Summary
Electron crystallography is a powerful technique for the study of membrane protein structure and function in the lipid environment. When well-ordered two-dimensional crystals are obtained the structure of both protein and lipid can be determined and lipid-protein interactions analyzed. Protons and ionic charges can be visualized by electron crystallography and the protein of interest can be captured for structural analysis in a variety of physiologically distinct states. This review highlights the strengths of electron crystallography and the momentum that is building up in automation and the development of high throughput tools and methods for structural and functional analysis of membrane proteins by electron crystallography.
Introduction
Electron crystallography is the only electron cryomicroscopy (cryo EM) technique capable of delivering atomic resolution information for membrane proteins. Here protein structure is determined within the context of a lipid bilayer that closely mimics the native environment, and in which lipids can exert their stabilizing effects on membrane protein structure. The pioneering work by Henderson and Unwin in the mid 1970’s revealed the first structure of a membrane protein (Henderson and Unwin, 1975). The three-dimensional (3D) structure of the purple membrane protein, bacteriorhodpsin (bR), produced from these studies showcased for the first time the transmembrane α-helices of a membrane protein. This work was extended over the years in conjunction with innovations in cryogenic specimen preparation and improvements in electron microscope design and technique to yield an atomic structure of bR in which native lipids surrounding and interacting with the protein were also identified (Grigorieff et al., 1996; Henderson et al., 1990; Mitsuoka et al., 1999).
Over the last decade, electron crystallography has been used to provide many important insights into the structure and function of several membrane proteins belonging to many different protein families (Figure 1). For example the first atomic structure of the human water channel aquaporin-1 (AQP1) (Murata et al., 2000). This structure was of particular importance not only because of the significance of water channels to life, but also because it allowed the investigators to propose the so called “proton exclusion mechanism” by which water permeation could occur at incredibly high rates while protons were strictly excluded from the pore (Murata et al., 2000). The structures of the rat AQP4 and sheep AQP0 were also determined by electron crystallography (Gonen et al., 2005; Tani et al., 2009), the latter to 1.9Å resolution setting a new benchmark for resolution achieved by cryo EM. Several other structures of membrane proteins have been determined by electron crystallography (Figure 1) including gap junctions (Unger et al., 1999) and acetyl choline receptor (Miyazawa et al., 2003), whose structures even at modest resolution provided tremendous insight into the diverse functional roles membrane proteins play in signaling, cell-cell communication, sensing and homeostasis.
Figure 1. Gallery of selected membrane protein structures determined by electron crystallography.
(top, left to right) The light-driven proton pump bacteriorhodopsin, represented by the first reported atomic structure (PDB 1BRD (Henderson et al., 1990)); the plant light-harvesting complex from photosystem II (Kuhlbrandt et al., 1994); aquaporin-1 as a representative of the aquaporin family of water channels (PDB 1FQY (Murata et al., 2000)); the membrane associated glutathione transferases, represented by microsomal glutathione S-transferase 1 (PDB 2H8A (Holm et al., 2006)); and the acetylcholine receptor (PDB 2BG9 (Unwin, 2005)). (bottom, left to right) The H+, K+ -ATPase (PDB 2XZB (Abe et al., 2011)); the major facilitator superfamily, represented by the oxalate sugar transporter (Hirai et al., 2002); the sodium coupled proton antiporter, NhaA (PDB 3FI1 (Appel et al., 2009)); the small multidrug transporter, EmrE (PDB 2I68 (Fleishman et al., 2006)); and gap junctions, represented by connexin-26 (PDB 3IZ1 (Oshima et al., 2011)).
Here we describe recent technical and methodological progress aimed at automating electron crystallography and streamlining structure determination by this technique. Structure determination by electron crystallography can be lengthy and challenging. Major bottlenecks include sample preparation, crystal growth, crystal screening, data collection and data processing. In X-ray crystallography, sophisticated robotics exist for crystallization, crystal growth screening, data collection, and in some cases even structure determination is automated. But in electron crystallography all of these steps are done manually and can take years to accomplish even when good crystals are available. To overcome these challenges a number of laboratories around the world have been diligently working towards the common goal of automating all aspects of structure determination by electron crystallography. The mutual emphasis is on strategies for high-throughput sample preparation and screening, high-resolution data collection, and routines for improved data processing and rapid structure determination.
This review takes the reader through an electron crystallographic experiment from start to end, highlighting along the way some of the challenges and recent developments in techniques, methodologies and automation aimed at making electron crystallography a high throughput tool for studying membrane protein structures.
Advances in electron crystallography
The typical electron crystallographic study begins with membrane protein expression, solubilization and purification. In rare cases enough membrane protein is expressed natively and the protein can be solubilized from native sources thus bypassing the protein expression step. Nevertheless, the same stringent biochemistry is applied for electron crystallographic studies as for X-ray crystallography: a vast array of conditions is screened for protein solubilization including many different detergents, buffers and temperatures. Protein purification follows using a variety of chromatographic columns with the aim of obtaining a monodispersed solution of the purified protein in detergent that is then used in crystallization assays.
Robotics for two-dimensional crystallization of membrane proteins
Two-dimensional crystallization is achieved by reconstitution of the purified detergent-solubilized membrane protein into a lipid bilayer. First the detergent-solubilized protein is mixed with detergent-solubilized lipids. The choice of lipid or lipid mixture is important and so is the lipid-to-protein ratio (LPR) which is measured in mg/mg lipid:protein. The LPR in reconstitution experiments for functional analysis is typically in the order of 100–200 while for 2D crystal growth it is often significantly lower at ~0.5 to allow extensive crystal contacts to form. The detergent is then removed from the sample or dropped below its critical micelle concentration (CMC) to allow the lipids to form a membrane and for the membrane protein to insert into the lipid bilayer and form planar arrays or two-dimensional (2D) crystals.
