is in purple. Hfq is represented as gray lines. Electron density of Hfq residues involved in RNA binding is shown as a gray mesh. Difference maps of Fo–Fc contoured at 2.2σ before inclusion of RNA and ADP are shown in red. Densities (2Fo–Fc) contoured at 1.0σ are shown in cyan. (D) Two closely packed asymmetric units were put together; one Hfq hexamer is presented as molecular surface and colored gray, and the other is presented as a ribbon in blue. ADP and AU6A are represented as sticks. Five out of six R sites on the distal side are occupied by ADPs, while the remaining R site is occupied by the 5′-end adenosine of AU6A (A28 on DsrA), which is bound mostly on the proximal side of another Hfq hexamer. (E) ADPs bind to R sites on the distal side of one Hfq hexamer and positions phosphate groups above the distal side. The α and β phosphates of one ADP on one Hfq hexamer (gray) contact the side chains of R16 and R17 on the proximal side of another Hfq hexamer (green). (F) The α phosphates of one ADP on one Hfq hexamer (gray) contact the side chains of Q41 on the proximal side of another Hfq hexamer (blue). (G) Superposition of the Ec Hfq–AU6A–ADP and Sa Hfq–AU5G complex structures. AU6A (yellow cartoon) recognition by the Ec Hfq (gray ribbon) proximal side differs from AU5G (blue cartoon) recognition in the Sa Hfq (blue ribbon)–AU5G complex.
Crystal structures show that both AU6A and ADP contact the proximal side and distal side of two different Hfq hexamers. In the Hfq65–AU6A–ADP complex structure, the region U29 to A35 of AU6A binds to the proximal side of the Hfq hexamer in the same asymmetric unit, while A28 inserts into the purine nucleotide-selective site (R site; so named due to its likely ability to bind both guanosine and adenosine) (Link et al. 2009) on the distal side of a closely packed Hfq hexamer. To enable this binding, the phosphodiester backbone extends from A28 toward the proximal side of Hfq and then curves annularly around the central pore in a way similar to AU5G when bound to Sa Hfq (Schumacher et al. 2002). The remaining five R sites on the distal side are filled by five ADP purines (Fig. 2D). ADPs bind to the R sites in a way very similar to adenines on poly(A). The purine ring stacks against the side chains of Y25, L26′, I30′, and L32′ (where prime denotes residues from a neighboring Hfq monomer in the same hexamer) (Supplemental Fig. S1E), while phosphate groups are positioned above the Hfq distal side. One ADP on each Hfq hexamer makes polar contact by phosphate groups to R16 and R17 or Q41 on the proximal side of another adjacent Hfq hexamer (Fig. 2E,F). In the Hfq65–ADP complex, the binding mode of ADP to Hfq is the same as in the Hfq65–AU6A–ADP complex. Four out of six R sites on the distal side of each Hfq hexamer are occupied by ADP molecules, indicating nonequivalence of the six R sites in ADP binding (Supplemental Fig. S1A). A similar inequality of ATP-binding sites has previously been reported (Bochtler et al. 2000). The density for the ribose and/or phosphate groups is not clear for several ADP molecules. Phosphate groups (with good density fit) of some ADPs also contact Q41 or R16 and R17 from other Hfq hexamers (Supplemental Fig. S1C,D). This ADP-binding mode is different from the previously postulated models in which ATP binds at the R site in a different orientation to position phosphate groups inside a nearby cleft (Arluison et al. 2007; Lazar et al. 2010). Contact of these proximal side residues with ADP phosphate groups observed in these two differently packed crystals (space groups I121 for Hfq65–ADP and P1 for Hfq65–AU6A–ADP) indicates the significance of this interaction. No direct interaction is observed between ADP and AU6A. The binding of AU6A on the proximal side of Hfq prominently differs from AU5G binding on Sa Hfq (Fig. 2G). Unlike the interaction between AU5G and Sa Hfq, the binding of AU6A and ADP onto Ec Hfq did not significantly change the protein structure. Approximately 0.55 Å root-mean-square deviation (RMSD) of backbone atoms between apo and the complex form is observed (Fig. 2A).
AU6A binding is unique
Although DsrADII (23∼60 nt) (Fig. 1A) is believed to bind to the proximal side on Hfq (Brescia et al. 2003; Updegrove et al. 2011), our crystal structure suggests that AU6A (28∼35 nt on DsrA), which is a part of DsrADII, inserts its 5′-end A28 into the R site on the distal face of another Hfq hexamer (Fig. 3A). A28 binds to the R site in a manner similar to poly(A) (Link et al. 2009). Its adenine ring stacks against the side chains of Y25, L26′, I30′, and L32′. Oɛ of Q52′ and Oγ of T61 form a hydrogen bond to the adenine N6 and N1 atoms, respectively. S60 and N28′ side chains make polar contact with the adenine N3 atom. Instead of directly forming a hydrogen bond between ribosyl 2′ hydroxyl and G29 backbone carbonyl oxygen like poly(A) at the R site or ADP, A28 used a water molecule to mediate this interaction. The 5′ ribosyl hydroxyl group makes polar contact with Q41 Nɛ from the proximal side and is positioned at a place similar to that of poly(A) adenine at the R site. This orientation of the 5′ end of AU6A favors the A-rich extension to the 5′ end of AU6A in Uex binding in a way similar to poly(A), occupying both the R and A (adenine-selective binding site) (Link et al. 2009) sites. The ribose and base of A28 take C2′-endo conformations and anti-conformations, respectively, similar to ADP and rA15 bound at the same site.
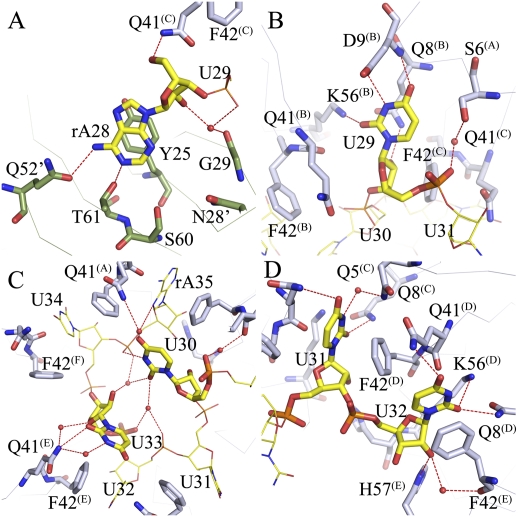
Figure 3.
