Abstract
Whole mount in situ hybridization (WISH) is a common technique in molecular biology laboratories used to study gene expression through the localization of specific mRNA transcripts within whole mount specimen. This technique (adapted from Albertson and Yelick, 2005) was used in an upper level undergraduate Comparative Vertebrate Biology laboratory classroom at Syracuse University. The first two thirds of the Comparative Vertebrate Biology lab course gave students the opportunity to study the embryology and gross anatomy of several organisms representing various chordate taxa primarily via traditional dissections and the use of models. The final portion of the course involved an innovative approach to teaching anatomy through observation of vertebrate development employing molecular techniques in which WISH was performed on zebrafish embryos. A heterozygous fibroblast growth factor 8 a (fgf8a) mutant line, ace, was used. Due to Mendelian inheritance, ace intercrosses produced wild type, heterozygous, and homozygous ace/fgf8a mutants in a 1:2:1 ratio. RNA probes with known expression patterns in the midline and in developing anatomical structures such as the heart, somites, tailbud, myotome, and brain were used. WISH was performed using zebrafish at the 13 somite and prim-6 stages, with students performing the staining reaction in class. The study of zebrafish embryos at different stages of development gave students the ability to observe how these anatomical structures changed over ontogeny. In addition, some ace/fgf8a mutants displayed improper heart looping, and defects in somite and brain development. The students in this lab observed the normal development of various organ systems using both external anatomy as well as gene expression patterns. They also identified and described embryos displaying improper anatomical development and gene expression (i.e., putative mutants).
For instructors at institutions that do not already own the necessary equipment or where funds for lab and curricular innovation are limited, the financial cost of the reagents and apparatus may be a factor to consider, as will the time and effort required on the part of the instructor regardless of the setting. Nevertheless, we contend that the use of WISH in this type of classroom laboratory setting can provide an important link between developmental genetics and anatomy. As technology advances and the ability to study organismal development at the molecular level becomes easier, cheaper, and increasingly popular, many evolutionary biologists, ecologists, and physiologists are turning to research strategies in the field of molecular biology. Using WISH in a Comparative Vertebrate Biology laboratory classroom is one example of how molecules and anatomy can converge within a single course. This gives upper level college students the opportunity to practice modern biological research techniques, leading to a more diversified education and the promotion of future interdisciplinary scientific research.
Keywords: Developmental Biology, Issue 49, in situ hybridization, genetics, development, anatomy, vertebrate, undergraduate, education, interdisciplinary
Protocol
1. Plasmid Transformation of Target cDNA
Part I: Transformation of Plasmid
(Time Required: 3 hours plus overnight incubation)
Warm SOC nutrient broth in a 42°C water bath (500 μL per reaction)
Thaw competent cells on ice
Add 1 μL plasmid to 25 μL competent cells
Place on ice for 20 minutes
Heat shock cells in 42°C water bath for 45 seconds
Immediately place cells on ice for 2 minutes
Add 500 μL of 42°C nutrient broth to each vial of cells
Shake at 37°C for 2 hours at 255 rotations per minute (rpm)
Plate 75 μL transformation mix on Luria Bertani (LB) agar plates
Incubate plates inverted at 37°C overnight
Part II: E. coli Culture
(Time Required: 15 minutes plus overnight incubation)
For each reaction, aliquot 3 mL LB broth and 3 μL of 50 mg/ mL ampicillin into a culture tube
Scrape 1 bacteria colony from the agar plate and add to each culture tube
Shake culture tubes at 37°C at 255 rpm overnight
Part III: Plasmid Prep
(Time Required: 1.5 hours)
Isolate plasmid from overnight cultures using the 5 Prime FastPlasmid Mini Kit (Catalog #2300000)
