Abstract
Many molecular biology assays depend in some way on the polymerase chain reaction (PCR) to amplify an initially dilute target DNA sample to a detectable concentration level. But the design of conventional PCR thermocycling hardware, predominantly based on massive metal heating blocks whose temperature is regulated by thermoelectric heaters, severely limits the achievable reaction speed1. Considerable electrical power is also required to repeatedly heat and cool the reagent mixture, limiting the ability to deploy these instruments in a portable format.
Thermal convection has emerged as a promising alternative thermocycling approach that has the potential to overcome these limitations2-9. Convective flows are an everyday occurrence in a diverse array of settings ranging from the Earth's atmosphere, oceans, and interior, to decorative and colorful lava lamps. Fluid motion is initiated in the same way in each case: a buoyancy driven instability arises when a confined volume of fluid is subjected to a spatial temperature gradient. These same phenomena offer an attractive way to perform PCR thermocycling. By applying a static temperature gradient across an appropriately designed reactor geometry, a continuous circulatory flow can be established that will repeatedly transport PCR reagents through temperature zones associated with the denaturing, annealing, and extension stages of the reaction (Figure 1). Thermocycling can therefore be actuated in a pseudo-isothermal manner by simply holding two opposing surfaces at fixed temperatures, completely eliminating the need to repeatedly heat and cool the instrument.
One of the main challenges facing design of convective thermocyclers is the need to precisely control the spatial velocity and temperature distributions within the reactor to ensure that the reagents sequentially occupy the correct temperature zones for a sufficient period of time10,11. Here we describe results of our efforts to probe the full 3-D velocity and temperature distributions in microscale convective thermocyclers12. Unexpectedly, we have discovered a subset of complex flow trajectories that are highly favorable for PCR due to a synergistic combination of (1) continuous exchange among flow paths that provides an enhanced opportunity for reagents to sample the full range of optimal temperature profiles, and (2) increased time spent within the extension temperature zone the rate limiting step of PCR. Extremely rapid DNA amplification times (under 10 min) are achievable in reactors designed to generate these flows.
Keywords: Molecular Biology, Issue 49, polymerase chain reaction, PCR, DNA, thermal convection
Protocol
1. Basic Design of the Convective Thermocycler
We have constructed a simple convective thermocycling device consisting of interchangeable plastic reaction chamber "cartridges" that are sandwiched between two aluminum plates whose temperatures are independently controlled7 (Figure 2).
Cylindrical reactor wells are embedded by machining arrays of holes in polycarbonate blocks, with different combinations of hole diameter and plastic sheet thickness employed to achieve the desired aspect ratios (height/diameter = h/d). Two different reactor geometries are considered in the results presented here: h/d = 9: h = 15.9 mm, d = 1.75 mm, 38.2 μL volume; h/d = 3: h = 6.02 mm, d = 1.98 mm, 18.5 μL volume.
The bottom surface of the reactor is heated using an aluminum block containing cartridge heaters interfaced with a microprocesser driven temperature controller.
The temperature at the reactor's upper surface is regulated using an aluminum block connected to a recirculating water bath.
The entire assembly is clamped together using nylon screws to limit thermal conduction between the opposing aluminum blocks.
2. Preparation and Loading of the PCR Reaction Mixture
A typical 100 μL reaction mixture contains 10 μL of 10x buffer solution, 6 μL of 25 mM MgCl2, 10 μL of dNTPs (2 mM each), 41.2 μL of DI water, 10 μL of β-actin probe, 10 μL of β-actin forward primer, 10 μL of β-actin reverse primer, 2 μL of human genomic template DNA (10 ng/μL) and 0.8 μL of KOD DNA polymerase (2.5 units/ μL).
Before loading reagents, seal the bottom surface of the PCR chamber using a thin sheet of aluminum tape.
Rinse the reactor wells with a 10 mg/mL aqueous solution of bovine serum albumin followed by Rain-X Anti-Fog.
Pipette reagents into the reactor wells using long gel-loading pipette tips and seal the upper surface with FEP Teflon tape.
3. Running the Reaction in the Convective Thermocycler
Before beginning the reaction, preheat both aluminum blocks to the desired upper and lower surface temperatures.
Sandwich the plastic reactor loaded with PCR reagents between the aluminum heating blocks, and quickly clamp the assembly together using nylon screws.