Binding of AU6A to Hfq is unique. (A) The 5′-end adenosine of AU6A, A28, binds to the R site on the distal side of an adjacent Hfq hexamer, stacking against the side chains of Y25 and forming polar contacts with Q52, T61, S60, N28, and G29. Adenines on poly(A) bind to R sites in a similar way. (B) U29 binds to a novel uridine recognition site on the proximal side. Instead of stacking against the side chains of F42 and Q41, U29 uracil O3, O4, and N2 forms polar contacts to the D9, Q8, and K56 side chains. The uracil lies in a pocket in the vicinity of the N termini of the N-terminal α helix. (C) U30 and U33 sit above the proximal side of the central pore, making little contact with Hfq. These two bases are coordinated by a network of water-mediated hydrogen bonds. The interactions between these two nucleic acid residues may be the cause of usual conformation. (D) Canonical binding pattern at the proximal site. U31, U32, U34, and A35 bind in a way similar to that observed in the Sa AU5G Hfq complex.
Besides the novel A28 binding at the distal side of a neighboring Hfq hexamer, unique binding of nucleotides U29 to A35 to the proximal side is also observed. U29 is inserted into a novel uracil recognition site away from the central pore (Fig. 3B). Two bases, U30 and U33, float above the central pore, with few or no direct interactions with Hfq (Fig. 3C). The rest of the four nucleotides (U31, U32, U34, and A35) bind in a way similar to that previously reported (Fig. 3D; Schumacher et al. 2002). Unlike AU5G complexed with Sa Hfq, in which all nucleotides adopt a C2′-endo conformation for all riboses, four nucleotides—U31, U32, U34, and A35 in AU6A—take the C3′-endo pucker conformation, while U29, U30, and U33 take C2′-endo pucker conformation. All bases adopt anti-conformation, while the dihedral χ of U33 is unusually large (∼281°).
U29 binds to a formerly undiscovered uracil recognition site
Uracil of U29 binds in close proximity to the N-terminal of the α helix, leaving ribose near the canonical uracil stacking pocket formed between F42 and Q41′. N3 and O4 of uracil form hydrogen bonds to Oγ and backbone amide hydrogen of a highly conserved residue, D9, in the Hfq α helix. Uracil O3 makes polar contact with Nζ of K56. The 2′ ribosyl hydroxyl group forms a hydrogen bond to Nɛ of Q8 (Fig. 3B). In comparison, in a previously reported uridine-binding mode, Nɛ of Q8 and Nζ of K56 form a hydrogen bond to uracil O3, and D9 is not involved in binding.
U30 and U33 sit on opposite sides above the central pore
The only direct interaction between U33 and Hfq is that of uracil O2 to Nɛ of Q41. No direct interaction between U30 and Hfq was observed. N3 of U30 forms a direct hydrogen bond to the 5′ phosphate group of A35. Despite the scarcity of direct interaction with Hfq, U30 and U33 are coordinated by a network of water-mediated hydrogen bonds; namely, O2 of U30 to O2 and the 2′ hydroxyl group of U33, O4 of U30 to N7 of A8, and U33 uracil N3 and 2′ ribosyl hydroxyl to Nɛ of Q41. The conformation of U33 is especially interesting. It is well positioned by preceding and following two canonically bound bases to take up the canonical uracil-binding cleft. However, instead of filling in the ready pocket, U33 twists its ribose and uracil away, taking a relatively unstable conformation with dihedral χ ≈ 281°. The binding pocket for U33 is left empty. We assume that the interactions between U30 and U33 may be responsible for this unusual conformation (Fig. 3C).
U31, U32, U34, and A35 bind in a canonical way as AU5G complexed with Sa Hfq
Compared with the complex structure of AU5G and Sa Hfq, Ec Hfq Q41 and F42 side chains take the places of Sa Hfq K41 and Y42 in hydrophobic stacking and polar interactions with uracil or adenine bases. Q8 involved in the interaction with uracil O2 remains the same in both structures. Another residue involved in the uracil O2 interaction, K57 in Sa Hfq, is replaced by K56 in Ec Hfq. H57 of Ec Hfq works in contacting 2′ ribosyl hydroxyl in a way similar to H58 in Sa Hfq (Fig. 3D).
Both the distal and proximal sides as well as the outer rim of the Hfq hexamer are involved in AU6A and Uex binding in solution
To investigate the regions on Hfq that are involved in AU6A and Uex interaction in solution, NMR experiments were performed. NMR triple-resonance experiments were employed to achieve nearly complete assignment of backbone H, N, CO, Cα, and Cβ chemical shifts of Hfq65 (Supplemental Fig. S2A). Our assignment is highly consistent with the Hfq65 backbone assignment reported very recently by Beich-Frandsen et al. (2011).
Unfortunately, Hfq aggregates upon binding with RNA oligonucleotides, ADP, or ATP, preventing further solution NMR characterization of RNA–Hfq interaction. Inspired by the contact of R16/R17 with ADP phosphate groups revealed in our crystal structures (Fig. 2E; Supplemental Fig. S1C), we assumed that these positively charged residues may be responsible for Hfq aggregation upon nucleic acid binding. The R16A/R17A double mutant of Hfq65 was therefore generated. This mutant prevented the aggregation of Hfq upon binding to all of the oligonucleotides we investigated and enabled NMR characterization of these interactions. The majority of the backbone H–N resonances of the Hfq65 R16A/R17A double mutant remain unchanged compared with wild-type Hfq65, indicating that wild-type and mutant Hfq share a similar conformation (Supplemental Fig. S2B).
1H–15N HSQC spectra of Hfq65 R16A/R17A were recorded for each AU6A and Uex titration point. Selected regions of the titration spectrum are shown in Supplemental Figure S2C. A chemical shift differences histogram between the 0.1 mM apo Hfq hexamer and the last titration point with ∼0.43 mM AU6A or Uex (Hfq:RNA molar ratio of ∼1:4.3) is plotted (Fig. 4A) and mapped onto a surface presentation of the Hfq hexamer using a color gradient from blue to red (Fig. 4B).
