Abstract
The functional analysis of missense mutations can be complicated by the presence in the cell of the endogenous protein. Structure-function analyses of the BRCA1 have been complicated by the lack of a robust assay for the full length BRCA1 protein and the difficulties inherent in working with cell lines that express hypomorphic BRCA1 protein1,2,3,4,5. We developed a system whereby the endogenous BRCA1 protein in a cell was acutely depleted by RNAi targeting the 3'-UTR of the BRCA1 mRNA and replaced by co-transfecting a plasmid expressing a BRCA1 variant. One advantage of this procedure is that the acute silencing of BRCA1 and simultaneous replacement allow the cells to grow without secondary mutations or adaptations that might arise over time to compensate for the loss of BRCA1 function. This depletion and add-back procedure was done in a HeLa-derived cell line that was readily assayed for homologous recombination activity. The homologous recombination assay is based on a previously published method whereby a recombination substrate is integrated into the genome (Figure 1)6,7,8,9. This recombination substrate has the rare-cutting I-SceI restriction enzyme site inside an inactive GFP allele, and downstream is a second inactive GFP allele. Transfection of the plasmid that expresses I-SceI results in a double-stranded break, which may be repaired by homologous recombination, and if homologous recombination does repair the break it creates an active GFP allele that is readily scored by flow cytometry for GFP protein expression. Depletion of endogenous BRCA1 resulted in an 8-10-fold reduction in homologous recombination activity, and add-back of wild-type plasmid fully restored homologous recombination function. When specific point mutants of full length BRCA1 were expressed from co-transfected plasmids, the effect of the specific missense mutant could be scored. As an example, the expression of the BRCA1(M18T) protein, a variant of unknown clinical significance10, was expressed in these cells, it failed to restore BRCA1-dependent homologous recombination. By contrast, expression of another variant, also of unknown significance, BRCA1(I21V) fully restored BRCA1-dependent homologous recombination function. This strategy of testing the function of BRCA1 missense mutations has been applied to another biological system assaying for centrosome function (Kais et al, unpublished observations). Overall, this approach is suitable for the analysis of missense mutants in any gene that must be analyzed recessively.
Keywords: Cell Biology, Issue 48, BRCA1, homologous recombination, breast cancer, RNA interference, DNA repair
Protocol
1. Cell Line with an Integrated Recombination Substrate
The HeLa cells were stably transformed with pDR-GFP (plasmid7 gift from M. Jasin, Memorial Sloan-Kettering Cancer Center) and maintained in puromycin for selection of transformants.
The cells are passaged in DMEM/FBS, containing 10% fetal bovine serum, 2 mM GlutaMAX, 1 mM sodium pyruvate, penicillin/streptomycin (100 U/ mL/0.1 mg/ mL), and 1.5 ug/ mL puromycin.
The recombination substrate in the pDR-GFP plasmid, and in stable transformants, contains two inactive alleles of GFP (Figure 1). One allele is inactive due to the inclusion in its coding sequence of the 18 bp restriction enzyme recognition site for I-SceI. Expression of I-SceI in these cells creates a double-stranded DNA break (DSB). There is a second allele of inactive GFP on this DNA, but the portion of its sequence correlated with the I-SceI site on the first allele is wild-type. This second GFP allele can function as a sequence donor for homologous recombination repair of the DSB and would result in an active GFP allele, which is readily detected by flow cytometry. Alternatively, the DSB could be repaired by nonhomologous end-joining, which would destroy the I-SceI restriction site and would result in GFP-negative cells. The level of homologous recombination in a cell is determined by counting the percentage of GFP-positive cells.
This cell line, pHeLa-DR13-9, was selected among all clones for zero background GFP signal and a relatively high percentage of GFP-positive cells upon transfection with the I-SceI expressing plasmid, pCBASce. (The pCBASce and its vector control, pCAGGS, were both supplied by M. Jasin of Memorial Sloan-Kettering Cancer Center.) The HeLa-DR13-9 cell line is available from the authors upon request.