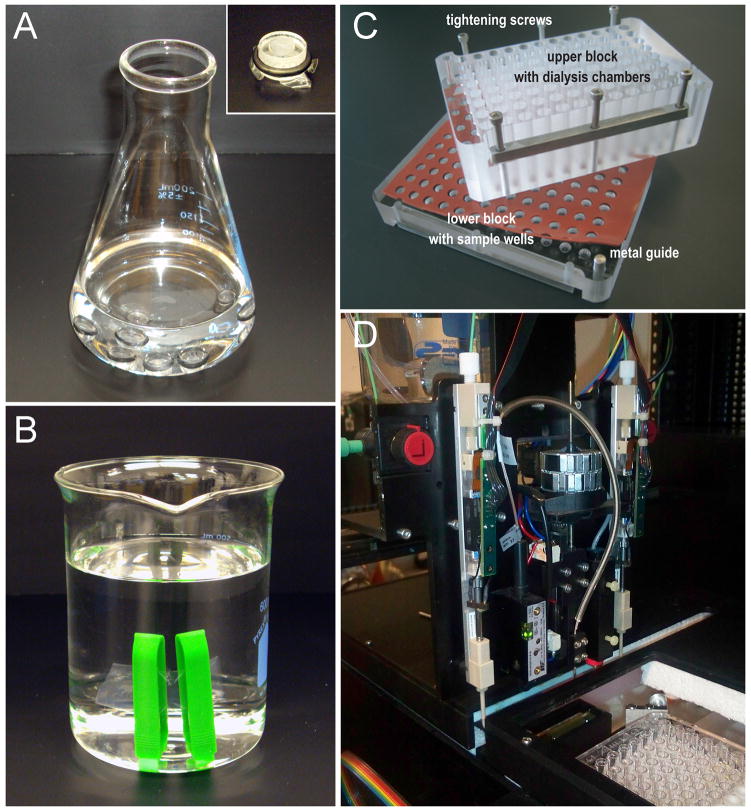
Dialysis is a widely used method for detergent removal and 2D crystallization (Andrews et al., 2008; Reichow and Gonen, 2009). Slow dialysis can be carried out in dialysis buttons or dialysis tubes (Figure 2A and B, respectively). This method is efficient in removing detergents with high CMC such as octyl-glucoside or decyl-maltoside. For low CMC detergents, addition of cyclodextrins has been shown to be effective in removing the detergent by forming detergent-cyclodextrin complexes (Signorell et al., 2007). Hydrophobic adsoption using polystyrene beads (Biobeads SM2) has also been used for 2D crystallization (Rigaud et al., 1997). Finally, the controlled dilution method to reduce the detergent concentration below its CMC can also produce 2D crystals (Remigy et al., 2003).
Figure 2. Dialysis and high-throughput robotics for 2D crystallization.
(A) Slow dialysis using dialysis buttons. Membrane protein/detergent/lipid mixtures are prepared in the 50μl buttons and sealed with a dialysis membrane. The detergent is gradually removed from the mixture by slow dialysis against buffer lacking the detergent. Inset, closeup view of an assembled dialysis button. (B) Dialysis tubing. Membrane protein/detergent/lipid mixture is placed in the dialysis tube between two plastic clamps that are assembled at either end. This set up is submerged in a buffer lacking the detergent and provides a faster dialysis rate than the buttons presented in panel A. (C) 96-well dialysis block for high-throughput crystallization screening. The samples of membrane protein/detergent/lipid mixtures are placed in the lower block. The dialysis membrane is sandwiched in between the lower block and the upper block together with silicone sheets to prevent leakage. The dialysis buffers are added to the upper block. Reprinted from (Vink et al., 2007). Copyright (2007) with permission from Elsevier. (D) The 2DX robot for automated addition of cyclodextrin for detergent removal (Iacovache et al., 2010). A dispenser adds cyclodexdrin to the samples of membrane protein/detergent/lipid mixtures in a 96-well microplate. The robot is equipped a light scattering detector to measure turbidity as an indicator of reconstitution and crystal growth. Picture courtesy of Prof. Andreas Engel (Case Western Reserve University).
A number of laboratories have developed robotics for automated detergent removal and 2D crystal growth. A 96-well block has been designed to handle parallel dialysis against 96 unique buffer conditions (Vink et al., 2007) (Figure 2C). The block is coupled to a liquid-handling system that can vary the reconstitution conditions for each of the 96-wells independently. Dialysis conditions against various pH values, different ionic compositions, and with or without inhibitors or substrates are automated. This system carries out buffer addition, exchange and removal, and also retrieves the samples at the end of the crystallization for evaluation by electron microscopy. This system still requires some user intervention during the setup stage but it has been successfully used for large-scale 2D crystallization trials (Kim et al., 2010).
The “2DX robot” is the first fully automated robot dedicated for 2D crystallization (Iacovache et al., 2010). It is based on the controlled addition of methyl-β-cyclodextrins for detergent removal (Figure 2D). Here, the detergent concentration within the protein solution is carefully measured in order to determine the concentration of cyclodextrin that is needed to remove the detergent and facilitate crystal growth. The user interface allows crystallization parameters such as the reaction volume, the amount of cyclodextrin added and temperature to be set and tightly controlled. The dispenser adds the cyclodextrin solution at a specified rate to the samples into a standard 96-well microplate. A light scattering device monitors the turbidity of the solution as an indicator of reconstitution and 2D crystal growth.
Automated 2D crystal evaluation
Once the reconstitution experiment is complete, the results are evaluated by electron microscopy (EM) of negatively stained samples. Traditionally the samples are prepared manually as described elsewhere (Ohi et al., 2004) but when hundreds of conditions are to be evaluated this process can be lengthy and inefficient. Significant progress has been made so far in automating 2D crystal evaluation starting with automated staining, automated grid loading onto the electron microscope and automated imaging of the crystals (Figure 3).
Figure 3. Automated grid preparation, screening and crystal evaluation.
(A) Automatic grid staining robot (Coudray et al., 2011). The multi-channel pipetting system in the robot allows sample deposit, blotting, washing and staining of EM grids in a 96-well microplate format (inset). Picture courtesy of Prof. Andreas Engel (Case Western Reserve University). Inset reprinted from (Coudray et al., 2011). Copyright (2007) with permission from Elsevier. (B) Robotic grid loading system. An EM grid is first picked up from a 96-well format grid tray by a vacuum system and transferred to a grid holder. Subsequently, the robot automatically loads the grid holder into the electron microscope. The operation of the robotic arm is controlled by software. Reprinted from (Cheng et al., 2007). Copyright (2007) with permission from Elsevier. (C) 2D crystallization data management and evaluation. The Laboratory Information Management System (LIMS) software archives EM images with the corresponding crystallization conditions and allows image viewing through the graphical user interface. Reprinted from (Hu et al., 2010). Copyright (2007) with permission from Elsevier. (Inset) Crystallization results are evaluated and sorted into different categories such as crystal lattice, planar sheets and tubular vesicles, proteoliposomes, protein aggregates, lipidic structures, and macroscopic precipitation. Reprinted from (Kim et al., 2010). Copyright (2010) with kind permission from Springer Science+Business Media B. V.
For automated high-throughput negative staining of samples, a liquid-handling robot in conjunction with a grid-staining block can be used (Vink et al., 2007). The block consists of 96 cubical aluminum platforms that are spaced as in a standard 96-well microplate. Each platform holds a single carbon coated grid and the entire block is then placed on the liquid-handling robot. The system then carries out the staining protocol simultaneously on all grids. More recently a high throughput strategy was developed to couple grid preparation with a liquid handling robot for negative staining of samples (Coudray et al., 2011) (Figure 3A). In this design, 96 grids are placed in a special magnetic holder that allows for plastic film deposit, carbon evaporation, and glow discharging of all the grids simultaneously. The liquid-handling robot then deposits the samples, washes, and stains all 96 grids.