To check for the presence of prepared plasmid, run the DNA eluted from the kit on a 1% Tris-Acetate-EDTA (TAE) gel stained with ethidium bromide
2. In Situ DIG-labeled RNA Probe Synthesis
Part I: Linearization of Plasmid
(Time Required: 2.5 hours)
- In a 1.5 mL microfuge tube, combine:
Prepared Plasmid 20 mL Restriction Enzyme* 2 mL Buffer** 10 mL 10X Bovine Serum Albumin (BSA) 10 mL Diethyl Pyrocarbonate (DEPC) Water 58 mL 100 mL Incubate at 37°C for 2 hours *Varies with plasmid **Varies with restriction enzyme
Part II: Transcription
(Time Required: 3 hours)
- In a 1.5 mL microfuge tube, combine:
Linear Plasmid 4 mL 10X Transcription Buffer** 4 mL Digoxigenin Label Mix 4 mL Polymerase* 2 mL RNase Inhibitor 2 mL DEPC Water 24 mL 40 mL Incubate at 37°C for 1 hour
Add 2 μL of polymerase*
Incubate at 37°C for 1 hour
Add 2 μL of DNase
Incubate at 37°C for 20 minutes *Varies with plasmid **Varies with polymerase
Part III: Precipitation
(Time Required: 5 minutes plus 2 hour to overnight incubation, 1 hour to centrifuge and re-suspend)
Add 4 μL of 0.2M EDTA
Add 5 μL of 4M lithium chloride
Add 150 μL of ice cold 100% ethanol
Incubate at -80°C for 2 hours to overnight
Centrifuge at 14,000 rpm for 30 minutes at 4°C, decant away supernatant
Dry pellet for 7 minutes
Re-suspend in 20 μL DEPC water
Incubate at 37°C for 5 minutes
Part IV: Fractionation - Perform only if probe size is greater than 0.6 kb
(Time Required: Varies based on probe size, usually no more than 20 minutes)
- In a 1.5 mL microfuge tube, combine:
RNA Probe 20 mL DEPC Water 12 mL Sodium Bicarbonate 4 mL Sodium Carbonate 4 mL 40 mL Incubate in a 60°C water bath. Base the incubation time on the equation: Time (min.) = (starting kb - desired kb) / (.11 x starting kb x desired kb) Average desired size = 0.6 kb
Part V: Final Precipitation
(Time Required: 5 minutes plus 2 hour to overnight incubation, 1 hour to centrifuge and re-suspend, 1 hour for gel electrophoresis)
Add 40 μL DEPC water
Add 8 μL Sodium Acetate
Add 1.04 μL Glacial Acetic Acid
Add 240 μL ice cold 100% ethanol
Incubate at -80°C for 2 hours to overnight
Centrifuge at 14,000 rpm for 30 minutes at 4°C, decant away supernatant
Dry pellet for 7 minutes
Re-suspend in 20 μL DEPC water
Vortex to mix
To check for the presence of riboprobe, run the re-suspended solution on a 1% TAE gel stained with ethidium bromide
3. Whole Mount In Situ Hybridization
Part I: Fixation of Embryos and Proteinase K Digestion
(Time Required: Fix overnight plus 4 hours the next day for storage, 2.5 hours from storage to Part II)
Collect staged zebrafish embryos and fix in 4% paraformaldehyde (PFA) overnight at 4°C
Wash fixed embryos in phosphate buffered saline containing 0.1% Tween-20 (PBSt) 3 times for 10 minutes each
To ensure exposure of the embryos to the experimental reagents, manually dechorionate embryos (if necessary) using fine-tipped forceps
Dehydrate embryos by washing in a graded series of 25% and 50% methanol in PBSt for one hour each wash, then store in 100% methanol at -20°C
When ready to use, rehydrate embryos in PBSt by washing in 50% (2 times) and 25% (1 time) methanol in PBSt for 10 minutes each. Finally wash 2 times for 10 minutes each in 100% PBSt. Exact timing is not important for PBSt/methanol washes
Bleach embryos in a 10% hydrogen peroxide solution in PBSt, if necessary, to remove dark pigments. Embryos should incubate in hydrogen peroxide solution for 10-20 minutes, depending on the age of the embryo, and the cap of the microfuge tube should remain open to prevent build-up of air pressure
Digest embryos with 50 mg/ mL Proteinase K diluted 1:5000 in PBSt for 3-15 minutes, depending on the age of the embryo
Re-fix embryos in 4% PFA for 30 minutes, and then wash 3 times in PBSt for 5 minutes each
Part II: Hybridization of Riboprobes
(Time Required: 3 hours plus overnight for hybridization, 1.5 hours the next day to Part III)
Prehybridize embryos with prehybridization solution (PHS) in a preheated 70°C water bath for 2-3 hours
Remove PHS, add 0.5 mL hybridization solution and 1.5 μL previously synthesized Digoxigenin labeled riboprobes, incubate at 70°C overnight. Temperature can fluctuate within a few degrees depending on the riboprobe target