2. Transfection to Deplete Endogenous BRCA1 and Add Back Mutant BRCA1.
The day before the first transfection (usually a Tuesday): Begin with a 10 cm dish of HeLa-DR13-9 cells at least 90% confluent.
Trypsinize in 1 mL and stop the reaction by adding 9 mL DMEM/FBS, mix well by pipetting.
Add 40 μL of trypsinized cells per well of a 24 well plate in 0.5 mL of DME/FBS.
Day 1 transfections (Wednesday): cells should be ~40-50% confluent, if much less start over. Transfect by mixing: 5 pmol siRNA + 0.3 ug BRCA1-mutant expressing plasmid DNA in 12.5 μL Opti-MEM; and 0.5 μL Lipofectamine 2000 in 12.5 μL Opti-MEM. Proceed with the transfection according to the Lipofectamine protocol (Invitrogen).
Replace the media 4-6 hours later. The siRNA we use to deplete BRCA1 is based on the sequence GCUCCUCUCACUCUUCAGU specifying BRCA1 3'-UTR sequences 80,780 to 80,798 (accession number AY273801).
Day 2: Transfer the cells by trypsinizing from the 24 well plate to a 6 well plate.
Day 3 transfections: 25 pmol siRNA + 0.75 ug BRCA1-mutant expressing plasmid DNA + 0.75 ug pCBASce (or pCAGGS vector control) in 62.5 μL Opti-MEM and 2.5 μL Lipofectamine 2000 in 62.5 μL Opti-MEM. Replace the media 4-6 hours later.
3. Analysis of Transfectants.
On day 6, trypsinize cells from each well and stop in 1 mL DMEM/FBS. Analyzing the cells for GFP expression can be done on days 5 or 7, but we find that day 6 works best.
Analyze 10,000 cells per well by flow cytometry. We use a Becton Dickinson FACSCalibur instrument in the Ohio State University Comprehensive Cancer Center Analytical Cytometry shared resource. Single, live cells are gated using the forward and side scatter parameters. GFP-positive cells are detected using the GFP fluorescence detector (FL1), and results are presented as a histogram (Figure 2, bottom).
Left over cells can be replated for photography by fluorescence microscopy if desired.
4. Representative Results:
BRCA1 regulates the pathway for repair of double stranded DNA breaks by homologous recombination6,11. Those cells which have undergone repair by homologous recombination are readily detected by fluorescence microscopy (Figure 2, top). Depletion of BRCA1 results in a decrease in the detected GFP-positive cells (Figure 2, right). The output from the FACSCalibur is a histogram with GFP intensity per cell on the X-axis and number of cells on the Y-axis (Figure 2, bottom). In a typical set of results from a single experiment, 15.5% of the cells were GFP-positive after I-SceI expression and control RNAi depletion (Figure 3). By comparison, depletion of BRCA1 and vector add back reduced GFP-conversion to 0.7%. Add-back of wild-type BRCA1 or of BRCA1(I21V) fully restored homologous recombination, as detected by obtaining 15-16% GFP-positive cells. Add-back of the BRCA1(M18T) mutant (Figure 3, lane 5) had similar effect as the add-back of the vector control. Since this variant expressed at similar levels as the endogenous protein9, we thus interpret the BRCA1(M18T) variant is non-functional in homologous recombination.
We typically have 10-20% of the cells convert to GFP-positive after transfecting the I-SceI expression plasmid. Depletion of BRCA1 reproducibly reduces the conversion of GFP-positive cells by about 8-10 fold. Though the actual percentages of GFP-positive cells may change from experiment to experiment, the ratio of GFP-positive cells following BRCA1 depletion relative to the control depletion within an experiment is quite consistent. We normalize results by setting the uninhibited conversion rate to 100% and express results with BRCA1 depletions and add-backs as percentages. In this way, we can combine multiple experiments, and the experimental variation is not high.
Add back of BRCA1 mutants have so far yielded robust results: each BRCA1 variant has either fully restored homologous recombination activity or has had no effect. An important consideration in obtaining these results is that our transfection conditions produce levels of BRCA1 protein that approximately equal the concentration of the endogenous protein in untreated cells. Too high of an overexpression of even a defective mutant of BRCA1 may result in some positive homologous recombination activity.
