Abstract
Imaging is a valuable technique that can be used to monitor biological processes. In particular, the presence of cancer cells, stem cells, specific immune cell types, viral pathogens, parasites and bacteria can be followed in real-time within living animals 1-2. Application of bioluminescence imaging to the study of pathogens has advantages as compared to conventional strategies for analysis of infections in animal models3-4. Infections can be visualized within individual animals over time, without requiring euthanasia to determine the location and quantity of the pathogen. Optical imaging allows comprehensive examination of all tissues and organs, rather than sampling of sites previously known to be infected. In addition, the accuracy of inoculation into specific tissues can be directly determined prior to carrying forward animals that were unsuccessfully inoculated throughout the entire experiment. Variability between animals can be controlled for, since imaging allows each animal to be followed individually. Imaging has the potential to greatly reduce animal numbers needed because of the ability to obtain data from numerous time points without having to sample tissues to determine pathogen load3-4.
This protocol describes methods to visualize infections in live animals using bioluminescence imaging for recombinant strains of bacteria expressing luciferase. The click beetle (CBRLuc) and firefly luciferases (FFluc) utilize luciferin as a substrate5-6. The light produced by both CBRluc and FFluc has a broad wavelength from 500 nm to 700 nm, making these luciferases excellent reporters for the optical imaging in living animal models7-9. This is primarily because wavelengths of light greater than 600 nm are required to avoid absorption by hemoglobin and, thus, travel through mammalian tissue efficiently. Luciferase is genetically introduced into the bacteria to produce light signal10. Mice are pulmonary inoculated with bioluminescent bacteria intratracheally to allow monitoring of infections in real time. After luciferin injection, images are acquired using the IVIS Imaging System. During imaging, mice are anesthetized with isoflurane using an XGI-8 Gas Anethesia System. Images can be analyzed to localize and quantify the signal source, which represents the bacterial infection site(s) and number, respectively. After imaging, CFU determination is carried out on homogenized tissue to confirm the presence of bacteria. Several doses of bacteria are used to correlate bacterial numbers with luminescence. Imaging can be applied to study of pathogenesis and evaluation of the efficacy of antibacterial compounds and vaccines.
Protocol
1. Pulmonary Infection by Intratracheal Intubation
Weigh mice and, optionally, marks can be made on the ears for easy identification.
Anesthetize the mice with ketamine (100 μg per g of mouse weight) and xylazine (10 μg per g of mouse weight) by intraperitoneal inoculation.
Place mice in cages until fully anesthetized. Squeeze the pads of their feet to check pedal reflex. Mice should display reduced or no reflex reaction.
Place the mouse on the intubation stand lying on its back.
Fix a rubber band to intubation stand and then place it under the upper incisors of the mouse. Use tape to fix legs and arms onto the intubation stand.
Gently move tougue out of the mouth using forceps.
Insert speculum with octoscope inside of the mouth and look into oropharynx until larynx opening is visible.
Advance guide wire covered with cathether into trachea until catether hub is at the incisors. Remove guide wire. Put air into cathether to observe chest expansion and confirm correct intubation.
Inject 50 μL of bacterial (in the images shown we used bacillus Calmette Guerin (BCG) expressing click beetle red luciferase (CBRLuc), constructed in our laboratory, but any luminescent bacterial strain can be used) solution through cathether using a 1 mL syringe.
Place mouse in a cage and observe it during recovery from anesthesia.
2. Animal Anesthetization and Preparation for Bioluminescence Imaging
Mice are anesthetized with isoflurane using the XGI-8 Gas Aneshesia System.
Before operating the XGI-8 anesthesia system, weigh each charcoal filter canister and write the weight and date on it. If the weight is 50 gram over of the initial weight, replace it with a new canister.
Check the isoflurane level in the vaporizer and fill it if necessary.
Turn on the evacuation pump in the front of the XGI-8 anesthesia system and set to 8 lpm (liters per minuite).
