Abstract
Insecurity in the supply of fossil fuels, volatile fuel prices, and major concerns regarding climate change have sparked renewed interest in the production of fuels from renewable resources. Because of this, the use of biodiesel has grown dramatically during the last few years and is expected to increase even further in the future. Biodiesel production through the use of microbial systems has marked a turning point in the field of biofuels since it is emerging as an attractive alternative to conventional technology. Recent progress in synthetic biology has accelerated the ability to analyze, construct, and/or redesign microbial metabolic pathways with unprecedented precision, in order to permit biofuel production that is amenable to industrial applications. The review presented here focuses specifically on the role of synthetic biology in the design of microbial cell factories for efficient production of biodiesel.
1. Introduction
Global warming and the continued depletion of nonrenewable fuel resources are two major problems that entangle our planet today and demand immediate solutions [1]. The extensive use of fossil fuels has caused greenhouse gas emissions and damage to the environment, and has also led to the current instability of oil supplies and continuous fluctuations in prices. These factors, which revolve around economic, environmental and geopolitical issues, are central to the continued interest seen in renewable energy sources [2]. An entire branch of biotechnology, referred to as “white biotechnology” [3], centers on the bioproduction of fuels and chemicals from renewable sources. For biofuels, delicate optimization, and fine tuning of these processes to maximize productivity and yield is of particular concern, as the viability of any biofuel process is extremely sensitive to factors related to both raw material supply and production costs [4].
About 90% of the current biofuel market is represented by biodiesel and bioethanol. However, bioethanol is not seen as an ideal biofuel for the future because of its low energy density and incompatibility with the existing fuel infrastructure [5, 6]. On the contrary, biodiesel is already better established [7] and is preferable to petrodiesel in terms of several characteristics, such as environmental friendliness, renewability, reduced emissions, higher combustion efficiency, improved lubricity, and higher levels of safety [8]. Chemically, biodiesel comprises a mixture of fatty acid alkyl esters (FAAEs). The most commonly used method to produce biodiesel is the in vitro transesterification process, where triacylglycerides (TAGs) of vegetable oils are combined with methanol to form fatty acid methyl esters (FAMEs) and the byproduct glycerol (Figure 1). Alkalies (e.g., sodium hydroxide, potassium hydroxide, sodium metoxide, and potassium metoxide) [9–12], acids (e.g., sulfuric acid) [13], or enzymes can be used to catalyze this reaction [14]. However, issues related to high cost and limited availability of vegetable oils have become growing concerns for large-scale commercial viability of biodiesel production [15]. Also, the in vitro transesterification reaction presents some unresolved issues, such as the need to use large amounts of toxic compounds (sodium hydroxide, sulfuric acid, or methanol) and the high cost of isolation and immobilization of enzyme catalysts [16, 17]. Various approaches to addressing these problems have been explored. First, increasing interest in developing microbial processes for the production of biodiesel from a wide range of other raw materials may represent a promising alternative to the vegetable oils. Second, technologies now exist that use living cells to synthesize products that are more easily biodegradable, require less energy, and create less waste during production than those obtained by chemical synthesis. In order for a fermentation process to compete with existing petroleum-based processes, the target molecule must be produced at high levels of yield, titer, and productivity. These goals can be difficult to attain with naturally occurring microbes. While metabolic engineering has enabled extraordinary advances in the redesign of pathways for efficient target molecule production, including biofuels [5, 18–20], tools from synthetic biology make it possible to create new biological functions that do not exist in nature. Essentially, this is achieved either by heterologous expression of natural pathways or design of de novo pathways. This paper reviews approaches to microbial synthesis of biodiesel, focusing on the role of synthetic biology as an enabling technology in the design of optimal cell factories.
Figure 1.
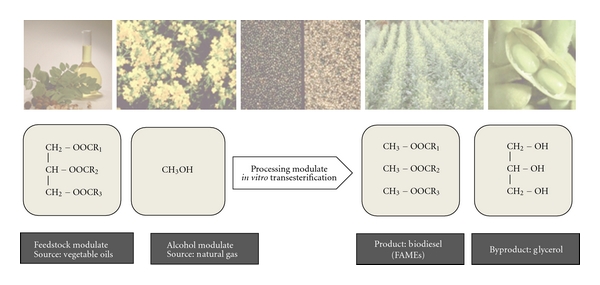
Conventional process for biodiesel production.
2. Biofuel Feedstocks: Potential Contributions of Synthetic Biology
Because of its abundance and renewable nature, biomass has the potential to produce extensive supplies of reliable, affordable, and environmentally sound biofuels to replace fossil fuels. Many biomass feedstocks, which include lignocellulosic agricultural residues as well as edible and nonedible crops, can be used for the production of biofuels. In Figure 2, biofuel feedstocks are listed according to their levels of environmental and economical sustainability.
Figure 2.
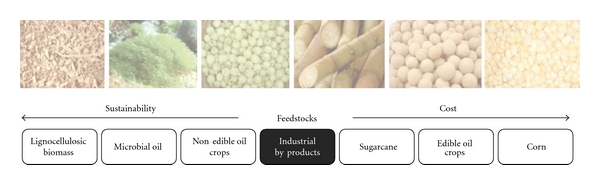
Different feedstocks for biofuel generation arranged according to their levels of environmental and economical sustainability.