Once the samples are deposited on an EM grid they are inserted one at a time into the transmission electron microscope (TEM) for evaluation. At least two robotic systems have been developed to insert grids into the electron microscope (Hu et al., 2010; Potter et al., 2004). In either case, a robotic arm is programmed to select a grid from the 96-well grid tray, place it into the specimen holder and load it into the microscope (Figure 3B). Alternatively, a carousel that holds all 96 grids simultaneously can be mounted on an electron microscope (Coudray et al., 2011). Sophisticated software controls grid exchange and manages multi-scale image acquisition from each grid for crystal evaluation (Suloway et al., 2005).
Software was also developed for automated high-throughput imaging of 2D crystals for initial evaluation of the crystallization assays. The Leginon software conducts a systematic survey through the entire grid, identifies areas of suitable stain, and records overview images at a number of different magnifications (Cheng et al., 2007). The images are stored in a database and the program provides the user with a web-based graphical interface for viewing the data. Users can then browse through the acquired libraries of images and identify targets for further in-depth analysis.
Similar software was more recently reported in which high-resolution images of 2D crystals can also be recorded (Coudray et al., 2011). Here, low magnification images are recorded initially for grid quality assessment. Once suitably stained areas are identified a second set of images, this time at medium magnification, is recorded and used for sample image characterization. The new algorithms can select regions of interest by identifying crystal shape (Karathanou et al., 2010). The program then switches to high magnification mode for high-resolution image acquisition. Fourier transforms are subsequently calculated to assess crystal quality. A Laboratory Information Management System (LIMS) known as Sesame (Zolnai et al., 2003) was adopted to store the recorded images in folders corresponding to the crystallization conditions. The user may view these images through a graphical user interface (Hu et al., 2010) (Figure 3C). Imaging results from crystallization trials can be evaluated using a crystallization scoring function to assign a quality grade to every condition (Kim et al., 2010) (Figure 3C, inset).
Data collection in electron crystallography
Once 2D crystals of suitable quality are identified they are prepared for analysis and data collection under cryogenic conditions as described elsewhere (Andrews et al., 2008). Unfortunately, as with X-ray crystallography, none of the steps required for the successful freezing of crystals are automated. No single success formula exists and the operator simply needs to assay freezing conditions manually by testing different embedding conditions, different grids, sample back injection, carbon sandwich, slow or fast freezing, etc.
Data collection in electron crystallography is largely manual and can involve imaging of the crystals and/or the collection of electron diffraction data. Images of 2D crystals yield both amplitude and phase information. Recording a complete 3D data set from images to high resolution is challenging even when a microscope equipped with a field emission gun (FEG) electron source is used and the images recorded with low electron dose. This is because image quality and resolution are affected by temperature fluctuation and acoustic and mechanical vibration. A top-entry electron microscope can minimize these problems because the grid is dropped into the microscope column where it is isolated from the outside environment (Fujiyoshi, 1998). But even under such stringent conditions loss of resolution still occurs especially at higher tilt angles because of beam-induced specimen movement and various charging effects leading to image shift and loss of resolution perpendicular to the tilt axis (Glaeser et al., 2011; Gyobu et al., 2004). Moreover, extracting accurate phases to high resolution from images of highly tilted crystals is still a challenge because it is difficult to accurately determine the defocus for such images. The carbon sandwich grid preparation (Gyobu et al., 2004), spot scanning (Downing, 1991), and cooling of the samples to helium temperatures (Fujiyoshi, 1998) have been shown to minimize the beam-induced specimen movement and charging effects in a number of examples.
Electron diffraction data can be collected when large and well-ordered 2D crystals are available. Such crystals can yield amplitude information to atomic resolution. The diffraction data is not affected by stage and temperature instabilities or by the charging affects. As a result, data can be collected from highly tilted 2D crystals resulting in a data set where the missing cone is significantly minimized and the data is more complete (Fujiyoshi, 1998; Glaeser and Downing, 1993). Furthermore, electron diffraction data yields information on a continuous lattice line along the z* axis rather than the discrete sampling at a rate of 1/c as in X-ray crystallography. The continuous sampling in electron diffraction offers more complete structural information and better map quality than those obtained by X-ray crystallography at comparable resolutions. The downside of electron diffraction is that only amplitudes are measured and therefore phases need to be determined by other methods. Never the less, a number of recent studies where electron diffraction data was used without imaging yielded structures at resolution levels that rival X-ray crystallographic studies (Gonen et al., 2005; Gonen et al., 2004; Hiroaki et al., 2006; Hite et al., 2010; Jegerschold et al., 2008; Tani et al., 2009).
Image and/or diffraction data are collected either on photographic film or digitally. Film is gradually being phased out in favor of digital recording media. This is in part because the price of charge couple device (CCD) cameras has dropped significantly over the last decade while the size of the recordable area has increased. Moreover, new digital media based on direct detection (Faruqi and Henderson, 2007) have been developed in recent years where the quality of the image approach that of the photographic film. The monolithic active pixel sensors (MAPS) are based on complementary metal oxide semiconductor (CMOS) technology and have faster readout time than the CCD cameras (Deptuch et al., 2007). Performance wise, the back-thinned MAPS detector is comparable to film for radiation-sensitive samples at 300 keV (McMullan et al., 2009). The hybrid pixel detectors (HPD) such as Medipix2 (McMullan et al., 2007) have a reported zero noise and are suitable for use at lower energies or with very low count rates (Faruqi and Henderson, 2007). Since data can be collected on electronic media the potential for high throughput and automation is within reach.
Data processing and high resolution structure determination
Data processing software for electron crystallography originated from the work on bacteriorhodopsin (Henderson et al., 1990; Henderson et al., 1986) at the Medical Research Council (MRC). The original work evolved into the MRC suite of programs for processing electron microscopy images and diffraction data (Crowther et al., 1996). Software packages such as 2dx and XDP have been recently developed primarily to interface with the MRC suite (although in the case of 2dx new algorithms were also included). A common feature to these newly developed packages is the focus on a graphical user interface for executing the various MRC scripts and commands. Image processing can be done in a semi-automatic way in the 2dx user interface (Gipson et al., 2007b) (Figure 4A). For diffraction data processing, XDP provides a graphical user interface to execute the MRC program codes (Hirai et al., 1999). More recently the IPLT software has been developed to use entirely new codes for processing both images and electron diffraction data (Philippsen et al., 2007; Philippsen et al., 2003) (Figure 4B).
Figure 4. Newly developed electron crystallographic data processing suites.