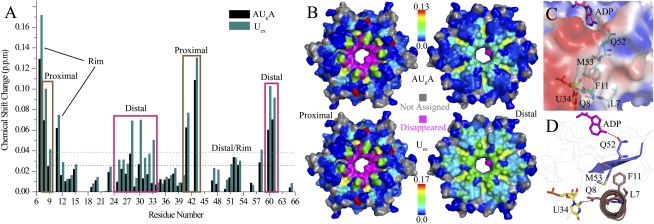
Figure 4.
AU6A and Uex bind to both the Hfq distal and proximal sides in solution NMR. (A) The chemical shift change between the apo Hfq and the last titration point are presented in the column bars. Black and blue bars correspond to AU6A and Uex titrations, respectively. Chemical shift perturbations on both the proximal and distal sides were observed. Columns enclosed in purple boxes correspond to residues on the distal side, while brown boxes enclose the proximal side residues. F42 and H57 disappeared upon titration and are not presented in this graph. Two residues, L7 and F11, on the outer rim (side) of the hexameric ring, exhibited a large chemical shift change. (B) The chemical shift perturbation by AU6A (top) and Uex (bottom) titration is mapped in a blue-to-red color gradient on the proximal (left) and distal (right) sides of the Hfq protein surface. Residues that disappeared during titrations are colored magenta. Those that cannot be confidently assigned are colored gray. (C) Hfq is represented in an electrostatic surface generated using APBS tools (Baker et al. 2001) in PyMol. The side chains of L7, F11, and M53 cluster around a relatively hydrophobic cleft on the outer rim of Hfq. This cleft could be an undiscovered nucleotide-binding site. (D) A possible cross-talk pathway between the U-rich RNA bond at the proximal side and the A-rich sequence bond at the distal side. Q8 adjacent to L7 forms a direct hydrogen bond to uridine at the proximal side. The side chains of F11 and M53 interact via hydrophobic stacking. Q52 just adjacent to M53 makes direct polar contact with some adenines bound at the R site on the distal side.
Chemical shifts of residues around the proximal binding site are affected by AU6A and Uex binding in a similar pattern. The disappearance of resonance peaks of F42 and H57, which are involved in hydrophobic packing with uracil and forming hydrogen bonds with the ribose 2′-hydroxyl group, is most likely due to a slow or intermediate exchange on the NMR time scale. Residues Q8, Q41, and V43, which form direct or indirect polar contacts with uracils, experienced large chemical shift changes. The chemical shift of D9, which interacts specifically with U29, underwent a notable chemical shift change despite the low population (one of six) of this interaction on the Hfq hexamer, in good agreement with the direct hydrogen bond formed between backbone amide and O4 of U29 uracil (Fig. 3B). The chemical shift changes caused by Uex binding on the corresponding sites are slightly larger than those affected by AU6A, with an average factor of 1.4 ± 0.2, indicating a common cause of population difference of bound Hfq due to higher Uex–Hfq binding affinity.
The chemical shifts of residues on the distal side are influenced by AU6A and Uex in different ways. Residues S60, T61, and N28, whose side chains are involved in forming hydrogen bonds with adenine rings at the R site (Fig. 3A), exhibited larger chemical shift changes induced by Uex than AU6A by a factor of 1.6 ± 0.3, indicating a slight increase in the population of the occupied R site. However, much larger chemical shift changes induced by Uex than AU6A, with a factor of 2.4 ± 0.3, were observed for residues I30, K31, and L32, whose backbone amides are near the A site of Hfq. This increase is an indication of the occupied A sites in the Hfq–Uex complex. In accordance, the chemical shift of Q33, whose backbone amide forms a hydrogen bond to N7 of the adenine bound at the A site (Fig. 2B in Link et al. 2009), exhibited prominent change upon Uex but not AU6A binding.
Most interestingly, besides the distal and proximal sites that are known to be involved in RNA interaction, we observed that a cluster of residues around the outer rim of the Hfq hexamer is affected by AU6A and Uex binding. Actually, residue L7, which lies on the path to the rim of the Hfq hexamer, experienced the largest chemical shift perturbation of all residues during titration. Moreover, large chemical shift perturbations on L7 and F11 are observed not only in AU6A and Uex titrations, but also in ADP and A7 titrations (Supplemental Fig. S3). Perturbations on Q52 and M53 are also notable in oligonucleotide titrations (data not shown). In crystal structures, side chains of L7, F11, and M53 cluster around a hydrophobic site on the outer rim (Fig. 4C). Q52 adjacent to M53 makes polar contact with N6 of ADP at the R site, while Q8, adjacent to L7, forms a hydrogen bond with uracil O2 or ribose hydroxyl groups of uridine. The side chain of F11 in the N-terminal helix of each Hfq subunit makes hydrophobic contact with the side chain of M53 on strand β4 of the neighboring subunit (Fig. 4D). The above structural information suggests that these perturbed residues may constitute a path for the cross-talk of the distal and proximal binding sites. Another possibility is that an as-yet-unidentified nucleic acid-binding pocket is formed near these residues. Alteration in Hfq N termini flexibility could also contribute to the observed chemical shift change. Clearly, further investigations are necessary for understanding the roles of these residues in Hfq–RNA interactions.
Uex and full-length DsrA simultaneously bind to different Hfq hexamers in solution
The NMR chemical shift perturbation experiments show that both the proximal and distal sides of the Hfq hexamer are involved in AU6A and Uex binding. That Uex is not long enough to wind around the outer rim of Hfq to reach both the distal and proximal sides of one hexamer (Supplemental Fig. S4) indicates that the A-rich and U-rich segments of Uex are associated with different Hfq hexamers. To validate whether Uex and the full-length DsrA sRNA simultaneously binds to two different Hfq hexamers, we used PRE experiments in solution NMR and FRET experiments to detect the interaction of different Hfq hexamers upon RNA binding.