After the reaction has proceeded for the desired time, switch off the heaters and place the lower (hot) surface of the device on top of a chilled metal block to rapidly cool it and stop the convective flow.
Remove the plastic reactor and pipette the products out of the wells (using the long gel loading tips) for subsequent analysis.
4. Performing Gel Electrophoresis Analysis
Prepare a 2 wt% agarose gel by heating 10 g of agarose with 500 mL of 1x buffer on a stirring hot plate until the solution becomes clear.
Load the agarose gel into the casting tray and insert the comb. Let the gel set for ~ 30 min.
Remove the comb and add 1x TAE buffer until the gel is submerged.
Fluorescently stained DNA samples contain 2 μL 100x SYBR Green I solution, 2 μL DNA sample, 2 μL 6x Orange Loading Dye, and 4 μL TAE buffer.
Add the DNA samples into the wells and run the separation at 60 V for 1 h with a 100 bp DNA ladder sizing marker.
Remove the gel and photograph it under UV light to obtain the result.
5. Representative Results:
Optimal design of convective thermocyclers involves choosing the correct reactor geometry that will generate a circulatory flow capable of transporting reagents through the key temperatures involved in the PCR process. The geometric parameters that can be varied in the cylindrical reactors considered here are the height (h) and diameter (d), or equivalently the aspect ratio (h/d). We have explored the 3-D flow fields inside convective PCR reactors over a range of different aspect ratios using computational fluid dynamics (CFD), and found that unexpectedly complex patterns can arise. More importantly, our analysis has uncovered a subset of these complex flow fields that significantly accelerate the reaction12.
These geometric effects can be clearly seen by comparing the flow fields in reactors at high and low aspect ratios. In the high aspect ratio case (h/d = 9; a tall, skinny cylinder), fluid elements are advected along trajectories tracing out paths that are essentially closed loops, and they are locked-in to follow the same paths for long periods of time (Figure 3a). Consequently, there is little opportunity for exchange between flow trajectories that expose reagents to the optimal sequence of thermal conditions for PCR (oscillating between the annealing and denaturing extremes at the reactor's top and bottom surfaces) and the much larger ensemble of remaining trajectories that do not contribute to amplification (localized closer to the center of the reactor).
The flow field is much different at a smaller aspect ratio (h/d = 3; a shorter, wider cylinder), becoming more chaotic in the sense that the fluid element trajectories no longer follow closed paths (Figure 3b). Thus, even though reagents are subjected to a more complex temperature profile, fluid elements are able to explore a much wider range of trajectories so that more of the reagent volume has an opportunity to experience optimal thermal profiles. These results counterintuitively suggest that while individual flow trajectories at h/d = 9 may appear to be more favorable for PCR by producing temperature profiles that "look" similar to those employed in a conventional thermocycler, the chaotic nature of the flow field at h/d = 3 ultimately dominates on a global scale by promoting enhanced exchange so that reagents do not become trapped in unfavorable trajectories for very long.
To test this hypothesis and determine which reactor geometry was more favorable for PCR, we used both of them to perform PCR replication of a 295 bp target associated with the β-actin gene from a human genomic DNA template. Remarkably, we routinely achieved amplification of the correct target product in only 10 min at h/d = 3 (Figure 4a), while the same reaction required at least 20 min before detectable products were observed at h/d = 9 (Figure 4b). The reaction specificity is also much greater at h/d = 3 where a single PCR product is obtained, while multiple nonspecific products are generated at h/d = 9 where the flow field traps fluid elements in unfavorable thermal trajectories for extended periods of time.
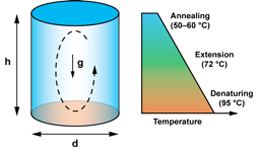 Figure 1. Thermal convection in a cylindrical chamber whose top and bottom surfaces are maintained at different fixed temperatures. If the temperature at the bottom surface is higher than at the top, a vertical density gradient is established within the enclosed fluid that is capable of generating a circulatory flow pattern. With the right choice of geometric parameters (height h and diameter d), the convective flow field can be harnessed to actuate PCR thermocycling when the top and bottom surfaces are manintained near annealing and denaturing temperatures, respectively (gravity acts vertically downward).