A problem that we needed to overcome was the toxicity of transfection. Depletion of BRCA1 itself can slow cell growth, and the balance of Lipofectamine-2000 and plasmid DNA relative to the number of cells on the dish must be held at a consistent level. Too much Lipofectamine or plasmid DNA will reduce cell numbers, and the number of cells will be insufficient to obtain reliable flow cytometry results. The balance of the plasmid and Lipofectamine reagents versus the number of cells transfected is quite important for maintaining the viability of the cells.
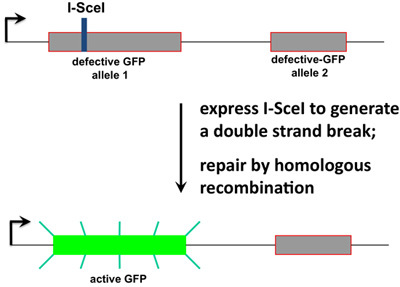 Figure 1. The recombination substrate. The strategy that had been developed by the group of M. Jasin7,12 is depicted. The pDR-GFP plasmid contains two defective alleles of GFP, one of which contains an I-SceI restriction endonuclease site. A HeLa derived cell line contains this DNA substrate integrated at a single site in the genome9, and active repair of the double-stranded DNA break at the I-SceI site by homologous recombination results in conversion of one allele to GFP-positive.
Figure 1. The recombination substrate. The strategy that had been developed by the group of M. Jasin7,12 is depicted. The pDR-GFP plasmid contains two defective alleles of GFP, one of which contains an I-SceI restriction endonuclease site. A HeLa derived cell line contains this DNA substrate integrated at a single site in the genome9, and active repair of the double-stranded DNA break at the I-SceI site by homologous recombination results in conversion of one allele to GFP-positive.
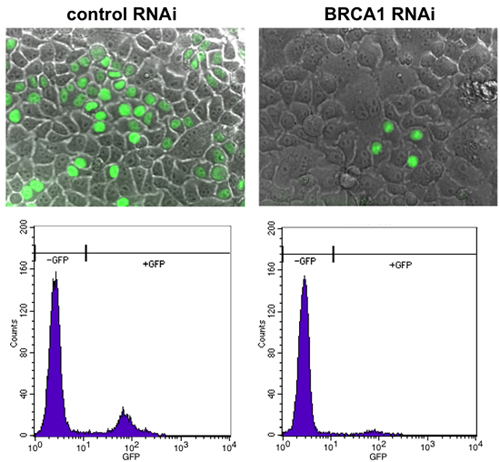 Figure 2. Detection of GFP-positive cells by fluorescence microscopy. Cells were tested as per this protocol, and control-depleted cells were active in homologous recombination, as detected by a high percentage of cells with GFP expression (left). Depletion of BRCA1 by RNAi results in a sharp decrease in the number of GFP-positive cells (right).
Figure 2. Detection of GFP-positive cells by fluorescence microscopy. Cells were tested as per this protocol, and control-depleted cells were active in homologous recombination, as detected by a high percentage of cells with GFP expression (left). Depletion of BRCA1 by RNAi results in a sharp decrease in the number of GFP-positive cells (right).
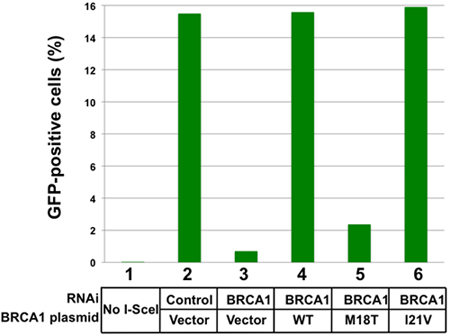 Figure 3. Results of an add-back of BRCA1 missense mutants. Results from a single experiment are shown in this histogram. In the absence of transfected I-SceI expressing plasmid (lane 1) there are no GFP-positive cells detected. The results in lanes 2-6 were all taken from cells in which the I-SceI expressing plasmid was transfected on day 3. In cells that were depleted for an irrelevant gene (luciferase) and with vector add-back, 15.5% of the cells were GFP-positive (lane 2). Depletion of BRCA1 and vector add-back reveals the effect of loss of BRCA1 on homologous recombination (lane 3). The effects of depletion of BRCA1 and add-back of wild-type BRCA1 (lane 4), BRCA1(M18T) (lane 5), and BRCA1(I21V) (lane 6) are shown. These results are from a single experiment, and would be combined with at least two more repeat experiments in order to obtain a measure of the homologous recombination supported by a given mutant BRCA1 protein.