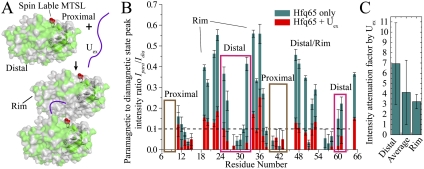
In the PRE experiments, the proximal side residue S6 of Hfq65 R16A/R17A was mutated to cysteine. Nitroxide paramagnetic probe MTSL ([1-oxy-2,2,5,5-tetramethyl-D-pyrroline-3-methyl]-methanethiosulfonate) was site-specifically labeled to this residue on a U-[15N]-labeled sample (the MTSL-labeled Hfq65 S6C R16A/R17A protein is denoted as Hfq65M) (Fig. 5A). A histogram of the ratio of the paramagnetic to the diamagnetic state peak intensities of Hfq65M with and without Uex is shown (Fig. 5B). The intensities of most proximal side resonances of Hfq65M are attenuated to <10% of the corresponding resonances in the reduced (diamagnetic) sample. Resonances on the distal side and rim of the hexamer are much less affected. Upon addition of Uex RNA to a half concentration of the Hfq65M hexamer, in addition to the proximal side resonances, all resonance intensities on the distal side are weakened to <10% of intensities in the reduced diamagnetic sample. Several residues on the rim of the hexamer remained >10% of intensity. The distance between paramagnetic center and spin with 90% intensity loss is roughly estimated to be ∼15.3 Å (Supplemental Material). To evaluate the effect of Uex binding to signal intensities on different regions of Hfq, the intensity attenuation factor by Uex (calculated as the ratio of the intensity ratios shown in Fig. 5B between Hfq-only and Hfq with Uex samples) is averaged for distal side residues, residues on the rim, and all residues (Fig. 5C). The standard deviation of signal attenuation for all residues is large, indicating that the interhexameric interaction has an unequal influence on different sites. Residues on the rim experienced relatively small and uniform attenuation, indicating that in the interhexamer interaction, they are uniformly relatively away from the proximal side spin label on another hexamer. The average attenuation of distal side residues is large with a wide distribution, indicating that some residues in the distal side come into closer proximity to the spin label in the proximal side of the interacting hexamer than other residues. These results are consistent with our observation in the Hfq65–AU6A–ADP crystal structure and indicated that upon RNA addition, the cooperation between hexamers prefers a distal side to a proximal side interaction orientation. Orientation preference suggests that the observed hexamer interaction is not likely caused by random nonspecific aggregation.
Figure 5.
The proximal and distal sides from different Hfq65 hexamers cooperate in Uex binding. (A) Spin label MTSL was site-specifically conjugated to the proximal side residue Cys6 (red spheres) of Hfq65 S6C R16A/R17A. Hfq is presented in the molecular surface. Residues retaining >10% of intensity in the paramagnetic state are labeled green. Residues not assigned or with <10% intensity in the paramagnetic state are labeled gray. MTSL purged most resonance signals on the proximal side. When Uex was added to half of the molar concentration (50 μM) of the Hfq hexamer (0.1 mM), resonances on the distal side of Hfq were also significantly attenuated. Resonance peaks of a few residues on the rim of Hfq retained >10% intensity. This demonstrates that upon Uex binding, the distal side and proximal side from different hexamers approximate to bind the Uex molecule. (B) Histogram of the resonance intensity ratio of the paramagnetic state to the diamagnetic state of apo Hfq (blue) and Hfq bound to Uex (red) samples. The bound state ratios are plotted on top of (overlapping) the apo state columns. Errors are estimated by the signal-to-noise ratio of the NMR spectra. (C) Intensity attenuation factors induced by Uex binding (calculated as the ratio of the intensity ratios shown in B between Hfq-only and Hfq + Uex samples) averaged for the distal side, outer rim, and all residues. Resonances with <10% residue signal intensity in the paramagnetic state in the Hfq-only sample were not used in this calculation to avoid large error.
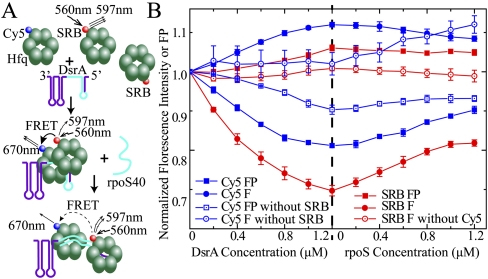
PRE experiments were performed with Hfq65 and Uex to avoid the excessive molecular weight of the full-length Hfq–DsrA complex, which would limit NMR spectrum quality. However, directly biologically significant interaction between full-length Hfq (HfqFL) and full-length DsrA RNA may differ from the above truncated constructs. To address this question, we performed FRET experiments with HfqFL and full-length DsrA. To observe the cooperation of two different Hfq hexamers in binding DsrA RNA, Cy5 maleimide was site-specifically labeled to C6 of the S6C mutant HfqFL (Cy5-Hfq). Sulforhodamine B (SRB) fluoride was labeled to the lysine side chain of wild-type HfqFL (SRB-Hfq). Lysine residues existed in different regions of the Hfq surface, so SRB may be conjugated to different sites of Hfq. SRB-Hfq and Cy5-Hfq were mixed, and DsrA RNA was added to the mixture. Donor SRB was excited at 560 nm, and both fluorescence intensity and FP of the donor and acceptor (Cy5) were monitored (Fig. 6A). Upon DsrA titration, the fluorescence intensity of SRB drops while Cy5 intensity increases, accompanied by a decrease of Cy5 FP and an increase of SRB FP (Fig. 6B). These phenomena clearly demonstrate dipolar-mediated energy transfer (FRET) between these two fluorophores (Xu et al. 2005; Gilmore et al. 2011). The intensity of SRB reached the lowest level when DsrA was titrated to ∼67% of the Hfq hexamer molar concentration. Subsequently, an ssRNA segment constituting the pairing region on rpoS for DsrA (rpoS40, nucleotides 440∼479) was titrated into the DsrA–HfqFL complex. The FRET effect induced by DsrA was reversed partially (Fig. 6B). As a control, SRB HfqFL mixed with an identical concentration of nonlabeled HfqFL exhibited very little change of fluorescence during the same titration process. Cy5 HfqFL mixed with same concentration of nonlabeled HfqFL showed (excited directly at 645 nm) very little change of fluorescence intensity during DsrA titration. A moderate increase of Cy5 intensity is seen upon rpoS40 titration, possibly due to a change in the fluorophore local environment. However, the decrease of Cy5 intensity in the same rpoS40 titration process in the presence of the donor SRB indicates that the reversal of FRET dominates the change of Cy5 fluorescence intensity in the process. A very slight decrease of Cy5 FP is observed even without the SRB donor, indicating some flexibility alteration around the fluorophore upon RNA binding. The short RNA segment Uex, which was used in the PRE studies, is also capable of inducing FRET between differently labeled HfqFL hexamers (Supplemental Fig. S5). Although not providing information on the orientation preference of the interhexameric interaction, these results demonstrate that upon DsrA binding, two different HfqFL hexamers approach. This is highly consistent with the Hfq65–AU6A–ADP crystal structure and the PRE results. As the Hfq-binding site on DsrA is also involved in base-pairing with rpoS mRNA, the annealing of rpoS with DsrA will cause disruption of DsrA-induced Hfq hexamer cooperation. This may explain the partial reversal of DsrA-induced FRET upon rpoS40 titration. However, the addition of rpoS40 did not completely reverse the FRET, suggesting that the annealing of rpoS to DsrA is not sufficient for complete dissociation of Hfq.