Turn on the oxygen supply from the high pressure cylinder and set it to 55 psig.
Turn on the oxygen toggle (green) on the front of the XGI-8 anesthesia system.
Turn on the gas flow to the imaging chamber to set the level of gas flow. Set to 0.25 lpm and then turn off the gas flow.
Turn on the gas flow to the anesthesia induction chamber to set the level of gas flow. Set to 1.5 lpm and then turn off gas flow.
Turn on the isoflurane with the vaporizer and set to 2-2.5 percent. The isoflurane level can be adjusted depending upon the number of animals being used and weight.
Place mice into the induction chamber and close the lid. Turn on the gas flow to induction chamber. Place mice in the chamber for 5 - 10 minuites until they are anesthetize completely.
Apply optical ointment to the eyes to protect the mouse eyes during imaging and place the mice in the imaging chamber in one of the nose cones on the anesthesia manifold. Use light baffle between mice to prevent the reflection of light onto adjacent animal subjects.
Inject luciferin (150 μg/gram of body weight) by intraperitoneal inoculation.
3. Bioluminescence Imaging
Start the Living Imaging Software.
If the system is not initialized, initialize the IVIS Imaging system.
Set the parameters for imaging by clicking sequence setup in IVIS acquisition control panel. Select luminescence and photograph in Imaging Mode. If the option to carry out DLIT 3D reconstruction is desired, select photographic, luminescent and structural light images as part of the image sequence. Set exposure time from 0.5 sec to 10 minutes. Set Binning and F/stop based on expected brightness of sample. Set excitation filter to block and emission filter to open, unless planning to acquire only specific wavelengths of light. In the case of DLIT 3D constitution, set multiple emission filters of different wavelengths to allow accurate localization of source. Set FOV from A to D depending on the number of mice or region of animal to be imaged. Select A and B for 1 mouse, C for 2-3 mice, and D for 4-5 mice.
Click add in the Image Wizard in the Acquisition Control Panel to add the sequence setup.
Start imaging sequence by clicking acquire. In the case of DLIT 3D construction, luminescent images will be acquired for multiple emission filters at different wavelengths. Photographic and structured light images will also be acquired.
During acquisition the image and edit image levels panels will be visible. Fill in as detailed information as possible for each experiment in edit image to ensure easy tracking of images at later times and save your images.
ReRemove the mice from IVIS imaging chamber and return them to their cages. Observe recovery from anesthesia to ensure mice were not significantly affected by the procedure. Animals should be monitored constantly until they fully recover from anesthesia (usually 1-2 min).
4. ex vivo Imaging and CFU Analysis for the Quantification of Bacteria in the Lungs
Inject mice with luciferin intraperitoneally in the same manner as for imaging, just prior to euthanasia.
Euthanize the mice by intraperitoneal injection of 100 mg/kg pentobarbitol 5 min after luciferin injection.
Explant the lungs from the mice and place in sterile petri-dishes using sterile forceps and scissors.
Place petri-dish containing explanted organ in imaging chamber and acquire bioluminescence images in the same manner as for whole mouse.
After imaging, remove explanted organ from petri dish using sterile forceps and homogenize in 1 mL PBS.
Make dilutions and plate tissue on the appropriate selective media. Homogenized tissue can be stored -80°C for re-plating should it be necessary. Alternatively, homogenized tissue can be used for RNA or DNA extraction to quantify bacterial numbers by qPCR.
5. Analysis of Luminescence Imaging
Start "Living Imaging Software" and browse the image files.
Use "Tool Palette' to adjust images. Click Image Adjust to modify correction/filtering settings and min/max for individual color scales.
Use ROI tools in pulldown list for the quantification of light intensity. Select ROI shape, number and size. Drag ROI frame to region of interest on image. Ensure all ROI are the same size and shape. Click ROI measurement and save, copy and/or export quantitative data.