The next day, wash embryos at 70°C in graded solutions of 75%, 50%, and 25% PHS in 2X saline-sodium citrate (SSC) for 10 minutes each, then wash in 0.2X SSC for 30 minutes at 68°C
Wash in Maleic Acid Buffer (MAB) 2 times for 10 minutes each at room temperature
Part III: Anti-Digoxigenin (α-DIG) Antibody Incubation
(Time Required: 3 hours plus overnight to block, 2.5 hours the next day to Part IV)
Transfer embryos to a 12 well plate
Pre-block embryos in 1-2 mL blocking solution for at least 3 hours at room temperature
Simultaneously, pre-block the antibody by preparing a second volume of blocking solution and diluting the anti-digoxigenin antibody 1:2000 in this solution
Remove pre-block and add 1-2 mL pre-blocked α-DIG solution, incubate overnight at 4°C
The next day, wash embryos in MAB. Allow the embryos to incubate in MAB for 5 minutes first, then perform buffer changes and incubate for two 10 minute, one 30 minute and one 60 minute interval. Exact timing is not necessary
Wash embryos 3 times for 5 minutes each in Alkaline Phosphatase (AP) buffer
Part IV: Staining and Final Processing
(Time Required: 1 hour to overnight for staining, depending on the riboprobe used, 4 hours to beginning of glycerol washes, 6-10 hours per glycerol wash, may be stored in glycerol)
Add 1-2 mL staining solution to the embryos, wrap the plate in foil, and check staining at regular intervals (about every 20 minutes) until staining is sufficient
Wash embryos with PBSt 2 times for 5 minutes each to stop the staining reaction
Dehydrate embryos using 10 minute washes in 25% (1 time) and 50% (2 times) methanol in PBSt, then in 100% methanol, to remove background staining
Allow embryos to incubate in 100% methanol for at least 2 hours at room temperature
Rehydrate in PBSt using 10 minute washes in 50% (2 times) and 25% (1 time) methanol in PBSt, then wash in 100% PBSt 2 times
Transfer stained embryos to an 80% glycerol solution in PBSt, using a graded series of washes, and store at 4°C.
Recipes:
LB agar plates- 10 g LB agar + 250 mL distilled water, autoclave, when cool to the touch add 250 μL ampicillin, pour 15-20 mL warm solution into each petri dish, allow agar to solidify
LB broth- 12.5 g LB Broth + 200 mL distilled water, autoclave, allow to cool before use
1% TAE gel- 0.4 g agarose, 40 mL 1X TAE, 2 μL ethidium bromide; load 7 μL plasmid (Plasmid Transformation of Target cDNA) or 3 μL riboprobe and 4 μL DEPC water (In situ DIG-labeled RNA Probe Synthesis) + 1 μL loading dye, 5 μL DNA ladder
Prehybridization solution- 50% formamide, 5X SSC, 9.2mM citric acid, 1% Tween-20
Hybridization solution- prehybridization solution plus 500 μg/ mL tRNA and 50 μg/ mL Heparin
MAB- 100mM Maleic Acid, 150mM NaCl, 0.2M NaOH, 0.1% Tween-20, pH to 7.5
Blocking Solution- 3 parts MAB, 1 part 10% Boehringer blocking reagent in MAB, 1 part heat deactivated lamb serum
AP buffer- 60mM Tris-HCl pH to 9.5, 60mM NaCl, 30mM MgCl2, 0.1% Tween-20
Staining Solution- 5-Bromo-4-chloro-3-indolyl phosphate (BCIP) and Nitro blue tetrazolium (NBT) in AP buffer
4. REPRESENTATIVE RESULTS
When performed correctly, the reaction between the NBT, BCIP, and alkaline phosphatase will form a purple precipitate that should appear on the zebrafish embryo as a purple stain. Riboprobes should be previously synthesized from cDNA corresponding to the gene of interest. Therefore, it can be concluded any stain visualized represents areas of the zebrafish in which the gene of interest has been transcribed at that particular developmental stage. For the purposes of this course, riboprobes were synthesized from aldh1a2 (previously raldh2; Begemann et al., 2001; Figure 1); fgf8a (Reifers et al., 1998; Figure 2); deltaC (Oates et al., 2005); myod1 (Weinberg et al., 1996); shha (Krauss et al., 1993), pax2a (Brand et al., 1996; Figure 3) and myl7 (previously cmlc2; Yelon et al., 1999) cDNA. Staining was expected in the midline and in anatomical structures including the somites, tailbud, myotome, brain, and heart. Ace/fgf8a mutants were expected to have defects in many of these structures. Staining was easily visualized using a standard dissecting microscope. Additional sources contain more information and troubleshooting on WISH techniques similar to those described here (Clark, 2003; D'Costa et al., 2009; Schmoldt et al., 2009; Schoenwolf, 2009).