Figure 6.
Different HfqFL hexamers cooperate in binding to DsrA sRNA. (A) Cy5 maleimide was site-specifically labeled to C6 of HfqFL S6C, and SRB was coupled to the lysine side chain. SRB could conjugate to all exposed lysines on Hfq. About 1.2:1 dye:Hfq hexamer labeling efficiency, on average, for both labeled proteins was obtained. The excitation wavelength was 560 nm, and the emission maximum of SRB and Cy5 were 597 nm and 670 nm, respectively. When different Hfq hexamers cooperate to bind one single DsrA, the FRET between SRB and Cy5 was observed due to approximation of different hexamers. rpoS40 constitutes the DsrA pairing region on rpoS mRNA. The rpoS40 and rpoS pairing region on DsrA is colored cyan. Upon annealing of rpoS40 to DsrA, the interaction of Hfq–DsrA is destabilized, resulting in an increase in Hfq hexamer distance. (B) SRB-labeled Hfq (0.6 μM) was mixed with 1.2 μM Cy5-labeled Hfq to give a total Hfq concentration of 1.8 μM. Fluorescence intensity (F, filled circles) and FP (filled rectangles) of both SRB (red) and Cy5 (blue) during titration of DsrA and, subsequently, rpoS40 were recorded. Control was made by mixing 0.6 μM SRB-labeled Hfq (or nonlabeled Hfq) with 1.2 μM nonlabeled HfqFL (or Cy5-labeled HfqFL, excited at 645 nm with the SRB-free sample). (Open circle) Fluorescence intensity; (open rectangles) FP. A decrease of donor fluorescence (SRB) and acceptor fluorescence (Cy5) polarization accompanied by an increase of acceptor fluorescence demonstrates energy transfer between the two fluorophores upon DsrA binding. Annealing of rpoS40 to DsrA partially reversed the FRET effect induced by DsrA.
Discussion
Variable U-rich sequence recognition on the Hfq proximal side
The complex structure of AU5G bound to Sa Hfq provided the first structural insight into the U-rich sequence recognition pattern of the Hfq proximal side (Schumacher et al. 2002). The recently reported U6 bound to St Hfq demonstrated interesting recognition of the 3′-end poly-U sequence by Hfq (Sauer and Weichenrieder 2011). Although the binding pockets for each nucleotide in these two structures are very similar, the phosphoribose backbone conformations of RNA in these two structures are quite different. The Hfq65–AU6A–ADP structure that we report here provided the first insight into the recognition of an internal segment of a specific Hfq substrate, DsrA sRNA. An utterly unique binding pattern was observed. A previously unidentified nucleotide-binding site was observed (Fig. 3B). Only four instead of all six known nucleotide-binding pockets on each Hfq hexamer are occupied (Fig. 2G). The significant variations in the binding patterns of the U-rich sequence to Hfq indicates a flexible recognition mode on the proximal side. We determined the dissociation constants of CU6C (3.2 ± 0.4 nM) and GU6G (7.7 ± 0.7 nM) with Ec Hfq65 using FP experiments (Supplemental Table S2). The affinity of CU6C and GU6G with Hfq is moderately higher than AU6A, indicating a small effect of flanking bases on the affinity of the U-rich segment with Hfq. Actually, Hfq has been reported to associate with the 3′ end as well as internal U-rich stretches with variations in the U-rich sequence length and interleaving or flanking nucleotide bases (Lease and Woodson 2004; Sun and Wartell 2006; Salim and Feig 2010; Hopkins et al. 2011; Sauer and Weichenrieder 2011; Updegrove et al. 2011; Vogel and Luisi 2011). The variation of the U-rich RNA recognition in the Hfq complex structures suggests that the differences in nucleoside-binding sites, the number of occupied binding pockets, and the phosphoribose conformations may be important in the recognition of different U-rich substrates and modulation of binding affinity.
The cooperation of Hfq hexamers in binding DsrA
The multimeric form of the Hfq hexamer (Hfq6) in binding DsrA and other RNAs has long been a puzzling problem. A differnce in binding stoichiometries observed in various methods and experiment conditions has been reported. A 2:1 Hfq6:DsrA binding ratio has been observed at submicromolar Hfq6 concentrations (Lease and Woodson 2004; Mikulecky et al. 2004; Sun and Wartell 2006), while 1:1 stoichiometry has also been reported (Brescia et al. 2003; Hwang et al. 2011; Updegrove et al. 2011). Our knowledge is still very limited due to the lack of atomic resolution structural information. In the crystal structure of the Hfq65–AU6A–ADP complex, we observed binding of AU6A to the distal and proximal sides from two different Hfq hexamers. Both the distal and proximal sides of the Hfq hexamer are involved in AU6A and Uex binding in the NMR titration experiments. The approximation of the distal and proximal sides from different Hfq hexamers upon Uex binding was observed in our PRE experiments. Moreover, the FRET between two differently labeled HfqFL hexamers was detected upon full-length DsrA and Uex binding. An approximately 2:1 apparent binding stoichiometry of Uex to Hfq in the FP experiments was also observed. We also observed that the 5′ A-rich region of DsrA SLI, Aex, binds to Hfq with an affinity similar to short poly(A) sequence A7 (Fig. 1B). Based on these results, we propose that the high-affinity linker Uex between DsrA SLI and SLII recruits two Hfq hexamers to DsrA sRNA, with the 5′ A-rich region bound to the distal side of one Hfq and the 3′ U-rich region bound to the proximal side of another Hfq hexamer. Aex is brought near the Hfq distal side where Uex is binding and may possibly bind to Hfq. This binding mode of DsrA to two Hfq hexamers may facilitate the exposure of DsrA regions for annealing with rpoS, which is consistent with the increased RNase T1 digestion at DsrA G18 and G21 upon Hfq binding (Lease and Woodson 2004). Simultaneous binding of DsrA and rpoS to different sides of Hfq may facilitate annealing of the two RNAs (Soper and Woodson 2008; Vogel and Luisi 2011). In our model of DsrA binding, each of the two Hfq hexamers associated with DsrA retained either the distal side or the proximal side unoccupied, presumably for association with Hfq-binding sequences on rpoS to facilitate the annealing.