(A) Graphical user interface of the 2dx program. Fourier transform of 2D crystal images can be indexed by automated lattice determination in 2dx_image and merged in 2dx_merge modules. Image parameters are displayed in the interface and can be easily entered or modified. The reconstructed projection map can be viewed through the interface. Reprinted from (Gipson et al., 2007a). Copyright (2007) with permission from Elsevier. (B) Graphic user interface of the IPLT program. Both image and electron diffraction data can be processed with IPLT. Several functions in IPLT include lattice search in a power spectrum or diffraction pattern, lattice refinement based on 2D Gaussian profile fitting, integration of diffraction peaks, tilt geometry determination, and import/export of the CCP4 mtz file format. Image courtesy of Prof. Andreas Engel (Case Western Reserve University).
As stated earlier, high resolution imaging of 2D crystals delivers both amplitude and phase information. The data can then be processed and a 3D density map calculated into which a model can be built and refined against. The various artifacts that are described above make high resolution imaging quite challenging and a complete data set can take years to be collected and processed. Therefore, when large 2D crystals are obtainable electron diffraction is favorable because these artifacts do not come into play and far better resolution can be obtained rapidly. However, sometimes images of the 2D crystals are necessary for sorting data sets and determining unit cell dimensions. This became very important in the case of AQP4 double-layered 2D crystals that had variable distances between crystal layers (Hiroaki et al., 2006). Moreover, if only small coherent patches of 2D crystals exist, electron diffraction will not be able to deliver any meaningful data, but imaging such patches coupled with crystal unbending procedures (Kunji et al., 2000) can result in meaningful structures as illustrated by the work on the H+, K+-ATPase (Abe et al., 2009) (Figure 5A).
Figure 5. Advances in strategies for structure determination by electron crystallography.
(A) Structure determination of H+, K+-ATPase from very small patches of coherent arrays. (left) Negatively stained mosaic 2D crystal of the H+, K+-ATPase shows small coherent areas of well-ordered lattice (arrows) as well as vesicle aggregation (arrowhead). These 2D crystals are not suitable for electron diffraction but could be used for structural analysis by imaging. (middle) The density map of the H+, K+-ATPase 2D crystal after image processing shows that the 2D crystals consist of two membrane layers (bar lines). One αβ-protomer (dark blue) is shown in the dashed box. Reprinted by permission from Macmillan Publishers Ltd. from (Abe et al., 2009), Copyright (2009). (right) Model of the H+, K+-ATPase αβ-protomer in ribbon representation. The transmembrane domain is indicated by the bar lines. (B) Fragment-based phase extension method for phasing high-resolution electron diffraction. The example of AQP0 at 1.9 Å is presented. (top) Phase data at 6 Å resolution served as the starting point for fragment positioning. The σA-weight 2Fobs-Fcalc density maps (with the corresponding protein models overlaid) were gradually improved at the end of cycles 1 and 2 of the phase extension procedures allowing model building and refinement. (bottom) The starting map at 6 Å resolution did not reveal densities for the lipid or water molecules, but after cycles 1 and 2 as phases were extended to 1.9 Å resolution, the density for lipid and water molecules became apparent and well-defined. In the final map, the densities for the lipid and water molecules became more accurate and appeared similar to the previously published study (PDB 2B6O (Gonen et al., 2005)). Reprinted from (Wisedchaisri and Gonen, 2011). Copyright (2011) with permission from Elsevier.
If large and well-ordered 2D crystals exist then electron diffraction may be used exclusively for structure determination. Phases can be obtained by molecular replacement, or by phase extension as described recently (Wisedchaisri and Gonen, 2011). Molecular replacement (MR) has become indispensable in crystallography and was utilized recently in a number of electron crystallographic studies (Gonen et al., 2005; Gonen et al., 2004; Hiroaki et al., 2006; Hite et al., 2010; Jegerschold et al., 2008; Tani et al., 2009). Here phase information is deduced from atomic models of homologous proteins. For example, the structures of sheep AQP0 (Gonen et al., 2005; Gonen et al., 2004) and rat AQP4 (Hiroaki et al., 2006) were determined by MR using the X-ray structure of the bovine AQP1 (Sui et al., 2001) as a search model. Recent advances in MR procedures focus on automated model preparation and parallel search pipeline (Keegan and Winn, 2008; Long et al., 2008). In addition, the density- and energy-guided protein structure optimization approach has been developed for cases where only distantly related homologous models (20–30% identity) are available for MR searches (DiMaio et al., 2011).
A newly reported method called “fragment-based phase extension” promises rapid structure determination of novel membrane proteins by electron crystallography without relying on the existence of a homology model and without the use of high-resolution imaging (Wisedchaisri and Gonen, 2011) (Figure 5B). In this method, high quality phases are obtained by imaging 2D crystals but only to low resolution. This can be done quite rapidly if only ~6 – 8Å resolution is sought. Next, electron diffraction data is collected to the best possible resolution. Polyalanine α-helical fragments are fit into the low-resolution density map and their coordinates are refined against the high-resolution diffraction amplitudes. The low-resolution phases are then extended to high resolution by phase combination with newly calculated phases originating from the fragments. The new phases are further improved using density modification, which employs solvent flattening and histogram matching procedures. Model building and refinement using various crystallographic programs follow together with several iterations of the procedure until the structural model is complete. When developed and tested against data sets of three different membrane proteins, the resulting structures determined by this method had ~1Å r.m.s. deviation from the previously determined structures (Wisedchaisri and Gonen, 2011). The phase accuracy at the end of the procedure was sufficient to reveal density features for lipids, ligand, and water molecules (Figure 5B). Efforts are currently underway to completely automate this approach.
Unraveling the mechanisms of membrane transport by electron crystallography
Perhaps one of the most important aspects of electron crystallography is that membrane protein structures are determined from protein that is reconstituted in the near native environment of the lipid bilayer. Membrane proteins are dynamic and often undergo conformational changes to carry out their biological functions. A theme that is emerging in electron crystallography is that crystal contacts are mediated by lipids – not by direct lateral protein-protein interactions (Gonen et al., 2005; Hite et al., 2008; Mitsuoka et al., 1999; Tani et al., 2009; Zheng et al., 2010). In a way the lipids act as a buffer surrounding the protein, allowing it to undergo conformational changes within the 2D crystals without destroying the crystal lattice. Thus 2D crystals are robust, and once produced, can be used to determine membrane protein structure under a large variety of physiologically important functional states simply by incubating the crystalline membranes with activators, inhibitors, changing pH or by adding a variety of substrates. Functional as well as structural analysis of the protein of interest can therefore be carried out from the very same preparations (Figures 6 and 7).
Figure 6. Unraveling membrane transport mechanisms by electron crystallography at increasing resolutions.