More than 95% of global biodiesel production now begins from virgin edible vegetable oils [21], which account for about 80% of the total production cost. However, the socioeconomic impacts of large-scale biodiesel production from edible feedstocks can be significantly lowered by the use of alternative feedstocks such as nonedible oils or lignocellulosic biomass [22]. The use of nonedible vegetable oils is especially significant for biodiesel production in developing countries [9, 23, 24], because of the tremendous demand for edible oils as food. Increasing attention is also now being given to the use of microbial oils as biodiesel feedstock, which are produced by certain oleaginous microorganisms [25]. Lignocellulosic biomass, on the other hand, is the largest known renewable source of carbohydrates. It generally consists of about 25% lignin and 75% carbohydrate polymers (cellulose and hemicellulose) [22]. These polymers, upon complete hydrolysis, yield a mixture of hexose (glucose, galactose, and mannose) and pentose (arabinose and xylose) [26]. Synthetic biodiesel can be produced from this renewable carbon source using the Fischer-Tropsch process [27]. Although conversion of lignocellulose into biofuels appears simple in theory, the techniques used in this field are not fully established. The main reason for this lag is the recalcitrant nature of cellulose [28] and the toxic nature of the products of lignin degradation. Several prokaryotes and eukaryotes that have cellulolytic properties and are tolerant to the toxic products of lignin degradation have been identified [5, 29]. However, the yield and productivity of biofuels synthesized in this way are not sufficient to meet current energy demands. An efficient cellulolytic organism should be able to hydrolyze lignocellulose completely, ferment all sugars of lignocellulosic hydrolysate simultaneously, and tolerate toxic compounds of lignin without compromising productivity [30]. Therefore, for cost-effective production of biofuels, the fuel-producing hosts must be designed.
In terms of ethanol production, starches (maize, wheat, barley, etc.) and sugar-rich biomass (grasses, maize leaves, beets, sugar cane, etc.) have been the feedstocks most commonly used for their bioconversion [31]. However, advances in metabolic engineering and synthetic biology have provided new tools for creating desirable phenotypes for the production of ethanol from lignocellulosic biomass [32–36]. Since ethanol is one of the substrates used for in vivo synthesis of biodiesel, advances in terms of maximization of two-carbon alcohol production from the most economically viable feedstocks will be discussed later in this review.
3. Synthetic Biology and Biodiesel Production
Synthetic biology emerged around the year 2000 as a new biological discipline, and many different definitions have been applied to this field. However, one commonly used way to describe synthetic biology is as the design and construction of new biological functions that are not found in nature [37]. Synthetic biology is a discipline encompassing contributions from many fields [38–40], but this review places particular emphasis on the design of microbes, either by modification of existing pathways or by heterologous expression of natural pathways, in order to allow efficient production of biodiesel. In this connection, the synthesis of biodiesel using microbes is currently a highly promising alternative to conventional technologies. Microbial biodiesel production has been approached from two different angles: (1) by indirect synthesis from microbial oils, which are produced and harvested for use in the conventional in vitro transesterification process, and (2) by direct biodiesel synthesis using redesigned cell factories to increase production of alcohols and/or FFAs, which are subsequently used for in vivo synthesis of biodiesel. In the following sections, both approaches are reviewed.
3.1. Indirect Synthesis of Biodiesel from Microbial Oils
It is well known that many microbes, including certain types of microalgae, bacteria, filamentous fungi, and yeasts, can accumulate intracellular lipids, primarily TAGs, with these representing a large proportion of their biomass [41–53]. Oils derived from these oleaginous microbes represent promising raw materials for biodiesel production through transesterification using the plant-based process [54, 55] (Figure 3).
Figure 3.
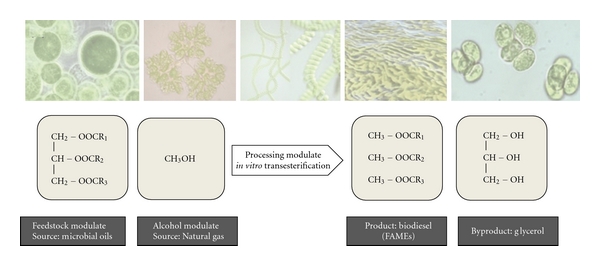
Synthesis of biodiesel from microbial oils.
The use of microbial oils offers several advantages when compared to plant-derived oils [41]. However, oleaginous microbes have varying prospects in the biodiesel industry. For example, microalgae are photoautotrophic microorganisms that can convert CO2 directly to lipids, which can then be used for biofuel production, particularly for biodiesel [42, 56, 57]. The oil content of microalgae usually ranges between 15 and 70% by weight of the dry biomass [41–45]. Scaling-up process for autotrophic microalgae is complex, however, since light is needed during cultivation. Although it is known that algae could be grown in dedicated artificial ponds for generating biodiesel [58], the harvesting of miles and miles of algae growth is required in order to generate substantial amounts of biodiesel. Thus, while the microbiological aspects of this approach are extremely promising, the engineering aspects pose the greatest challenge. Tools from synthetic biology have been effectively used to convert certain autotrophic microalgae into heterotrophic microorganisms [59]. Essentially, this consists of the introduction of nonnatural metabolic pathways into the autotrophic microalgae, thereby, allowing cultivation using an organic carbon source instead of photosynthesis from sunlight.
3.2. Direct Synthesis of Biodiesel Using Cell Factories
Methanol, conventionally used as part of in vitro transesterification, is largely derived from nonrenewable natural gas and is also both toxic and hazardous. On the contrary, ethanol can be naturally produced from renewable resources, while exhibiting low levels of toxicity and a higher degree of biodegradability. Ethanol produced endogenously can therefore be used for in vivo synthesis of fatty acid ethyl esters (FAEEs) with exogenously added FFAs. Similarly, microbial FFAs can be used as feedstock for in vivo production of biodiesel, instead of TAGs from vegetable oils. Therefore, critical aspects of metabolic engineering and synthetic biology used for maximizing biosynthesis of ethanol and FFAs, and subsequent generation of biodiesel from these, are discussed next.