Despite the long lifetime of tightly associated Hfq–RNA complexes, Hfq has to be recycled rapidly in the large sRNA regulation network. A stepwise active competing mechanism between RNAs for Hfq has been proposed to reconcile this “strong binding–high turnover” paradox (Fender et al. 2010). The annealing of sRNA and target mRNA on Hfq is an effective method for Hfq dissociation from bound RNA (Hopkins et al. 2011; Hussein and Lim 2011). Since most nucleotides of Uex, which binds to Hfq, are also involved in DsrA annealing with rpoS, base-pairing between DsrA and rpoS competes with Hfq–Uex binding and would cause partial dissociation of DsrA from Hfq. This is consistent with our observation in a FRET experiment where addition of rpoS40 to the Hfq–DsrA complex increased the average distance between the two Hfq hexamers bound to DsrA. As Uex binds to the distal and proximal sides on the two different Hfq hexamers, the dissociation of Hfq from DsrA due to rpoS competition may be stepwise. The “two Hfq hexamers to one DsrA sRNA” binding model seems to provide a structural basis for recent interesting findings. Apparently, more high-resolution structures of the Hfq–DsrA complex and Hfq–rpoS complex as well as biochemical data are required for further understanding and validation of this interaction model.
Chemical shift perturbations on the outer rim of Hfq
In the NMR titration, we found that several residues on the outer rim of the Hfq hexamer experienced a large chemical shift perturbation upon binding of all of the RNA oligonucleotides that we tested as well as ADP (Fig. 4; Supplemental Fig. S3). L7 experienced even larger perturbation than residues on already known binding sites. F11 on the rim, far away from both the distal and proximal binding sites, exhibited prominent chemical shift differences. The NMR chemical shift perturbation in the titration experiments may be induced by a binding interaction or a shift in the kinetic or thermodynamic properties of exchanging conformations (Niu et al. 2007; Kalodimos and Tzeng 2011). At this point, these perturbations may correspond to potential unidentified RNA-binding sites. A change in the dynamics of this region of protein, a way of signal transduction between the distal and proximal binding sides, is also possible. Clearly, further investigations are required to understand the mechanism behind this interesting observation.
Unexpected ADP binding
Hfq has been reported to have weak ATPase activity (Sukhodolets and Garges 2003). However, the currently reported chaperoning activity of Hfq is independent from ATPase activity (Brennan and Link 2007). Studies on possible ADP and ATP recognition by Hfq have lead to a model structure of Hfq–ATP interactions (Arluison et al. 2007; Lazar et al. 2010). In these postulated structure models, Y25 plays an important role, and the phosphate groups of ATP make contact with nearby polar residues such as Q33, K31, and S23. However, in our Hfq–AU6A–ADP and Hfq–ADP crystal structures, ADP bound on the distal side with the adenine ring flipped ∼180° compared with the postulated structure. The Y25 side chain is involved in stacking with the ADP purine ring. The phosphate groups of ADP are exposed above the distal binding sites. In both structures, polar residues R16 and R17 or Q41 of a different Hfq hexamer have been observed to contact the phosphate groups of bound ADP (Fig. 2E,F; Supplemental Fig. S1C, D). These observations suggest R16, R17, and Q41 may play an important role as Y25 for ATPase activity of Hfq.
Materials and methods
Hfq purification and crystallization
Hfq65 (residues 1∼65) wild type and mutants were constructed into pET28a (Novagen) expression vector without a tag. HfqFL wild type and mutants were cloned into a modified pET28a vector with an upstream sequence coding for a His6 tag followed by a TEV protease site. The expression vectors were transformed in BL21 DE3 bacteria for overexpression. Bacteria were grown in LB to an OD600 of 0.6 and induced with 1 mM IPTG for 6 h at 37°C. U-[15N]- and U-[15N, 13C, 2H]-labeled samples were grown in LR medium supplemented with 15NH4Cl, 15NH4Cl, and U-[2H, 13C] glucose in D2O, respectively. For Hfq65, bacteria harvested after induction were lysed in 1 M NaCl, 50 mM sodium phosphate, and 1 mM EDTA (pH 7.5) with 10 μg/mL DNase I and RNase A. After heating the lysate for 10 min at 85°C and centrifugation at 14,000 rpm for 30 min, the supernatant was dialyzed against QFFB (50 mM PB, 400 mM NaCl, 1 mM EDTA at pH 7.8). Dialyzed protein was passed through the QFF column (Pharmacia) and then concentrated and subjected to Superdex S200 16/60 (Pharmacia) size-exclusive gel filtration. The supernatant of the HfqFL bacteria lysate was applied to a Hitrap chelating column (Pharmacia) and eluted as recommended by the manufacturer. After TEV protease digestion overnight at 16°C, the sample was further purified on Superdex S200 16/60 (Pharmacia). Hfq-containing fractions were pooled and dialyzed against NMRB (40 mM PB, 40 mM NaCl, 1 mM EDTA at pH 6.8) or DB (20 mM Tris, 100 mM NaCl at pH 8.0, for ATPase assays). Hfq65 hexamer (0.2 mM) was mixed with 0.1–1.2 mM AU6A together with 3 mM ADP, mixed with an equal volume of crystallization buffer (100 mM NaCl, 100 mM Cacodylate, 20% PEG 8000), and crystallized by hanging drop vapor diffusion. Hfq65 hexamer (0.2 mM) with 5 mM ADP was crystallized by mixing an equal volume of 200 mM NH4Ac, 100 mM NaAc, and 22% PEG4000 (pH 4.2). Hfq65–AU6A–ADP took the P1 space group and diffracted it to 1.7 Å resolution, and Hfq65–ADP took the I121 space group and diffracted it to 2.0 Å. X-ray intensity data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) using beamline BL17U and were merged and scaled with MOSFLM and SCALA in the CCP4 suite (Supplemental Table S1; Leslie 1992; Bailey 1994)
Structure determination and refinement
Ec Hfq (Protein Data Bank ID 1hk9) (Sauter et al. 2003) was used as a search model in molecular replacement, using Phaser (McCoy et al. 2007) for phase determination in solving both the Hfq65–ADP and Hfq65–AU6A–ADP complex structures. TLS refinement was followed by restrained refinement using Refmac5. Composite omit maps were generated by FFT in the CCP4 package (Bailey 1994). The Hfq65–AU6A–ADP model was refined to Rwork and Rfree values of 16.5% and 20.5%, respectively, while the Hfq65–ADP model was refined to Rwork and Rfree values of 19.6% and 23.8%, respectively. Statistics of refinements are listed in Supplemental Table S1. No residue was found in the disallowed region on the Ramachandran plot.