(A) Substrate specific transport by the trimeric E. coli sugar transporter (GalP) in crystalline vesicles. (left, top) Electron micrograph of GalP crystalline vesicles. (left, bottom) 2D-projection structure of GalP at 18 Å reveals a lattice composed of GalP trimers. (right) The crystalline vesicles were shown to selectively transport glucose over lactose in assays using the fluorescent glucose analogue 2-NBDG (Zheng et al., 2010). Reprinted from (Zheng et al., 2010). Copyright (2010) with permission from Elsevier. (B) Gating mechanism of the acetylcholine (Ach) receptor at millisecond resolution. (left) Tubular crystals of the Ach receptor were embedded on an electron microscopy (EM) grid and sprayed with Ach less then 5 ms before plunge freezing into liquid ethane. (right) The Ach receptor in its closed conformation; colored by secondary structure (Unwin, 2005). Binding of Ach at a conserved site in the extracellular domain (residue in red) induces a rotation of the transmembrane helical domain that opens the channel pore allowing ion conductance. (C) Light driven proton transport in bacteriorhodopsin (bR) determined at atomic resolution. (left) Structural overlay of native bR (white) and a bR mutant that mimics the open state of the proton transport cycle (red) (Subramaniam and Henderson, 2000). (middle) Zoom view of bR with the retinal chromophore (RET) shown in cyan. The acidic residues (E194 and D212 shown as stick representation) and two waters (or hydroxonium ions, shown in purple) involved in the proton transport pathway of bR are highlighted. (right) Positive charges were directly visualized from the |Fo-Fc| difference maps obtained from low and high-resolution diffraction data, shown as red cage (arrow) (Mitsuoka et al., 1999). Reprinted from (Mitsuoka et al., 1999). Copyright (1999) with permission from Elsevier.
Figure 7. Lipid-protein interactions in two-dimensional crystals.
(A) Density map of bacteriorhodopsin (bR) showing the native purple membrane lipid bilayer (Mitsuoka et al., 1999) (B) Density map of aquaporin-0 (AQP0) showing the phosphatidylcholine (PC) lipid bilayer (Gonen et al., 2005). A complete annular lipid shell was modeled from the observed electron density for both bR and AQP0. (C) Hexagonal lattice of bR trimers with protein (grey) and lipids (purple). Specifically bound lipids were observed within the trimeric 3-fold axis and at the monomer-monomer interface. (D) Square lattice of AQP0 tetramers with protein (grey) and lipids (purple). Bulk lipids were observed at the 4-fold tetramer interface. (E) Box plot comparing the protein crystallographic B-factors of bR and AQP0 obtained by cryo EM (in blue) (PDB 2AT9 (Mitsuoka et al., 1999) and PDB 2B6O (Gonen et al., 2005)) and X-ray crystallography (in red) (PDB 1C3W (Luecke et al., 1999) and PDB 1YMG (Harries et al., 2004)). Each box indicates the median B-factor and the first and third quartile B-factor values for all atoms of each protein. The whiskers represent the minimum and maximum values. The B-factors obtained by cryo EM are overall lower compared to structures determined by X-ray crystallography (Hite et al., 2008). (F) Comparison of AQP0 monomer (grey) with annular PC lipids (PC1 – 7) (Gonen et al., 2005) and E. coli lipids (EL1 – 7) (Hite et al., 2010). The AQP0 structure is nearly identical in both structures. The PC and EL lipids bind in similar positions, but adopt unique conformations that adapt to the protein surface.
A classic example is the demonstration of water permeability of the human red-blood cell water channel aquaporin-1 (AQP1) from highly ordered 2D crystals (Walz et al., 1994). The crystalline AQP1 vesicles were shown to be active for water transport by loading the vesicles with carboxyfluorescein and monitoring fluorescence quenching upon change in osmolality by stopped-flow techniques (Walz et al., 1994). The first atomic structure of a water channel was determined from those 2D crystals revealing the aquaporin fold and the mechanism of proton exclusion (Murata et al., 2000).
Many membrane proteins form oligomeric assemblies that are important for the function and/or regulation of the protein. Sometimes these oligomeric structures are destroyed during solubilization with detergent but are reformed upon reconstitution back into a membrane. Structural information provided by 2D-projection analysis from 2D crystals, even at low-resolution, can therefore offer important insights into how a membrane protein is organized within the membrane. Recently a homologue of the human glucose transporter from E. coli (the H+/galactose permease, GalP) was characterized by electron crystallography (Zheng et al., 2010). GalP is a member of the Major Facilitator Superfamily (MFS) of secondary transporters. GalP appeared trimeric in reconstituted vesicles although such an assembly was not described before for other MFS members. As with AQP1 2D crystals, GalP 2D crystals also contained fully functional transporter that was shown to selectively transport the monosaccharide glucose over the disaccharide lactose (Figure 6A). It is still unclear why GalP forms trimers although it is possible that the trimeric organization is important for protein stability and/or in the allosteric regulation of protein function.
Electrospray coupled with fast freezing can be used for rapidly capturing various conformational states of membrane proteins within pre-formed crystalline arrays, and those in turn can be used in electron crystallographic studies for structure determination. This is illustrated beautifully by the work on the acetylcholine (Ach) receptor and bacteriorhodopsin (bR). Electrospray and fast freezing was used to capture the substrate induced conformational changes of the Ach receptor in response to acetylcholine in the millisecond time-scale (Unwin, 1995). The Ach receptor belongs to a family of neurotransmitter gated ion channels, and is a membrane protein that forms an ion channel that rapidly opens in response to acetylcholine (opening rate constant ~20μs). Once the channel opens it deactivates within ~50 – 100ms under the continued presence of Ach. Tubular crystals of the Ach receptor were laid on electron microscopy grids and then sprayed with Ach less then 5 ms prior to plunge freezing in liquid ethane. Electron crystallography was used to determine the structure of the non-activated as well as the Ach-activated channel, and in this way the substrate-binding site, as well as channel conformational changes were characterized at high resolution (Unwin, 1995, 2005) (Figure 6B). A similar approach has been used to study the light-induced conformational changes of bR, by illuminating crystals less then 1ms prior to plunge freezing (Subramaniam et al., 1993; Subramaniam et al., 1999). The activation of many other membrane proteins, receptors and transporters can occur transiently in nature, at rates that may be impossible to capture for structural analysis by other methods. Electron crystallography therefore provides a truly unique approach into the structural analysis of fast channel activation/deactivation and an exceptional opportunity to characterize such mechanisms within the physiologically relevant environment of the membrane.