3.2.1. Biosynthesis of Ethanol
The yeast Saccharomyces cerevisiae and the bacterium Zymomonas mobilis have long been used in the brewing industry to produce ethanol from 6-C sugars (glucose, galactose, and mannose), but these microorganisms are unable to ferment 5-C sugars (arabinose and xylose). On the other microorganisms such as the bacteria Escherichia coli and Klebsiella oxytoca, as well as the yeast Pichia stipitis, are in fact able to produce ethanol from pentose sugars, but they cannot do so naturally at sufficient yields and productivity levels. However, tools from metabolic engineering and synthetic biology have been effectively used to enhance ethanol production, through the use of two different strategies. The first essentially consists of the expression of pentose catabolic pathways in ethanologenic microorganisms such as S. cerevisiae [33, 34] and Z. mobilis [60]. Two natural enzymatic pathways for xylose catabolism are known, and both of these have been independently transferred to S. cerevisiae. In one pathway, the conversion of D-xylose to D xylulose is performed by a bacterial heterologous xylose isomerase (XYLA) (Figure 4). However, this approach generally failed because xylose isomerase is strongly inhibited by xylitol, favoring an isomerization equilibrium towards xylose formation. A second natural pathway for xylose catabolism, which involves a xylose reductase (XR) and a xylitol dehydrogenase (XDH), has been found in certain fungi and yeast. Introduction of this pathway from the xylose-fermenting yeast P. stipitis by engineering [61, 62], enables S. cerevisiae to consume xylose [63] (Figure 4). This strategy, however, was unsuccessful since this process also results in the excretion of xylitol, again favoring xylose formation [64].
Figure 4.
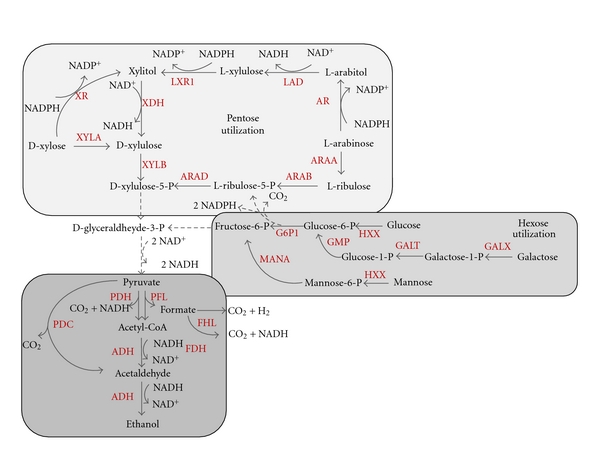
Redesigned pathways for microbial production of ethanol from carbohydrates. The dashed lines indicate multiple steps. Abbreviations: ADH: alcohol dehydrogenase; AR: aldose reductase; ARAA: L-arabinose isomerase; ARAB: L-ribulokinase; ARAD: L-ribulose-5-P-4-epimerase; FDH: formate dehydrogenase; FHL: formate hydrogen lyase; LAD: L-arabitol 4-dehydrogenase; LXR1: L-xylulose reductase; PDC: pyruvate decarboxylase; PDH: pyruvate dehydrogenase; PFL: pyruvate formate lyase; XDH: xylitol dehydrogenase; XR: xylose reductase; XYLA: xylose isomerase; XYLB: xylulokinase.
Arabinose is another pentose sugar obtained through decomposition of biomass. Bacterial and fungal pathways for arabinose catabolism exist in nature, and both pathways have been independently expressed in S. cerevisiae [65, 66]. Recombinant strains engineered with the heterologous fungal pathway showed only low levels of growth on L-arabinose [66]. However, after introduction of the bacterial pathway for arabinose utilization from Lactobacillus plantarum, mutant yeast produced an ethanol yield of 0.43 g per g of carbohydrate consumed, and a specific ethanol production rate of 0.29 g/g/h.
As mentioned, Z. mobilis is an ethanologenic bacterium that is able to produce ethanol only from 6-C sugars. However, xylose metabolism was introduced into Z. mobilis CP4 (pZB5) by expression of E. coli genes encoding for xylose isomerase (XYLA) and xylulokinase (XYLB) (Figure 4). This resulted in xylose catabolism with an ethanol yield of 86% [60].
Arabinose metabolism was also introduced in Z. mobilis ATCC39676 (pZB206) by expression of E. coli genes encoding for L-arabinose isomerase (ARAA), L-ribulokinase (ARAB), and L-ribulose-5-P-4-epimerase (ARAD) (Figure 4). This results in growth on arabinose with an ethanol yield of 98% [67]. Xylose or arabinoses are both first converted to xylulose-5-P. Xylulose-5-P then, by means of the pentose phosphate pathway, yields D-glyceraldehyde-3-P, which is an intermediate product in the Embden-Meyerhof-Parnas (EMP) pathway (Figure 4). However, the recombinant Z. mobilis can produce ethanol only at low pentose concentrations, which limits its potential industrial applications [68].