Figure 1. Thermal convection in a cylindrical chamber whose top and bottom surfaces are maintained at different fixed temperatures. If the temperature at the bottom surface is higher than at the top, a vertical density gradient is established within the enclosed fluid that is capable of generating a circulatory flow pattern. With the right choice of geometric parameters (height h and diameter d), the convective flow field can be harnessed to actuate PCR thermocycling when the top and bottom surfaces are manintained near annealing and denaturing temperatures, respectively (gravity acts vertically downward).
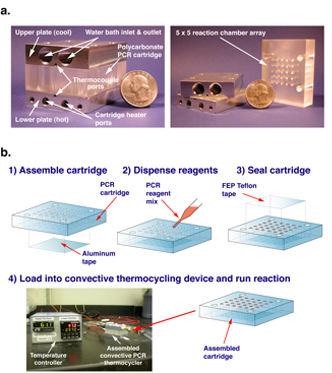 Figure 2. (a) Key components of a convective flow PCR thermocyler. A reactor cartridge consists of a polycarbonate block containing an array of cylindrical chambers. The block is sandwiched between a top plate designed to be connected to a circulating water bath and a bottom plate incorporating embedded heaters. (b) The PCR reagents are loaded and sealed inside the reactor cartridge, after which the device is assembled to perform the reaction. Upon completion, the procucts can be easily pipetted out for subsequent analysis.
Figure 2. (a) Key components of a convective flow PCR thermocyler. A reactor cartridge consists of a polycarbonate block containing an array of cylindrical chambers. The block is sandwiched between a top plate designed to be connected to a circulating water bath and a bottom plate incorporating embedded heaters. (b) The PCR reagents are loaded and sealed inside the reactor cartridge, after which the device is assembled to perform the reaction. Upon completion, the procucts can be easily pipetted out for subsequent analysis.
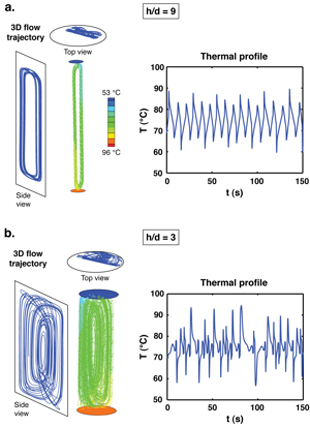 Figure 3. Computational simulations of 3-D convective flow fields generated inside cylindrical reactors with temperatures of 53 and 96 °C imposed at the top and bottom surfaces, respectively. Results are shown at aspect ratios of (a) h/d = 9 (38.2 μL reactor volume) and (b) h/d = 3 (18.5 μL reactor volume). A representative trajectory followed by a fluid element traveling through the 3D flow field is shown at the left along with top- and side view projections of the path; the corresponding plot of temperature versus time following a fluid element is shown at the right. The thermal profile at h/d = 9 appears close to that of a conventional thermocycler, but the chaotic flow field at h/d = 3 is counterintuitively more favorable for PCR.
Figure 3. Computational simulations of 3-D convective flow fields generated inside cylindrical reactors with temperatures of 53 and 96 °C imposed at the top and bottom surfaces, respectively. Results are shown at aspect ratios of (a) h/d = 9 (38.2 μL reactor volume) and (b) h/d = 3 (18.5 μL reactor volume). A representative trajectory followed by a fluid element traveling through the 3D flow field is shown at the left along with top- and side view projections of the path; the corresponding plot of temperature versus time following a fluid element is shown at the right. The thermal profile at h/d = 9 appears close to that of a conventional thermocycler, but the chaotic flow field at h/d = 3 is counterintuitively more favorable for PCR.
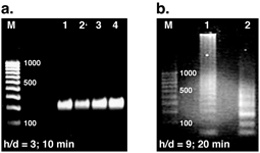 Figure 4. DNA replication results obtained in the reactor geometries shown in Figure 3 (upper and lower surfaces were maintained at 53 and 96 °C, respectively). (a) PCR is greatly accelerated at h/d = 3, as evident by strong products visible after only 10 min of reaction time (M: 100 bp ladder, lanes 1-4: products from parallel reactions in 4 different cylindrical chambers). (b) The same reaction performed at h/d = 9 requires at least 20 min before visible products are observed, and multiple bands are evident indicating replication of non-specific target DNA in addition to the desired product (M: 100 bp ladder, lanes 1-2: products from parallel reactions in 2 different cylindrical chambers).