Some transporters couple the transfer of protons or ions to the movement of substrate across the membrane. Protons are not visible in X-ray crystallographic studies unless resolution better than 1Å is achieved while small ions are generally indistinguishable from water in density maps. However, the difference in scattering factors of electrons caused by charged atoms compared with neutral atoms can be significant, especially at low resolution (< 5 Å) (Hirai et al., 2007). Electron crystallography can therefore be used to characterize the ionic charge state of membrane proteins and visualize the transport pathway of protons and charged ions. The first illustration of this was provided by work on the light driven proton pump bR (Mitsuoka et al., 1999). Light induced isomerization of the retinal chromophore in bR induces a series of conformational changes in the protein that are associated with proton transfer from the cytoplasm to the periplasm. A number of acidic residues in bR have been implicated in the proton transport pathway but by using electron crystallography the atomic charge states in the protein were unambiguously identified (Mitsuoka et al., 1999) (Figure 6C). While the ability to detect charge states on atoms by electron crystallography has not yet been widely appreciated, this technique could be generally applied to many classes of ion channels, and proton coupled transporters to characterize their transport pathways.
Lipid-protein interactions probed by electron crystallography
Membrane proteins exist within a membrane where they are surrounded by lipids. It is not surprising that lipids can, and often do, affect membrane protein structure and function (Lee, 2005; Reichow and Gonen, 2009). Some lipids interact directly with the protein forming an annular shell, while bulk lipids are those that are not bound to the protein. Membrane proteins can interact with annular lipids non-specifically or with very high specificity and high affinity. Examples of high affinity lipid-protein interactions have been reported in X-ray crystallographic studies of membrane proteins that have co-crystallized with a few specifically bound lipids (Hunte and Richers, 2008; Lee, 2003). While these studies have been highly informative, they fall short in describing a complete lipid bilayer. Moreover, most lipid-protein interactions in nature are non-specific and occur transiently as annular and bulk lipids exchange rapidly in the fluid mosaic environment of the biological membrane.
Electron crystallography is the only structural biology technique currently available where the membrane protein of interest is crystallized within a membrane in which lipids form a complete bilayer around the protein establishing an intricate system of lipid-protein interactions that mimic biological membranes. If sufficiently high resolution is achieved (better then ~3Å) the structures of both protein and its surrounding membrane can be determined (Gonen et al., 2005; Hite et al., 2010; Mitsuoka et al., 1999; Tani et al., 2009). Complete lipid bilayers were identified, modeled and structurally refined for a number of electron crystallographic studies. In the case of bR (Mitsuoka et al., 1999) and AQP0 (Gonen et al., 2005) a complete belt of annular lipids was modeled around the protein (Figure 7A– D). Additionally, a few bulk lipids were also seen in the AQP0 study. In the bR structure, all lipids were native and co purified with the protein. Additional lipids were embedded within the bR trimer, at the 3-fold axis, and in between bR monomers (Figure 7C). These native lipids act like glue to hold the bR trimer together, and not surprisingly they are required for protein function and stability. In fact, when the protein was delipidated, the trimer was destroyed and no proton transport detected (Mukhopadhyay et al., 1996; Sternberg et al., 1992). Not surprisingly, a deeper crystallographic analysis showed that bR and AQP0 are more stable when surrounded by lipids rather then by detergent (Hite et al., 2008) (Figure 7E).
The success of electron crystallography in determining structures of water channels is now used to understand the fundamental mechanisms by which membrane proteins interact with lipids. The structure of the lens specific aquaporin-0 (AQP0) has now been determined in the presence of two different lipid compositions: one made entirely of the synthetic lipid dimyristoyl-phosphatidylcholine (PC) (Gonen et al., 2005) and one made out of a mixture of native E. coli lipids (EL) (Hite et al., 2010). Although the structures of AQP0 in the two membranes were the same, the conformations adopted by the different types of lipids varied greatly. Seven distinct annular lipids could be modeled in both structures as well as additional bulk lipids (Figure 7D). Despite the longer average acyl chain length in the EL lipids compared with PC (16 carbons versus 14) both formed bilayers of overall similar thickness around AQP0 (Figure 7F). The longer acyl chains of the EL lipids interdigitate between the spaces of the acyl chains in the apposing leaflet, thereby covering more of the hydrophobic surface of the protein.
Gaining momentum
Electron crystallography is a powerful tool for studying membrane protein structures at atomic resolution in an environment that closely mimics native membranes. Recent studies showcase the ability of electron crystallography in delivering structures at resolutions that rival those achieved by X-ray crystallography. Understanding lipid-protein interactions at the atomic level has been but a dream a few years ago but now is the subject of active study in a number of laboratories. The ability to directly couple structure and function from the very same crystalline membranes is indeed very powerful. Coupled with the ability to visualize the charged state of the protein, electron crystallography is quickly becoming instrumental in deciphering how ion channels, symporters and antiporters function. As more and more tools and methods are developed for high throughput crystal growth, data collection and structure determination it is expected that more structures will be determined by this technique. These are exciting times for being an electron crystallographer!
Acknowledgments