A second strategy to enhance ethanol biosynthesis by fermentation is based upon construction of synthetic pathways in hosts that are naturally able to utilize pentose. Wild-type E. coli can make use of a wide range of substrates, including all of the lignocellulosic sugars [22]. However, ethanol yield is low because under fermentative conditions E. coli produces lactic, acetic, formic, and succinic acids [68]. Cleavage of pyruvate into acetyl CoA and formic acid by pyruvate formate lyase (PFL) begins the pathway to ethanol production for E. coli (Figure 4). Reduction of acetyl-CoA to ethanol proceeds by means of the multienzyme protein and requires two NADHs [69] while glycolysis to pyruvate only requires one NADH in conversion of glyceraledehyde-3-P to 1,3-bisphosphoglycerate. Under such circumstances, production of ethanol is balanced by that of other more oxidized products such as acetic acid, which no NADH consumes. To circumvent the redox limitation of the endogenous ethanol pathway, pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH) enzymes from Z. mobilis were expressed in E. coli (Figure 4), via a plasmid bearing an artificial pet operon containing the pdc and adhB genes [70]. This has resulted in an ethanol yield of 95%. On the other hand, Kim et al. [32] reported homoethanol fermentation from xylose and glucose with yields of up to 82%, by combining the activity of pyruvate dehydrogenase and the alcohol dehydrogenase from native E. coli genes.
3.2.2. Biosynthesis of Fatty Acids
Most of the fatty acids produced in a microbial cell are utilized for the synthesis of phospholipids and TAGs, but some are also degraded through the β-oxidation pathway. Because of this, an improvement in the production of FFAs could be achieved by elimination of β-oxidation, along with overexpression of several genes related to fatty acid biosynthesis [71–75]. To this end, the fadE gene was initially disrupted in the E. coli strain, which was then incapable of degrading fatty acids and fatty acyl-CoA (Figure 5). The enforced fatty acid synthesizing ability was then accomplished through the overexpression of several genes encoding enzymes including thioesterase (tesA), acyl-CoA synthase (fadD), acetyl-CoA carboxylase (accABCD), and fatty acid synthase (fabH, fabD, fabG, and fabF) (Figure 5). To further enhance fatty acid production, expression of genes aceEF has been suggested for a production host, accompanied by the attenuation of glycerol-3-phosphate dehydrogenase (gpsA), glycerol-3-phosphate O-acyltransferase (plsB), phosphate acyltransferase (pta), and acetate kinase (ackA) (Figure 5). The chain length of the fatty acids can also be made to vary from C8 to C18 by alternative expression of thioesterase in the bacterial cytoplasm [76]. Along with these modifications, the expression of the acyl-acyl carrier protein (ACP) thioesterase from the plant Umbellularia resulted in the production of medium chain fatty acids [75].
Figure 5.
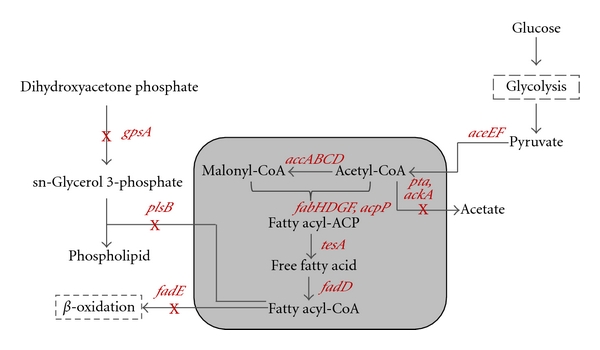
Metabolically engineered Escherichia coli strain for microbial production of free fatty acids. Deleted or attenuated genes are indicated by crosses. Gene/enzyme names: fadE (genes encoding for β-oxidation pathway); tesA (thioesterase); fadD (acyl-CoA synthase); accABCD (acetyl-CoA carboxylase); fabH, fabD, fabG, fabF (fatty acid synthase); aceEF (pyruvate dehydrogenase and pyruvate formate lyase); gpsA (glycerol-3-phosphate dehydrogenase); plsB (glycerol-3-phosphate O-acyltransferase); pta (phosphate acyltransferase); ackA (acetate kinase).
3.2.3. Biosynthesis of Fatty Acid Ethyl Esters (Biodiesel)
The development of in vivo biodiesel synthesis using endogenously produced alcohols has marked a turning point in the field of biofuel production during the last decade. Two model organisms, E. coli and S. cerevisiae, have been effectively used to develop this approach. Direct synthesis of biodiesel from microbes produces primarily FAEEs, which show better performance as a fuel than FAMEs. There are therefore multiple benefits associated with the production of FAEEs by using redesigned microbial cell factories. An important discovery related to this was a novel bifunctional wax ester synthase/acyl-CoA: diacylglycerol acyltransferase (WS/DGAT) isolated from Acinetobacter baylyi strain ADP1. This enzyme can synthesize wax esters from alcohols and fatty acids [77]. The idea of wax ester production was first applied effectively by Kalscheuer et al. [78], who reported on in vivo biodiesel synthesis from redesigned E. coli. Heterologous coexpression of the WS/DGAT gene (atfA) from Acinetobacter baylyi strain ADP1 with the ethanol synthesis pathway from Z. Mobilis (Figure 6) led to the production of 1.28 g/L of FAEEs after the addition of exogenous FFAs. More recently, Steen et al. [79] have reported on their engineering of E. coli to produce FAEEs, fatty alcohols, and waxes directly from simple sugars.
Figure 6.