Coordinates
Coordinates and structure factors for the Hfq65–AU6A–ADP and the Hfq65–ADP complexes have been deposited with the Protein Data Bank under the accession codes 3RER and 3RES.
NMR
Backbone resonances of Hfq65 were assigned using 0.3 mM U-[2H, 15N, 13C]-labeled hexamer in NMRB with 10% D2O using TROSY-version triple-resonance experiments HNCA, HN(CO)CA, HNCO, HN(CA)CO, HNCACB, and HN(CO)CACB (Salzmann et al. 1998, 1999) on a Bruker DMX 600 spectrometer equipped with a cryoprobe at 42°C. Backbone amide assignments were transferred to the Hfq65 R16A/R17A mutant for resonance peaks not prominently shifted by the mutation. 1H–15N HSQC spectra were recorded on a Bruker DMX 500 spectrometer equipped with a cryoprobe to follow RNA or ADP titration to U-[15N]-labeled Hfq65 R16A/R17A protein at 42°C in NMRB with 10% D2O.
In vitro transcription of full-length DsrA and rpoS40
For preparation of full-length DsrA, a dsDNA sequence with sense strand sequence 5′-TAATACGACTCACTATAGGAACACATCAGATTTCCTGGTGTAACGAATTTTTTAAGTGCTTCTTGCTTAAGCAAGTTTCATCCCGACCCCCCTAGGGTCGGGATTTAAAT-3′ was cloned in pUC18 plasmid. For transcription of rpoS40, a double-strand template was acquired by PCR using forward primer 5′-CAGGAAGCTTTAATACGACTCATATAGGCATTTTGAAATTCGTTACAAGG-3′ and backward primer 5′-CTAAGAATTCCCCGGGTTTACGGATTTCCCCTTGTAACGAATTTC-3′ and inserted into pUC18.
A plasmid template was digested to completion with SmiI (Fermentas) prior to in vitro transcription with T7 RNA polymerase. Transcription products were purified by denaturing polyacrylamide gel electrophoresis and dialyzed into DEPC-treated water.
PRE
Because Hfq contains no intrinsic cysteine, the nitroxide spin label MTSL was attached via a disulfide bond to Cys6 of a U-[15N]-labeled Hfq65 S6C R16A/R17A mutant protein. MTSL was added from a concentrated stock in DMSO at a molar ratio of 5:1 MTSL:Hfq65 monomer and incubated for 8 h at 37°C. Excess MTSL reagent was removed by extensive dialysis against NMRB. TROSY-15N-HSQC (Pervushin et al. 1997) spectra of 0.1 mM MTSL-labeled Hfq hexamer with and without 50 μM Uex were recorded on a Varian 700M spectrometer equipped with a cryoprobe. Three microliters of 400 mM ascorbic acid was added to each 400-μL sample and incubated for 1 h at room temperature for reduction of spin label before acquisition of diamagnetic spectra. The ratio of the paramagnetic against the diamagnetic resonance peak height was used for distance estimation.
FRET assays
HfqFL was incubated with approximately threefold molar excess SRB 2-fluoride (Fluka) in conjugation buffer (CB; 100 mM NaHCO3, 200 mM NaCl at pH 9.2) overnight at 4°C. HfqFL S6C was treated with Cy5 maleimide (GE Healthcare), as recommended by the manufacturer. Excess dye was removed by gel filtration. The labeling efficiency of purified protein (molar ratio of dye to Hfq monomer) was estimated from absorbance at 280 nm and 576 nm for SRB Hfq (∼21.2%), and 280 nm and 650 nm for Cy5 Hfq (20.6%). SRB Hfq and Cy5 Hfq (or nonlabeled HfqFL for control) were mixed to a final hexamer concentration of 0.6 μM and 1.2 μM (1.8 μM total hexamer concentration) in 250 μL of NB3 (20 mM sodium phosphate, 150 mM NaCl at pH 7.0). Full-length DsrA and rpoS40 were dissolved in DEPC-treated water to 50 μM concentration. One microliter of DsrA was titrated six times into the 250-μL system followed by six 1-μL titrations of rpoS40. The emission spectrum with excitation at 560 nm was scanned from 570 nm to 710 nm on a SHIMADZU RF-5301PC spectrofluorophotometer. Peak heights at 597 nM and 670 nm were read from spectrums for SRB and Cy5 fluorescence intensity. Cy5 was directly excited at 645 nm in control samples free of SRB. FP was measured on SpectraMax M5 (Molecular Devices) with an identical reaction system and titration process. Uex was made to 150 μM stock, and 1-μL titration was performed six times. All titrations were repeated for at least three times.
FP
Lyophilized 5′-FAM-labeled RNA fragments were purchased form Takara Bio, Inc., and dissolved in DEPC-treated water to a final concentration of 100 μM. Stock (100 μM) was diluted to 1 μM in NMRB or Tris buffer (20 mM Tris, 100 mM NaCl at pH 8.0). Concentrated Hfq protein stocks were diluted in corresponding buffer to 20 times the highest concentration used in the binding system. This 20× protein stock is diluted in 1/2 series to the lowest desired concentration. The 1/2 series with a different starting Hfq stock concentration was used when denser data points are required. Before the assay, 1 μM labeled RNA was diluted to 42 nM in corresponding buffer, and190 μL of this diluted RNA solution was mixed with 10 μL of 1/2 series protein stock and incubated for 5 min. Samples were excited at 490 nm, and fluorescence was detected at 526 nm. FPs were collected in a SpectraMax M5 (Molecular Devices) plate reader at 22°C. The data were fitted as described in the Supplemental Material.