Figure 4. DNA replication results obtained in the reactor geometries shown in Figure 3 (upper and lower surfaces were maintained at 53 and 96 °C, respectively). (a) PCR is greatly accelerated at h/d = 3, as evident by strong products visible after only 10 min of reaction time (M: 100 bp ladder, lanes 1-4: products from parallel reactions in 4 different cylindrical chambers). (b) The same reaction performed at h/d = 9 requires at least 20 min before visible products are observed, and multiple bands are evident indicating replication of non-specific target DNA in addition to the desired product (M: 100 bp ladder, lanes 1-2: products from parallel reactions in 2 different cylindrical chambers).
Discussion
The interplay between flow and chemical reaction can change dramatically under the influence of chaotic advection. But these complex flow states are challenging to study, particularly in realistic geometries where 3D effects become important. Rayleigh-Bénard convection at h/d > 1 not only provides a convenient platform to explore these phenomena, their application to perform PCR also enables the coupling between flow and chemical reactions to be probed. This unique capability makes it possible to select optimal flow states where chaotic effects act (counterintuitively) to accelerate the rate of DNA replication.
Disclosures
No conflicts of interest declared.
Acknowledgments
We gratefully thank T. Ragucci, B. Watkins, S. Wong, and B. Wallek at Lynntech, Inc. for technical assistance and many helpful discussions. This work was supported by the US National Science Foundation (NSF) under grant CBET-0933688.
References
- Spitzack KD, Ugaz VM. Microfluidic Techniques: Reviews and Protocols. In: Minteer SD, editor. Methods in Molecular Biology. Humana Press; 2005. Vol. 321. [Google Scholar]
- Krishnan M, Ugaz VM, Burns MA. PCR in a Rayleigh-Bénard convection cell. Science. 2002;298:793–793. doi: 10.1126/science.298.5594.793. [DOI] [PubMed] [Google Scholar]
- Braun D, Goddard NL, Libchaber A. Exponential DNA replication by laminar convection. Physical Review Letters. 2003;91:158103–158103. doi: 10.1103/PhysRevLett.91.158103. [DOI] [PubMed] [Google Scholar]
- Chen Z, Qian S, Abrams WR, Malamud D, Bau H. Thermosiphon-based PCR reactor: experiment and modeling. Analytical Chemistry. 2004;76:3707–3715. doi: 10.1021/ac049914k. [DOI] [PubMed] [Google Scholar]
- Wheeler EK. Convectively driven polymerase chain reaction thermal cycler. Analytical Chemistry. 2004;76:4011–4016. doi: 10.1021/ac034941g. [DOI] [PubMed] [Google Scholar]
- Krishnan M, Agrawal N, Burns MA, Ugaz VM. Reactions and fluidics in miniaturized natural convection systems. Analytical Chemistry. 2004;76:6254–6265. doi: 10.1021/ac049323u. [DOI] [PubMed] [Google Scholar]
- Ugaz VM, Krishnan M. Novel convective flow based approaches for high-throughput PCR thermocycling. Journal of the Association for Laboratory Automation (JALA) 2004;9:318–323. [Google Scholar]
- Yao DJ, Chen JR, Ju WT. Micro-Rayleigh-Benard convection polymerase chain reaction system. Journal of Micro-Nanolithography MEMS and MOEMS. 2007;6:043007–043007. [Google Scholar]
- Agrawal N, Hassan YA, Ugaz VM. A Pocket-sized Convective PCR Thermocycler. Angewandte Chemie International Edition. 2007;46:4316–4319. doi: 10.1002/anie.200700306. [DOI] [PubMed] [Google Scholar]
- Yariv E, Ben-Dov G, Dorfman KD. Polymerase chain reaction in natural convection systems: A convection-diffusion-reaction model. Europhysics Letters. 2005;71:1008–1014. [Google Scholar]
- Allen JW, Kenward M, Dorfman KD. Coupled flow and reaction during natural convection PCR. Microfluidics and Nanofluidics. 2009;6:121–130. [Google Scholar]
- Muddu R, Hassan YA, Ugaz VM. Accelerated Biochemistry by Chaotic Stirring. 2010. Forthcoming.