In the case of DLIT 3D reconstructions, load image sequence including the structured light image. Click Surface topography in the Tool Platte. In pulldown list, click Reconstruct tab to generate the surface topography. A surface mess will then appear in the window.
Select DLIT 3D reconstruction in the Tool Platte. In pulldown list, click the Properties tab to set tissue properties and source spectrum. Muscle is recommended under most circumstances. In the pulldown list, click the Analyze tab to select the wavelength for analysis. Click the Parameter tab and confirm default values or adjust DLIT parameters if necessary. Click Reconstruct tab to start 3D analysis.
When 3D reconstruction is finished, the 3D view will display the results of 3D reconstruction. Click results tab to see analysis data for photon density, voxels and DLIT algorithm parameters.
Save and/or export the results of the 3D reconstruction analyses into figures and data files.
6. Representative Results:
The bioluminescence images of mice infected with bioluminescent bacteria along with an uninfected mouse control are shown in Figure 1. The mice pulmonary infected with bioluminescent bacteria produce significant signal from the lungs (Figure 1). Luminescence intensity is quantified as total flux within a R.O.I (region of interested) (Figure 2). The quantitative data for light intensity is normalized for colony forming units (CFU) of bacteria obtained from lungs to confirm that the signal is produced from the bioluminescent bacteria and can be compared to negative control. The location and intensity of signal can be further analyzed by DLIT 3D reconstruction of the luminescence source based on tomography of the mouse surface11. These analyses allow quantification and localization of the bioluminescent signal produced. The results of 3D reconstruction of a luminescent source in infected mice demonstrates that light is produced from lungs of the mouse (Figure 3). The ex vivo images of the resulting mouse lungs confirm that luminescence is emitted from the lungs, rather than some other closely juxtaposed tissue or organ (Figure 2C).
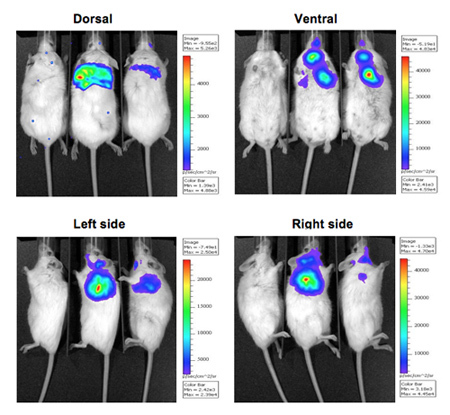 Figure 1. Luminescence imaging of pulmonary infected mice with bioluminescent bacteria tagged with CBRluc. Uninfected control mouse is on the left and two infected mice are on the right. Mice were infected with bacteria expressing CBRluc (n=2) via the intratracheal route. 10 minutes after luciferin injection, luminescence images were acquired for 10 min at 4 positions: dorsal, ventral, left side and right side.
Figure 1. Luminescence imaging of pulmonary infected mice with bioluminescent bacteria tagged with CBRluc. Uninfected control mouse is on the left and two infected mice are on the right. Mice were infected with bacteria expressing CBRluc (n=2) via the intratracheal route. 10 minutes after luciferin injection, luminescence images were acquired for 10 min at 4 positions: dorsal, ventral, left side and right side.