Acknowledgments
We thank Professor Ke Ruan, Zhiyong Zhang, Changlin Tian, Dr. Fangmin Wu, Zhijun Liu, Jianping Liu, Bo Wu, Zhenwei Song, Su Qin, Yu Qiu, and Lei Liu for helpful discussion; Dr. Fudong Li, Minhao Wu, Yiwei Liu, and Yongxing He for assistance in reflection data collection; Mr. Chunlei Pu for assistance in purchasing reagents; F. Delaglio and A. Bax for providing the NMRPipe software; T.D. Goddard and D. Kneller for Sparky; A.T. Brünger for CNS; and W.L. DeLano for PyMol. This research was supported by grants 2011CB966302 and 2011CB911104 from the National Basic Research Program of China (973 Program), grant 30830031from the Chinese National Natural Science Foundation, and grant 2006AA02A315 from the National High-Tech R&D program and “Outstanding Technical Talent” project of the Chinese Academy of Sciences.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.16746011.
References
- Arluison V, Mutyam SK, Mura C, Marco S, Sukhodolets MV 2007. Sm-like protein Hfq: location of the ATP-binding site and the effect of ATP on Hfq–RNA complexes. Protein Sci 16: 1830–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S 1994. The CCP4 suite—programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beich-Frandsen M, Vecerek B, Konarev PV, Sjoblom B, Kloiber K, Hammerle H, Rajkowitsch L, Miles AJ, Kontaxis G, Wallace BA et al. 2011. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res 39: 4900–4915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R 2000. The structures of HslU and the ATP-dependent protease HslU–HslV. Nature 403: 800–805 [DOI] [PubMed] [Google Scholar]
- Brennan RG, Link TM 2007. Hfq structure, function and ligand binding. Curr Opin Microbiol 10: 125–133 [DOI] [PubMed] [Google Scholar]
- Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD 2003. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA 9: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG 2010. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev 24: 2621–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore MG, Williams D, Okawa Y, Holguin B, James NG, Ross JA, Aoki KR, Jameson DM, Steward LE 2011. Depolarization after resonance energy transfer (DARET): a sensitive fluorescence-based assay for botulinum neurotoxin protease activity. Anal Biochem 413: 36–42 [DOI] [PubMed] [Google Scholar]
- Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ 2006. Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol 71: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Regnier P 2000. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci 97: 1501–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev 66: 373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins JF, Panja S, Woodson SA 2011. Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res 39: 5193–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein R, Lim HN 2011. Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci 108: 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W, Arluison V, Hohng S 2011. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res 39: 5131–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalodimos CG, Tzeng SR 2011. Protein dynamics and allostery: an NMR view. Curr Opin Struct Biol 21: 62–67 [DOI] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96: 375–387 [DOI] [PubMed] [Google Scholar]
- Lazar P, Kim S, Lee Y, Lee KW 2010. Computational approach to ensure the stability of the favorable ATP binding site in E. coli Hfq. J Mol Graph Model 29: 573–580 [DOI] [PubMed] [Google Scholar]
- Lease RA, Belfort M 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc Natl Acad Sci 97: 9919–9924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Woodson SA 2004. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol 344: 1211–1223 [DOI] [PubMed] [Google Scholar]
- Lease RA, Cusick ME, Belfort M 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci 95: 12456–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography, No. 26
- Link TM, Valentin-Hansen P, Brennan RG 2009. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci 106: 19286–19291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci 95: 12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 11: 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR 2004. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol 54: 905–920 [DOI] [PubMed] [Google Scholar]
- Morita T, Aiba H 2011. RNase E action at a distance: degradation of target mRNAs mediated by an Hfq-binding small RNA in bacteria. Genes Dev 25: 294–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler A, Fischer D, Hengge-Aronis R 1996. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev 10: 1143–1151 [DOI] [PubMed] [Google Scholar]
- Niu XG, Chen Q, Zhang JH, Shen WQ, Shi YY, Wu JH 2007. Interesting structural and dynamical behaviors exhibited by the AF-6 PDZ domain upon Bcr peptide binding. Biochemistry 46: 15042–15053 [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K 1997. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci 94: 12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim NN, Feig AL 2010. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS ONE 5: e13028 doi: 10.1371/journal.pone.0013028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K 1998. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci 95: 13585–13590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K 1999. TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J Am Chem Soc 121: 844–848 [Google Scholar]
- Sauer E, Weichenrieder O 2011. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci 108: 13065–13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter C, Basquin J, Suck D 2003. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res 31: 4091–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq–RNA complex: a bacterial Sm-like protein. EMBO J 21: 3546–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield DG, Lynch M 2008. Evolutionary diversification of the Sm family of RNA-associated proteins. Mol Biol Evol 25: 2255–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper TJ, Woodson SA 2008. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14: 1907–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhodolets MV, Garges S 2003. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry 42: 8022–8034 [DOI] [PubMed] [Google Scholar]
- Sun XG, Wartell RM 2006. Escherichia coli Hfq binds A(18) and DsrA domain II with similar 2: 1 Hfq(6)/RNA stoichiometry using different surface sites. Biochemistry 45: 4875–4887 [DOI] [PubMed] [Google Scholar]
- Tharun S, He WH, Mayes AE, Lennertz P, Beggs JD, Parker R 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518 [DOI] [PubMed] [Google Scholar]
- Updegrove TB, Correia JJ, Chen Y, Terry C, Wartell RM 2011. The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA 17: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu QH, Wang S, Korystov D, Mikhailovsky A, Bazan GC, Moses D, Heeger AJ 2005. The fluorescence resonance energy transfer (FRET) gate: a time-resolved study. Proc Natl Acad Sci 102: 530–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AX, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50: 1111–1124 [DOI] [PubMed] [Google Scholar]