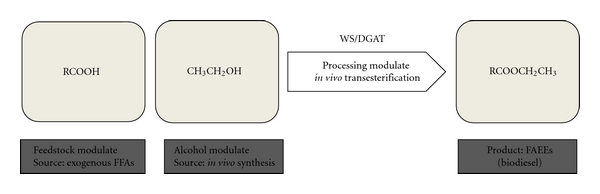
In vivo fatty acid ethyl esters synthesis using redesigned Escherichia coli. WS/DGAT: bifunctional wax ester synthase/acyl-CoA: diacylglycerol acyltransferase.
Given that ethanol is not naturally produced by E. coli, heterologous expression of a synthesis pathway for two-carbon alcohol is a prerequisite for any E. coli biodiesel producer. Because of this, a far better choice for a microbial cell factory for industrial production of biodiesel would be the yeast S. cerevisiae, which is a well-known organism already used in the production of ethanol through the fermentation of glucose [80]. Applying the same principle as used for E. coli, it has been reported that novel lipids, including FAEEs and fatty acid isoamyl esters, can be produced in S. cerevisiae H1246 after heterologous expression of the gram-negative bacterium Acinetobacter calcoaceticus ADP1 bifunctional WS/DGAT enzyme [81]. In vitro studies have confirmed that WS/DGAT is capable of utilizing a broad range of alcohols as substrates, including long-chain fatty alcohols like hexadecanol as well as short-chain alcohols like ethanol or isoamyl alcohol. These studies have demonstrated the highly unspecific acyltransferase activity of WS/DGAT from A. calcoaceticus ADP1, thereby indicating the broad biocatalytic potential of this enzyme for in vivo biotechnological production of a large variety of lipids in both prokaryotic and eukaryotic expression hosts.
A patent application by Schmidt-Dannert and Holtzapple [82] describes also a method for the production of biodiesel and wax esters, through heterologous expression of wax synthase (WS2) from Marinobacter hydrocarbonoclasticus in S. cerevisiae, after exogenous addition of fatty acids. Compared to the A. baylyi bifunctional WS/DGAT enzyme, WS2 shows higher wax synthase activity for ethanol and also does not have DGAT activity. It is important to point out that the WS2 does not catalyze the synthesis of TAGs from fatty acids, which could function as a competitive pathway for biodiesel production. Redesigned S. cerevisiae led to the successful production of 62 mg/L of ethyl oleate after exogenous addition of oleic acid. It is therefore envisioned that the WS2 from M. hydrocarbonoclasticus could be very suitable for the specific purpose of producing biodiesel.
In terms of endogenously produced fatty acids, an E. coli strain was manipulated not only for improvement of the biosynthesis of FFAs but also for derivatives thereof, including fatty esters [71, 72, 74]. Knothe [83] reported that by adding different types and/or amounts of alcohols to a medium containing the engineered fatty-ester-producing E. coli strain, it was possible to successfully change the properties of the biodiesel obtained, including cetane number, cloud point, viscosity, and lubricity.
4. Conclusions and Future Perspectives
As noted throughout this review, increasing interest in the development of microbial processes for the efficient production of biodiesel has emerged in recent years. Metabolic engineering and synthetic biology, as the latest approaches, have been essential for allowing new technologies to be developed, as evident in the design and refinement of microbial cell factories amenable to industrial applications. The research cited in this review clearly demonstrates the feasibility of direct production of biodiesel by microbes. The possibility of developing a synthetic host for efficient target-molecule production presents great opportunities for further biofuel research. However, investment of significant amounts of time and effort is still required in order to produce a better host, carrying novel metabolic pathways to lead to satisfactory biofuel production. This is because the complexity of the intertwined metabolic pathways creates substantial limitations. Development of a recombinant host that possesses a well-defined metabolic pathway, but which is also devoid of any competing pathways to hinder the production of a specific metabolite, can be seen as the most desirable goal. However, mere engineering of all of the genes needed to produce the desired pathway is not sufficient to confer a novel characteristic to a recombinant cell, since there are always many other unexplored pathways as well. Even a simple biocatalyst like E. coli is a complex system involving an estimated 4,603 genes, 2,077 reactions, and 1,039 unique metabolites [69, 84]. Fortunately, it appears likely that the formal integration of functional genomics and systems biology with synthetic biology and metabolic engineering will lead to enhancement of lipid accumulation and improved engineered pathways for in vivo biodiesel synthesis. Microorganisms may therefore become an ideal platform for the production of biodiesel in the future.
References
- 1.Atsumi S, Liao JC. Metabolic engineering for advanced biofuels production from Escherichia coli. Current Opinion in Biotechnology. 2008;19(5):414–419. doi: 10.1016/j.copbio.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schubert C. Can biofuels finally take center stage? Nature Biotechnology. 2006;24(7):777–784. doi: 10.1038/nbt0706-777. [DOI] [PubMed] [Google Scholar]
- 3.Schepens H. White biotechnology: gateway to a more sustainable future. EuropaBio, Lyon, France, 2003.