We thank Drs. Anchi Cheng (The Scripps Research Institute, La Jolla, USA), Andreas Engel (Case Western Reserve University, Cleveland, USA), Yoshinori Fujiyoshi (Kyoto University, Kyoto, Japan), David Stokes (New York University, New York, USA), and Iban Ubarretxena-Belandia (Mount Sinai School of Medicine, New York, USA) for providing us with high-resolution images of their work for inclusion in this review. We thank Yoshinori Fujiyoshi, Kazutoshi Tani (Kyoto University, Japan) and Kaoru Mitsuoka (Biomedicinal Information Research Center (BIRC) Tokyo, Japan) for kindly providing the data sets for aquaporin-4 and bacteriorhodopsin; and to Dr. Sriram Subramaniam (National Cancer Institute, Bethesda, USA) for sending us PDB coordinate files for the oxalate transporter. SLR is supported by the National Institutes for Health Ruth L. Kirschstein National Research Service Award (EY19768). The Gonen laboratory is supported by the National Institutes of Health (GM079233 and U54GM094598); the American Diabetes Association (1-09-CD-05) and by the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe K, Tani K, Fujiyoshi Y. Conformational rearrangement of gastric H(+), K(+)-ATPase induced by an acid suppressant. Nat Commun. 2011;2:155. doi: 10.1038/ncomms1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K, Tani K, Nishizawa T, Fujiyoshi Y. Inter-subunit interaction of gastric H+, K+-ATPase prevents reverse reaction of the transport cycle. EMBO J. 2009;28:1637–1643. doi: 10.1038/emboj.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S, Reichow SL, Gonen T. Electron crystallography of aquaporins. IUBMB life. 2008;60:430–436. doi: 10.1002/iub.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel M, Hizlan D, Vinothkumar KR, Ziegler C, Kuhlbrandt W. Conformations of NhaA, the Na/H exchanger from Escherichia coli, in the pH-activated and ion-translocating states. Journal of molecular biology. 2009;386:351–365. doi: 10.1016/j.jmb.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Cheng A, Leung A, Fellmann D, Quispe J, Suloway C, Pulokas J, Abeyrathne PD, Lam JS, Carragher B, Potter CS. Towards automated screening of two-dimensional crystals. Journal of structural biology. 2007;160:324–331. doi: 10.1016/j.jsb.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudray N, Hermann G, Caujolle-Bert D, Karathanou A, Erne-Brand F, Buessler JL, Daum P, Plitzko JM, Chami M, Mueller U, et al. Automated screening of 2D crystallization trials using transmission electron microscopy: a high-throughput tool-chain for sample preparation and microscopic analysis. Journal of structural biology. 2011;173:365–374. doi: 10.1016/j.jsb.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Deptuch G, Besson A, Rehak P, Szelezniak M, Wall J, Winter M, Zhu Y. Direct electron imaging in electron microscopy with monolithic active pixel sensors. Ultramicroscopy. 2007;107:674–684. doi: 10.1016/j.ultramic.2007.01.003. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Terwilliger TC, Read RJ, Wlodawer A, Oberdorfer G, Wagner U, Valkov E, Alon A, Fass D, Axelrod HL, et al. Improved molecular replacement by density- and energy-guided protein structure optimization. Nature. 2011;473:540–543. doi: 10.1038/nature09964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing KH. Spot-scan imaging in transmission electron microscopy. Science. 1991;251:53–59. doi: 10.1126/science.1846047. [DOI] [PubMed] [Google Scholar]
- Faruqi AR, Henderson R. Electronic detectors for electron microscopy. Curr Opin Struct Biol. 2007;17:549–555. doi: 10.1016/j.sbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, Harrington SE, Enosh A, Halperin D, Tate CG, Ben-Tal N. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. Journal of molecular biology. 2006;364:54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- Fujiyoshi Y. The structural study of membrane proteins by electron crystallography. Adv Biophys. 1998;35:25–80. doi: 10.1016/s0065-227x(98)90004-1. [DOI] [PubMed] [Google Scholar]
- Gipson B, Zeng X, Stahlberg H. 2dx_merge: data management and merging for 2D crystal images. J Struct Biol. 2007a;160:375–384. doi: 10.1016/j.jsb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson B, Zeng X, Zhang ZY, Stahlberg H. 2dx--user-friendly image processing for 2D crystals. J Struct Biol. 2007b;157:64–72. doi: 10.1016/j.jsb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Glaeser RM, Downing KH. High-resolution electron crystallography of protein molecules. Ultramicroscopy. 1993;52:478–486. doi: 10.1016/0304-3991(93)90064-5. [DOI] [PubMed] [Google Scholar]
- Glaeser RM, McMullan G, Faruqi AR, Henderson R. Images of paraffin monolayer crystals with perfect contrast: minimization of beam-induced specimen motion. Ultramicroscopy. 2011;111:90–100. doi: 10.1016/j.ultramic.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen T, Sliz P, Kistler J, Cheng Y, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004;429:193–197. doi: 10.1038/nature02503. [DOI] [PubMed] [Google Scholar]
- Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- Gyobu N, Tani K, Hiroaki Y, Kamegawa A, Mitsuoka K, Fujiyoshi Y. Improved specimen preparation for cryo-electron microscopy using a symmetric carbon sandwich technique. J Struct Biol. 2004;146:325–333. doi: 10.1016/j.jsb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Harries WE, Akhavan D, Miercke LJ, Khademi S, Stroud RM. The channel architecture of aquaporin 0 at a 2.2-A resolution. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14045–14050. doi: 10.1073/pnas.0405274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Downing KH, Lepault J, Zemlin F. Structure of Purple Membrane from Halobacterium-Halobium - Recording, Measurement and Evaluation of Electron-Micrographs at 3.5 a Resolution. Ultramicroscopy. 1986;19:147–178. [Google Scholar]
- Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Hirai T, Heymann JA, Shi D, Sarker R, Maloney PC, Subramaniam S. Three-dimensional structure of a bacterial oxalate transporter. Nature structural biology. 2002;9:597–600. doi: 10.1038/nsb821. [DOI] [PubMed] [Google Scholar]
- Hirai T, Mitsuoka K, Kidera A, Fujiyoshi Y. Simulation of charge effects on density maps obtained by high-resolution electron crystallography. J Electron Microsc. 2007;56:131–140. doi: 10.1093/jmicro/dfm019. [DOI] [PubMed] [Google Scholar]
- Hirai T, Murata K, Mitsuoka K, Kimura Y, Fujiyoshi Y. Trehalose embedding technique for high-resolution electron crystallography: application to structural study on bacteriorhoposin. J Electron Micros. 1999;48:653–658. doi: 10.1093/oxfordjournals.jmicro.a023731. [DOI] [PubMed] [Google Scholar]
- Hiroaki Y, Tani K, Kamegawa A, Gyobu N, Nishikawa K, Suzuki H, Walz T, Sasaki S, Mitsuoka K, Kimura K, et al. Implications of the aquaporin-4 structure on array formation and cell adhesion. J Mol Biol. 2006;355:628–639. doi: 10.1016/j.jmb.2005.10.081. [DOI] [PubMed] [Google Scholar]
- Hite RK, Gonen T, Harrison SC, Walz T. Interactions of lipids with aquaporin-0 and other membrane proteins. Pflugers Archiv : European journal of physiology. 2008;456:651–661. doi: 10.1007/s00424-007-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, Li Z, Walz T. Principles of membrane protein interactions with annular lipids deduced from aquaporin-0 2D crystals. EMBO J. 2010;29:1652–1658. doi: 10.1038/emboj.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm PJ, Bhakat P, Jegerschold C, Gyobu N, Mitsuoka K, Fujiyoshi Y, Morgenstern R, Hebert H. Structural basis for detoxification and oxidative stress protection in membranes. J Mol Biol. 2006;360:934–945. doi: 10.1016/j.jmb.2006.05.056. [DOI] [PubMed] [Google Scholar]
- Hu MH, Vink M, Kim C, Derr KD, Koss J, D’Amico K, Cheng AC, Pulokas J, Ubarretxena-Belandia I, Stokes D. Automated electron microscopy for evaluating two-dimensional crystallization of membrane proteins. J Struct Biol. 2010;171:102–110. doi: 10.1016/j.jsb.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Richers S. Lipids and membrane protein structures. Curr Opin Struct Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Iacovache I, Biasini M, Kowal J, Kukulski W, Chami M, van der Goot FG, Engel A, Remigy HW. The 2DX robot: a membrane protein 2D crystallization Swiss Army knife. Journal of structural biology. 2010;169:370–378. doi: 10.1016/j.jsb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Jegerschold C, Pawelzik SC, Purhonen P, Bhakat P, Gheorghe KR, Gyobu N, Mitsuoka K, Morgenstern R, Jakobsson PJ, Hebert H. Structural basis for induced formation of the inflammatory mediator prostaglandin E2. Proc Natl Acad Sci USA. 2008;105:11110–11115. doi: 10.1073/pnas.0802894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanou A, Coudray N, Hermann G, Buessler JL, Urban JP. Automatic TEM image analysis of membranes for 2D crystal detection. Adv Exp Med Biol. 2010;680:327–333. doi: 10.1007/978-1-4419-5913-3_37. [DOI] [PubMed] [Google Scholar]