Figure 3. Results of an add-back of BRCA1 missense mutants. Results from a single experiment are shown in this histogram. In the absence of transfected I-SceI expressing plasmid (lane 1) there are no GFP-positive cells detected. The results in lanes 2-6 were all taken from cells in which the I-SceI expressing plasmid was transfected on day 3. In cells that were depleted for an irrelevant gene (luciferase) and with vector add-back, 15.5% of the cells were GFP-positive (lane 2). Depletion of BRCA1 and vector add-back reveals the effect of loss of BRCA1 on homologous recombination (lane 3). The effects of depletion of BRCA1 and add-back of wild-type BRCA1 (lane 4), BRCA1(M18T) (lane 5), and BRCA1(I21V) (lane 6) are shown. These results are from a single experiment, and would be combined with at least two more repeat experiments in order to obtain a measure of the homologous recombination supported by a given mutant BRCA1 protein.
Discussion
The advantage of this method is that we can study the function of the BRCA1 protein in regulating DNA repair by homologous recombination using cells that have wild-type endogenously expressed BRCA1. We have adopted two separate established methods of studying the homologous recombination process and of utilizing RNAi-mediated depletion to obtain the novel method for testing BRCA1 variants for function in DNA repair. The key for the success of this method was to establish a readily transfectable cell line, such as HeLa, with the recombination substrate in the genome such that we obtain very high levels of GFP conversion. We had screened multiple clones of the original transformation in order to obtain one that had no detectable background GFP signal, but upon transfection of the I-SceI expression plasmid had a very high level of GFP conversion.
With this method, we are now investigating other specific BRCA1 amino acid residues for function in homologous recombination. Along with the biological insights from this work, these conclusions can be applied to clinical cancer genetics. A high percentage of BRCA1 missense mutations have unknown clinical significance since there are many rare variants and there is insufficient information for many of these to determine the cancer association by genetic segregation analysis3,10,13,14. Using this method, the consequences of a specific variant on the biological function of BRCA1 in regulating homologous recombination could be used to inform women who carry one of these variants in their genome.
Disclosures
No conflicts of interest declared.
Acknowledgments
We are grateful to the Ohio State University Comprehensive Cancer Center for providing funding for this work.
References
- Carvalho MA. Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res. 2007;67:1494–1501. doi: 10.1158/0008-5472.CAN-06-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FJ. Genetic epidemiology of BRCA1. Cancer Biol Ther. 2004;3:509–514. doi: 10.4161/cbt.3.6.840. [DOI] [PubMed] [Google Scholar]
- Couch FJ. Assessment of functional effects of unclassified genetic variants. Hum Mutat. 2008;29:1314–1326. doi: 10.1002/humu.20899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson GE. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci U S A. 2001;98:5134–5139. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Nakanishi K. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11:52–59. doi: 10.1016/s0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- Ransburgh DJ, Chiba N, Ishioka C, Toland AE, Parvin JD. Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 2010;70:988–995. doi: 10.1158/0008-5472.CAN-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snouwaert JN. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene. 1999;18:7900–7907. doi: 10.1038/sj.onc.1203334. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman AD. Clinically applicable models to characterize BRCA1 and BRCA2 variants of uncertain significance. J Clin Oncol. 2008;26:5393–5400. doi: 10.1200/JCO.2008.17.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet K, Senter L, Pilarski R, Wei L, Toland AE. Characterization of BRCA1 ring finger variants of uncertain significance. Breast Cancer Res Treat. 2009;119:737–743. doi: 10.1007/s10549-009-0438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