- 4.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Current Opinion in Biotechnology. 2008;19(6):556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315(5813):801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 6.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 7.Demirbaş A. Diesel fuel from vegetable oil via transesterification and soap pyrolysis. Energy Sources. 2002;24(9):835–841. [Google Scholar]
- 8.Demirbas A. Importance of biodiesel as transportation fuel. Energy Policy. 2007;35(9):4661–4670. [Google Scholar]
- 9.Budiman A, Kusumaningtyas RD, Rochmadi S, Purwono S. Second generation of biodiesel production from Indonesian jatropha oil by continuous reactive distillation process. AJChE. 2009;9(2):35–48. [Google Scholar]
- 10.Alamu OJ, Waheed MA, Jekayinfa SO, Akintola TA. Optimal transesterification duration for biodiesel production from Nigerian palm kernel oil. Agricultural Engineering International: CIGR Journal. 2007;9:1–11. [Google Scholar]
- 11.Rashid U, Anwar F, Ansari TM, Arif M, Ahmad M. Optimization of alkaline transesterification of rice bran oil for biodiesel production using response surface methodology. Journal of Chemical Technology and Biotechnology. 2009;84(9):1364–1370. [Google Scholar]
- 12.Singh AP, He BB, Thompson JC, van Gerpen JH. Process optimization of biodiesel production using alkaline catalysts. Applied Engineering in Agriculture. 2006;22(4):597–600. [Google Scholar]
- 13.Soriano NU, Venditti R, Argyropoulos DS. Biodiesel synthesis via homogeneous Lewis acid-catalyzed transesterification. Fuel. 2009;88(3):560–565. [Google Scholar]
- 14.Huang Y, Zheng H, Yan Y. Optimization of lipase-catalyzed transesterification of lard for biodiesel production using response surface methodology. Applied Biochemistry and Biotechnology. 2010;160(2):504–515. doi: 10.1007/s12010-008-8377-y. [DOI] [PubMed] [Google Scholar]
- 15.Miao X, Wu Q. Biodiesel production from heterotrophic microalgal oil. Bioresource Technology. 2006;97(6):841–846. doi: 10.1016/j.biortech.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Röttig A, Wenning L, Bröker D, Steinbüchel A. Fatty acid alkyl esters: perspectives for production of alternative biofuels. Applied Microbiology and Biotechnology. 2010;85(6):1713–1733. doi: 10.1007/s00253-009-2383-z. [DOI] [PubMed] [Google Scholar]
- 17.Colin VL, Baigorí MD, Pera LM. Mycelium-bound lipase production from Aspergillus niger MYA 135, and its potential applications for the transesterification of ethanol. Journal of Basic Microbiology. 2011;51(3):236–242. doi: 10.1002/jobm.201000232. [DOI] [PubMed] [Google Scholar]
- 18.Pleiss J. The promise of synthetic biology. Applied Microbiology and Biotechnology. 2006;73(4):735–739. doi: 10.1007/s00253-006-0664-3. [DOI] [PubMed] [Google Scholar]
- 19.Mukhopadhyay A, Redding AM, Rutherford BJ, Keasling JD. Importance of systems biology in engineering microbes for biofuel production. Current Opinion in Biotechnology. 2008;19(3):228–234. doi: 10.1016/j.copbio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Moya M, Gonzalez R. Systems biology approaches for the microbial production of biofuels. Biofuels. 2010;1(2):291–310. [Google Scholar]
- 21.Gui MM, Lee KT, Bhatia S. Feasibility of edible oil vs. non-edible oil vs. waste edible oil as biodiesel feedstock. Energy. 2008;33(11):1646–1653. [Google Scholar]
- 22.Aristidou A, Penttilä M. Metabolic engineering applications to renewable resource utilization. Current Opinion in Biotechnology. 2000;11(2):187–198. doi: 10.1016/s0958-1669(00)00085-9. [DOI] [PubMed] [Google Scholar]
- 23.Ghadge SV, Raheman H. Biodiesel production from mahua (Madhuca indica) oil having high free fatty acids. Biomass and Bioenergy. 2005;28(6):601–605. [Google Scholar]
- 24.Rashid U, Anwar F, Jamil A, Bhatti HN. Jatropha curcas seed oil as a viable source for biodiesel. Pakistan Journal of Botany. 2010;42(1):575–582. [Google Scholar]
- 25.Ma Y-L. Microbial oils and its research advance. Chinese Journal of Bioprocess Engineering. 2006;4(4):7–11. [Google Scholar]
- 26.Schädel C, Blöchl A, Richter A, Hoch G. Quantification and monosaccharide composition of hemicelluloses from different plant functional types. Plant Physiology and Biochemistry. 2010;48(1):1–8. doi: 10.1016/j.plaphy.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Sauciuc A, Dumitrescu L, Manciulea I, Zaha C. State of art of the first and the second generation of the biodiesel synthesis. Bulletin of the Transilvania University of Braşov. 2010;3(52):205–214. [Google Scholar]
- 28.Gowen CM, Fong SS. Exploring biodiversity for cellulosic biofuel production. Chemistry and Biodiversity. 2010;7(5):1086–1097. doi: 10.1002/cbdv.200900314. [DOI] [PubMed] [Google Scholar]
- 29.Pienkos PT, Zhang M. Role of pretreatment and conditioning processes on toxicity of lignocellulosic biomass hydrolysates. Cellulose. 2009;16(4):743–762. [Google Scholar]
- 30.Weber C, Farwick A, Benisch F, et al. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Applied Microbiology and Biotechnology. 2010;87(4):1303–1315. doi: 10.1007/s00253-010-2707-z. [DOI] [PubMed] [Google Scholar]