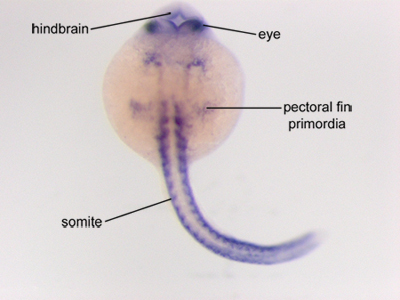 Figure 1. A zebrafish embryo 24 hours post fertilization, which has been hybridized with riboprobes specific for aldh1a2. Specific staining can be seen in the eyes, hindbrain, pectoral fin bud primordia, and somites. Anterior is to the top, posterior is to the bottom.
Figure 1. A zebrafish embryo 24 hours post fertilization, which has been hybridized with riboprobes specific for aldh1a2. Specific staining can be seen in the eyes, hindbrain, pectoral fin bud primordia, and somites. Anterior is to the top, posterior is to the bottom.
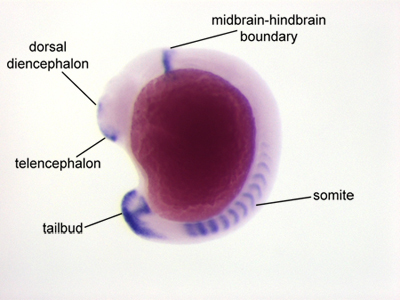 Figure 2. A zebrafish embryo at the 13 somite stage of development that has been hybridized with a probe specific for fgf8a. Specific staining is seen in the telencephalon, dorsal diencephalon, midbrain-hindbrain boundary, somites, and tailbud. Ventral is to the left, dorsal is to the right.
Figure 2. A zebrafish embryo at the 13 somite stage of development that has been hybridized with a probe specific for fgf8a. Specific staining is seen in the telencephalon, dorsal diencephalon, midbrain-hindbrain boundary, somites, and tailbud. Ventral is to the left, dorsal is to the right.
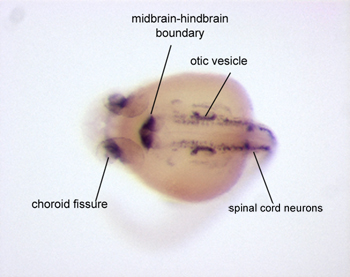 Figure 3. A 22 hours post fertilization zebrafish embryo that has been hybridized with a riboprobe specific for pax2a, a robust marker useful for visualizing the nervous system. Specific staining can be seen in the choroid fissure, midbrain-hindbrain boundary, otic vesicle, and spinal cord neurons. Dorsal view with anterior to the left.
Figure 3. A 22 hours post fertilization zebrafish embryo that has been hybridized with a riboprobe specific for pax2a, a robust marker useful for visualizing the nervous system. Specific staining can be seen in the choroid fissure, midbrain-hindbrain boundary, otic vesicle, and spinal cord neurons. Dorsal view with anterior to the left.