- Keegan RM, Winn MD. MrBUMP: an automated pipeline for molecular replacement. Acta Crystallogr. 2008;D64:119–124. doi: 10.1107/S0907444907037195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Vink M, Hu M, Love J, Stokes DL, Ubarretxena-Belandia I. An automated pipeline to screen membrane protein 2D crystallization. Journal of structural and functional genomics. 2010;11:155–166. doi: 10.1007/s10969-010-9088-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W, Wang DN, Fujiyoshi Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature. 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- Kunji ER, von Gronau S, Oesterhelt D, Henderson R. The three-dimensional structure of halorhodopsin to 5 A by electron crystallography: A new unbending procedure for two-dimensional crystals by using a global reference structure. Proc Natl Acad Sci USA. 2000;97:4637–4642. doi: 10.1073/pnas.080064697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AG. Lipid-protein interactions in biological membranes: a structural perspective. Biochimica et biophysica acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- Lee AG. Cell biology: a greasy grip. Nature. 2005;438:569–570. doi: 10.1038/438569a. [DOI] [PubMed] [Google Scholar]
- Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr. 2008;D64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 A resolution. Journal of molecular biology. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- McMullan G, Cattermole DM, Chen S, Henderson R, Llopart X, Summerfield C, Tlustos L, Faruqi AR. Electron imaging with Medipix2 hybrid pixel detector. Ultramicroscopy. 2007;107:401–413. doi: 10.1016/j.ultramic.2006.10.005. [DOI] [PubMed] [Google Scholar]
- McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, Fujiyoshi Y. The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution. Journal of molecular biology. 1999;286:861–882. doi: 10.1006/jmbi.1998.2529. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay AK, Dracheva S, Bose S, Hendler RW. Control of the integral membrane proton pump, bacteriorhodopsin, by purple membrane lipids of Halobacterium halobium. Biochemistry. 1996;35:9245–9252. doi: 10.1021/bi960738m. [DOI] [PubMed] [Google Scholar]
- Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biological procedures online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima A, Tani K, Toloue MM, Hiroaki Y, Smock A, Inukai S, Cone A, Nicholson BJ, Sosinsky GE, Fujiyoshi Y. Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels. Journal of molecular biology. 2011;405:724–735. doi: 10.1016/j.jmb.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen A, Schenk AD, Signorell GA, Mariani V, Berneche S, Engel A. Collaborative EM image processing with the IPLT image processing library and toolbox. Journal of structural biology. 2007;157:28–37. doi: 10.1016/j.jsb.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Philippsen A, Schenk AD, Stahlberg H, Engel A. Iplt--image processing library and toolkit for the electron microscopy community. J Struct Biol. 2003;144:4–12. doi: 10.1016/j.jsb.2003.09.032. [DOI] [PubMed] [Google Scholar]
- Potter CS, Pulokas J, Smith P, Suloway C, Carragher B. Robotic grid loading system for a transmission electron microscope. Journal of structural biology. 2004;146:431–440. doi: 10.1016/j.jsb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Reichow SL, Gonen T. Lipid-protein interactions probed by electron crystallography. Current opinion in structural biology. 2009;19:560–565. doi: 10.1016/j.sbi.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigy HW, Caujolle-Bert D, Suda K, Schenk A, Chami M, Engel A. Membrane protein reconstitution and crystallization by controlled dilution. FEBS letters. 2003;555:160–169. doi: 10.1016/s0014-5793(03)01105-0. [DOI] [PubMed] [Google Scholar]
- Rigaud JL, Mosser G, Lacapere JJ, Olofsson A, Levy D, Ranck JL. Bio-Beads: an efficient strategy for two-dimensional crystallization of membrane proteins. Journal of structural biology. 1997;118:226–235. doi: 10.1006/jsbi.1997.3848. [DOI] [PubMed] [Google Scholar]
- Signorell GA, Kaufmann TC, Kukulski W, Engel A, Remigy HW. Controlled 2D crystallization of membrane proteins using methyl-beta-cyclodextrin. Journal of structural biology. 2007;157:321–328. doi: 10.1016/j.jsb.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Sternberg B, L’Hostis C, Whiteway CA, Watts A. The essential role of specific Halobacterium halobium polar lipids in 2D-array formation of bacteriorhodopsin. Biochimica et biophysica acta. 1992;1108:21–30. doi: 10.1016/0005-2736(92)90110-8. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Henderson R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Lindahl M, Bullough P, Faruqi AR, Tittor J, Oesterhelt D, Brown L, Lanyi J, Henderson R. Protein conformational changes in the bacteriorhodopsin photocycle. J Mol Biol. 1999;287:145–161. doi: 10.1006/jmbi.1999.2589. [DOI] [PubMed] [Google Scholar]
- Sui HX, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tani K, Mitsuma T, Hiroaki Y, Kamegawa A, Nishikawa K, Tanimura Y, Fujiyoshi Y. Mechanism of aquaporin-4’s fast and highly selective water conduction and proton exclusion. Journal of molecular biology. 2009;389:694–706. doi: 10.1016/j.jmb.2009.04.049. [DOI] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999;283:1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- Unwin N. Acetylcholine receptor channel imaged in the open state. Nature. 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Vink M, Derr K, Love J, Stokes DL, Ubarretxena-Belandia T. A high-throughput strategy to screen 2D crystallization trials of membrane proteins. J Struct Biol. 2007;160:295–304. doi: 10.1016/j.jsb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz T, Smith BL, Zeidel ML, Engel A, Agre P. Biologically active two-dimensional crystals of aquaporin CHIP. J Biol Chem. 1994;269:1583–1586. [PubMed] [Google Scholar]
- Wisedchaisri G, Gonen T. Fragment-based phase extension for three-dimensional structure determination of membrane proteins by electron crystallography. Structure. 2011;19:976–987. doi: 10.1016/j.str.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Taraska J, Merz AJ, Gonen T. The prototypical H+/galactose symporter GalP assembles into functional trimers. J Mol Biol. 2010;396:593–601. doi: 10.1016/j.jmb.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolnai Z, Lee PT, Li J, Chapman MR, Newman CS, Phillips GN, Jr, Rayment I, Ulrich EL, Volkman BF, Markley JL. Project management system for structural and functional proteomics: Sesame. J Struct Funct Genomics. 2003;4:11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]