- 31.Zaldivar J, Nielsen J, Olsson L. Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Applied Microbiology and Biotechnology. 2001;56(1-2):17–34. doi: 10.1007/s002530100624. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y, Ingram LO, Shanmugam KT. Construction of an Escherichia coli K-12 mutant for homoethanologenic fermentation of glucose or xylose without foreign genes. Applied and Environmental Microbiology. 2007;73(6):1766–1771. doi: 10.1128/AEM.02456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 2008;72(3):379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsushika A, Inoue H, Kodaki T, Sawayama S. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Applied Microbiology and Biotechnology. 2009;84(1):37–53. doi: 10.1007/s00253-009-2101-x. [DOI] [PubMed] [Google Scholar]
- 35.Elkins JG, Raman B, Keller M. Engineered microbial systems for enhanced conversion of lignocellulosic biomass. Current Opinion in Biotechnology. 2010;21(5):657–662. doi: 10.1016/j.copbio.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Wen F, Sun J, Zhao H. Yeast surface display of trifunctional minicellulosomes for simultaneous Saccharification and fermentation of cellulose to ethanol. Applied and Environmental Microbiology. 2010;76(4):1251–1260. doi: 10.1128/AEM.01687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serrano L. Synthetic biology: promises and challenges. Molecular Systems Biology. 2007;3:p. 158. doi: 10.1038/msb4100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forster AC, Church GM. Synthetic biology projects in vitro. Genome Research. 2007;17(1):1–6. doi: 10.1101/gr.5776007. [DOI] [PubMed] [Google Scholar]
- 39.Greber D, Fussenegger M. Mammalian synthetic biology: engineering of sophisticated gene networks. Journal of Biotechnology. 2007;130(4):329–345. doi: 10.1016/j.jbiotec.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Morange M. A critical perspective on synthetic biology. Hyle. 2009;15(1):21–30. [Google Scholar]
- 41.Mata TM, Martins AA, Caetano NS. Microalgae for biodiesel production and other applications: a review. Renewable and Sustainable Energy Reviews. 2010;14(1):217–232. [Google Scholar]
- 42.Banerjee A, Sharma R, Chisti Y, Banerjee UC. Botryococcus braunii: a renewable source of hydrocarbons and other chemicals. Critical Reviews in Biotechnology. 2002;22(3):245–279. doi: 10.1080/07388550290789513. [DOI] [PubMed] [Google Scholar]
- 43.Illman AM, Scragg AH, Shales SW. Increase in Chlorella strains calorific values when grown in low nitrogen medium. Enzyme and Microbial Technology. 2000;27(8):631–635. doi: 10.1016/s0141-0229(00)00266-0. [DOI] [PubMed] [Google Scholar]
- 44.Khozin-Goldberg I, Cohen Z. The effect of phosphate starvation on the lipid and fatty acid composition of the fresh water eustigmatophyte Monodus subterraneus. Phytochemistry. 2006;67(7):696–701. doi: 10.1016/j.phytochem.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Chisti Y. Biodiesel from microalgae. Biotechnology Advances. 2007;25(3):294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Chen HC, Chang CC. Production of γ-linolenic acid by the fungus Cunninghamella echinulata CCRC 31840. Biotechnology Progress. 1996;12(3):338–341. [Google Scholar]
- 47.Čertík M, Baltészová L, Šajbidor J. Lipid formation and γ-linolenic acid production by Mucorales fungi grown on sunflower oil. Letters in Applied Microbiology. 1997;25(2):101–105. [Google Scholar]
- 48.Alvarez HM, Steinbüchel A. Triacylglycerols in prokaryotic microorganisms. Applied Microbiology and Biotechnology. 2003;60(4):367–376. doi: 10.1007/s00253-002-1135-0. [DOI] [PubMed] [Google Scholar]
- 49.Ratledge C, Wynn JP. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Advances in Applied Microbiology. 2002;51:1–44. doi: 10.1016/s0065-2164(02)51000-5. [DOI] [PubMed] [Google Scholar]
- 50.Liu B, Zhao Z. Biodiesel production by direct methanolysis of oleaginous microbial biomass. Journal of Chemical Technology and Biotechnology. 2007;82(8):775–780. [Google Scholar]
- 51.Angerbauer C, Siebenhofer M, Mittelbach M, Guebitz GM. Conversion of sewage sludge into lipids by Lipomyces starkeyi for biodiesel production. Bioresource Technology. 2008;99(8):3051–3056. doi: 10.1016/j.biortech.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 52.Gouda MK, Omar SH, Aouad LM. Single cell oil production by Gordonia sp. DG using agro-industrial wastes. World Journal of Microbiology and Biotechnology. 2008;24(9):1703–1711. [Google Scholar]
- 53.Meng X, Yang J, Xu X, Zhang L, Nie Q, Xian M. Biodiesel production from oleaginous microorganisms. Renewable Energy. 2009;34(1):1–5. [Google Scholar]
- 54.Li Q, Du W, Liu D. Perspectives of microbial oils for biodiesel production. Applied Microbiology and Biotechnology. 2008;80(5):749–756. doi: 10.1007/s00253-008-1625-9. [DOI] [PubMed] [Google Scholar]
- 55.Ahmad AL, Yasin NHM, Derek CJC, Lim JK. Microalgae as a sustainable energy source for biodiesel production: a review. Renewable and Sustainable Energy Reviews. 2011;15(1):584–593. [Google Scholar]
- 56.Beer LL, Boyd ES, Peters JW, Posewitz MC. Engineering algae for biohydrogen and biofuel production. Current Opinion in Biotechnology. 2009;20(3):264–271. doi: 10.1016/j.copbio.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Carioca JOB. Biofuels: problems, challenges and perspectives. Biotechnology Journal. 2010;5(3):260–273. doi: 10.1002/biot.200900137. [DOI] [PubMed] [Google Scholar]
- 58.Sheehan J, Dunahay T, Benemann J, Roessler P. A Look Back at the US Department of Energy’s Aquatic Species Program: Biodiesel From Algae. US Department of Energy, Washington, DC, USA: NREL; 1998. NREL/TP-580-24190. [Google Scholar]