Discussion
WISH was used in a Comparative Vertebrate Biology laboratory course to help students understand the roles of genetics in anatomical development through the visualization of known gene expression patterns. For the first part of the course, students performed dissections on organisms representing several different chordate taxa, allowing them ample time to study, understand, compare, and contrast vertebrate anatomy.
As an introduction to the second part of the course, students were given a formal lecture describing zebrafish development and anatomy. The methods and expected results of the WISH experiment were also discussed. Students were then given live zebrafish at somitogenesis and prim-6 stages of development, and at 2 and 5 days post fertilization (dpf) to examine under the dissecting microscope. This was to give students a better understanding of what zebrafish embryos look like and the types of morphological changes that occur over ontogeny.
In the next laboratory session, students were given zebrafish embryos in which WISH had been previously performed. They were asked to study and describe the gene expression patterns for each gene of interest (riboprobe used). Embryos used for WISH were derived from matings between members of a heterozygous ace/fgf8a line. Based on Mendelian inheritance, 25% of the embryos from the ace/fgf8a matings were expected to be homozygous mutants and to exhibit defects in many of the anatomical structures focused on in this course. Based on published reports and unpublished observations in the Albertson lab, defects in the brain and improper heart looping were expected, as well as defects in the somites (Brand et al., 1996; Albertson and Yelick, 2005; personal observations).
Students were asked to examine all specimens, wild type (heterozygous animals are indistinguishable from wild type siblings at early stages of development) and homozygous mutants, for each gene expression pattern presented. They were then asked to write lab reports describing their results, and based on their knowledge of anatomy and genetics, how defective gene expression may have precipitated anatomical malformations.
Students seemed to receive this laboratory exercise with excitement and curiosity. Most had never used WISH before and were extremely interested in this part of the course. Students found the different patterns of gene expression in the zebrafish embryos intriguing, some even described the staining patterns visualized by associating them with well-known designs and symbols, such as a smiley face. The resulting lab reports showed the students had a general understanding of the WISH protocol and the expression of genes in specific anatomical structures. Students were also required to understand the specific functions of the genes studied using WISH during the lab (Stickney et al., 2000; Huelsken et al., 2002; Geetha-Loganathan et al., 2008a; Geetha-Loganathan et al., 2008b). It was evident however, that certain students had limited background knowledge of the signaling pathways and genes of interest. More information about these concepts in the WISH introductory lecture may be a welcome addition to the use of WISH in future Comparative Vertebrate Biology courses.
Since the protocol generally takes four consecutive days, depending on the schedule of the course, students may only be able to complete a part of the experiment in class and the instructor must be responsible for the remainder. In our Comparative Vertebrate Biology class, students completed the staining reactions in the laboratory, while the Teaching Assistant performed all preceding steps. If it is preferred to have the students perform WISH in class, the protocol may be divided into subunits that can be performed over several laboratory sessions, depending on the time frame of the class. If it is not feasible for students to perform the entire protocol, as it was here due to the lab meeting only once a week, students can add the staining solution at the beginning of class and, depending on the riboprobe used, have the staining complete within an hour. The time it takes for the stain to develop varies greatly with each riboprobe and a variety of experimental conditions, and should be predetermined before class. Notably, if students will only be developing the stain in lab, instructors will be responsible for all preceding steps, which will require significant time and effort outside of the classroom. If desired, a shorter alternative to WISH could be immunohistochemistry, using antibody labels to visualize protein localization, however at this time, zebrafish-specific antibodies for developmental genes are not readily available. Another option would be to perform WISH on different vertebrate species and have the students compare the expression patterns of the same genes in different organisms (Pizard et al., 2004; Aramaki et al., 2007; Emerging Model Organisms, 2008; Emerging Model Organisms, 2010).
The overarching goal of using WISH in a Comparative Vertebrate Biology course was to demonstrate to students how molecular biological techniques are used to study anatomical development. It also provided an opportunity for students to speculate as to how altered gene expression may lead not only to developmental malformations but also to evolutionary change. Formalized as evolutionary developmental biology (often referred to as "evo-devo"), this rapidly growing field of study aims to link genotype and phenotype through development, and to elucidate potential mechanistic bases of evolutionary change. With the rise of this field, more ecologists, organismal biologists, and physiologists are employing molecular techniques in their research. We contend that the use of WISH in a Comparative Vertebrate Biology course will help to keep the curriculum up to date with current technological and conceptual advances in research, and to facilitate a better horizontal alignment of upper level biology courses by combining biological subfields. Moreover, this integrative approach will provide students the opportunity to learn an assortment of biological research techniques in one course, leading to a more diversified education and the promotion of future interdisciplinary scientific research.
Disclosures
No conflicts of interest declared.
Acknowledgments
The authors would like to acknowledge the Department of Biology at Syracuse University and Dr. Marilyn Kerr for their roles in the administration of the Comparative Vertebrate Biology course. The Albertson lab is supported by grant R21DE019223 from the National Institutes of Health/National Institute of Dental and Craniofacial Research, as well as grant R01AG031922 from the National Institutes of Health/National Institute on Aging.
References
- Albertson RC, Yelick PC. Roles for fgf8 signaling in left-right patterning of the visceral organs and craniofacial skeleton. Dev. Biol. 2005;283:310–321. doi: 10.1016/j.ydbio.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Aramaki M, Kimura T, Udaka T, Kosaki R, Mitsuhashi T, Okada Y, Takahashi T, Kosaki K. Embryonic expression profile of chicken CHD7, the ortholog of the causative gene for CHARGE syndrome. Birth Defects Res A Clin Mol Teratol. 2007;79:50–57. doi: 10.1002/bdra.20330. [DOI] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Brand M, Heisenberg CP, Jiang YJ, Beuchle D, Lun K, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, van Eeden FJ, Kane DA, Nüsslein-Volhard C. Mutations in zebrafish genes affecting the formation of the boundary between midbrain and hindbrain. Development. 1996;123:179–190. doi: 10.1242/dev.123.1.179. [DOI] [PubMed] [Google Scholar]
- Clark M. In situ Hybridization: Laboratory Companion. Wiley-VCH; 2003. [Google Scholar]
- D'Costa A, Shepherd IT. Zebrafish development and genetics: Introducing undergraduates to developmental biology and genetics in a large introductory laboratory class. Zebrafish. 2009;6:169–177. doi: 10.1089/zeb.2008.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerging Model Organisms: A Laboratory Manual, Volume 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- Emerging Model Organisms: A Laboratory Manual, Volume 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2010. [Google Scholar]
- Geetha-Loganathan P, Nimmagadda S, Scaal M. Wnt signaling in limb organogenesis. Organogenesis. 4:109–115. doi: 10.4161/org.4.2.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha-Loganathan P, Nimmagadda S, Scaal M, Huang R, Christ B. Wnt signaling in somite development. Ann Anat. 190:208–222. doi: 10.1016/j.aanat.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Behrens J. The Wnt signaling pathway. J Cell Sci. 2002;15:3977–3978. doi: 10.1242/jcs.00089. [DOI] [PubMed] [Google Scholar]
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Oates AC, Mueller C, Ho RK. Cooperative function of deltaC and her7 in anterior segment formation. Dev. Biol. 2005;280:133–149. doi: 10.1016/j.ydbio.2005.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizard A, Haramis A, Carrasco AE, Franco P, López S, Paganelli A. Whole-mount in situ hybridization and detection of RNAs in vertebrate embryos and isolated organs. Curr Protoc Mol Biol. 2004;Chapter 14 doi: 10.1002/0471142727.mb1409s66. [DOI] [PubMed] [Google Scholar]
- Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Schmoldt A, Forecki J, Hammond DR, Udvadia AJ. Exploring differential gene expression in zebrafish to teach basic molecular biology skills. Zebrafish. 2009;6:187–199. doi: 10.1089/zeb.2008.0574. [DOI] [PubMed] [Google Scholar]
- Schoenwolf GC. Laboratory Studies of Vertebrate and Invertebrate Embryos: Guide and Atlas of Descriptive and Experimental Development. 9th Edition. Upper Saddle River, NJ: Pearson Education- Benjamin Cummings; 2008. [Google Scholar]
- Stickney HL, Barresi MJ, Devoto SH. Somite development in zebrafish. Dev Dyn. 2000;219:287–303. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1065>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]