- 59.Miao XL, Wu QY. Bio-oil fuel production from microalgae after heterotrophic growth. Renewable Energy Resources. 2004;4(116):41–44. [Google Scholar]
- 60.Zhang M, Eddy C, Deanda K, Finkelstein M, Picataggio S. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science. 1995;267(5195):240–243. doi: 10.1126/science.267.5195.240. [DOI] [PubMed] [Google Scholar]
- 61.Walfridsson M, Anderlund M, Bao X, Hahn-Hägerdal B. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Applied Microbiology and Biotechnology. 1997;48(2):218–224. doi: 10.1007/s002530051041. [DOI] [PubMed] [Google Scholar]
- 62.Jin YS, Lee TH, Choi YD, Ryu YW, Seo JH. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae containing genes for xylose reductase and xylitol dehydrogenase from Pichia stipitis. Journal of Microbiology and Biotechnology. 2000;10(4):564–567. [Google Scholar]
- 63.Jeffries TW, Jin YS. Metabolic engineering for improved fermentation of pentoses by yeasts. Applied Microbiology and Biotechnology. 2004;63(5):495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- 64.Traäff-Bjerre KL, Jeppsson M, Hahn-Hägerdal B, Gorwa-Grauslund MF. Endogenous NADPH-dependent aldose reductase activity influences product formation during xylose consumption in recombinant Saccharomyces cerevisiae. Yeast. 2004;21(2):141–150. doi: 10.1002/yea.1072. [DOI] [PubMed] [Google Scholar]
- 65.Becker J, Boles E. A modified Saccharomyces cerevisiae strain that consumes L-arabinose and produces ethanol. Applied and Environmental Microbiology. 2003;69(7):4144–4150. doi: 10.1128/AEM.69.7.4144-4150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richard P, Verho R, Putkonen M, Londesborough J, Penttilä M. Production of ethanol from L-arabinose by Saccharomyces cerevisiae containing a fungal L-arabinose pathway. FEMS Yeast Research. 2003;3(2):185–189. doi: 10.1016/S1567-1356(02)00184-8. [DOI] [PubMed] [Google Scholar]
- 67.Deanda K, Zhang M, Eddy C, Picataggio S. Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering. Applied and Environmental Microbiology. 1996;62(12):4465–4470. doi: 10.1128/aem.62.12.4465-4470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Böck A, Sawers G. Fermentation. In: Niedhardt FC, Curtiss IR, Lin ECC, et al., editors. Escherichia Coli and Salmonella: Cellular and Molecular Biology. Washington, DC, USA: American Society for Microbiology; 1996. [Google Scholar]
- 69.Keseler IM, Bonavides-Martínez C, Collado-Vides J, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Research. 2009;37(3):D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ingram LO, Conway T, Clark DP, Sewell GW, Preston JF. Genetic engineering of ethanol production in Escherichia coli. Applied and Environmental Microbiology. 1987;53(10):2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keasling J, Hu Z, Somerville C, et al. Production of fatty acids and derivatives thereof. WO2007136762, 2007.
- 72.Hu Z, Valle F, et al. Enhanced production of fatty acids derivatives. WO2008119082, 2008.
- 73.Lu X, Vora H, Khosla C. Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metabolic Engineering. 2008;10(6):333–339. doi: 10.1016/j.ymben.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 74.Del Cardayre S, Cockrem M, Charles M. Systems and methods for the production of fatty esters. WO200900991, 2009.
- 75.Lennen RM, Braden DJ, West RM, Dumesic JA, Pfleger BF. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnology and Bioengineering. 2010;106(2):193–202. doi: 10.1002/bit.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fortman JL, Chhabra S, Mukhopadhyay A, et al. Biofuel alternatives to ethanol: pumping the microbial well. Trends in Biotechnology. 2008;26(7):375–381. doi: 10.1016/j.tibtech.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 77.Kalscheuer R, Steinbüchel A. A novel bifunctional wax ester synthase/acyl-CoA: diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. Journal of Biological Chemistry. 2003;278(10):8075–8082. doi: 10.1074/jbc.M210533200. [DOI] [PubMed] [Google Scholar]
- 78.Kalscheuer R, Stölting T, Steinbüchel A. Microdiesel: Escherichia coli engineered for fuel production. Microbiology. 2006;152(9):2529–2536. doi: 10.1099/mic.0.29028-0. [DOI] [PubMed] [Google Scholar]
- 79.Steen EJ, Kang Y, Bokinsky G, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 80.Bro C, Regenberg B, Förster J, Nielsen J. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metabolic Engineering. 2006;8(2):102–111. doi: 10.1016/j.ymben.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 81.Kalscheuer R, Luftmann H, Steinbüchel A. Synthesis of novel lipids in Saccharomyces cerevisiae by heterologous expression of an unspecific bacterial acyltransferase. Applied and Environmental Microbiology. 2004;70(12):7119–7125. doi: 10.1128/AEM.70.12.7119-7125.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schmidt-Dannert C, Holtzapple EK. Isoprenoid wax ester synthases, isoprenoid acyl CoA-synthetases, and uses thereof. US patent 2009/0117629, 2009.
- 83.Knothe G. “Designer” biodiesel: optimizing fatty ester composition to improve fuel properties. Energy and Fuels. 2008;22(2):1358–1364. [Google Scholar]
- 84.Feist AM, Henry CS, Reed JL, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Molecular Systems Biology. 2007;3:p. 121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]