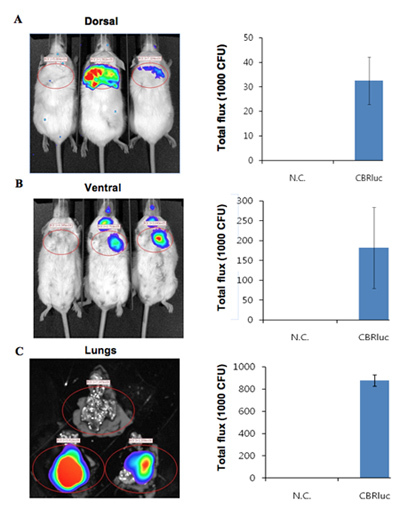 Figure 2. The quantitative light intensity from mice infected with bacteria expressing CBRluc. (A, B) Luminescence images of ROI analysis and the quantified total flux of light from R.O.I. in dorsal and ventral positions, respectively. Uninfected mice are on the left and two infected mice are on the right. Luminescence images were acquired from lungs of mice uninfected and infected with CBRlux (n=2). The light intensity around lungs was quantified by ROI analysis. C) ex vivo images of lungs from uninfected (top) and infected (bottom two sets of lungs) mice. Luciferin was injected 5 miniuts before of euthanasia and images were then acquired. The quantified values are normalized to CFU.
Figure 2. The quantitative light intensity from mice infected with bacteria expressing CBRluc. (A, B) Luminescence images of ROI analysis and the quantified total flux of light from R.O.I. in dorsal and ventral positions, respectively. Uninfected mice are on the left and two infected mice are on the right. Luminescence images were acquired from lungs of mice uninfected and infected with CBRlux (n=2). The light intensity around lungs was quantified by ROI analysis. C) ex vivo images of lungs from uninfected (top) and infected (bottom two sets of lungs) mice. Luciferin was injected 5 miniuts before of euthanasia and images were then acquired. The quantified values are normalized to CFU.
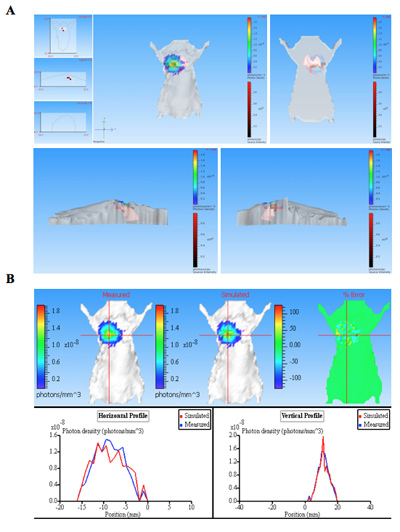 Figure 3. The 3D reconstruction of bioluminescence source(s) from a pulmonary infected mouse. The mouse was infected with bacteria expressing CBRluc by intratracheal injection. The image sequence was acquired using different emission filters of serial wavelengths from 540 nm to 700 nm. An image sequence was used for 3D reconstitution for luminescence source in animal subjects that contained a structured imag. A) The tomography for mouse is shown in different positions: front, back, left and right. Light sources are shown as voxels (red boxes within 3D mouse) that are located within lung as determined by reconstruction. B) Photon densiy map of measured and simulated data. Comparing the measured and simulated photon density curves provide the information of quality of reconstruction. Good quality reconstructions result in similar measured and simulated photon densities.
Figure 3. The 3D reconstruction of bioluminescence source(s) from a pulmonary infected mouse. The mouse was infected with bacteria expressing CBRluc by intratracheal injection. The image sequence was acquired using different emission filters of serial wavelengths from 540 nm to 700 nm. An image sequence was used for 3D reconstitution for luminescence source in animal subjects that contained a structured imag. A) The tomography for mouse is shown in different positions: front, back, left and right. Light sources are shown as voxels (red boxes within 3D mouse) that are located within lung as determined by reconstruction. B) Photon densiy map of measured and simulated data. Comparing the measured and simulated photon density curves provide the information of quality of reconstruction. Good quality reconstructions result in similar measured and simulated photon densities.
Discussion
Although following these protocols will usually result in high quality images, it is important to consider a few key issues in order to obtain accurate and consistent data from imaging studies. Luminescence images should be acquired that have counts from 600 to 60,000 to ensure that the signal is above background and the camera is not saturated. If signal obtained is less than 600 the exposure conditions should be adjusted to increase counts. If signal obtained is over 60,000 the camera is saturated in some regions. When the camera is saturated, quantification should not be attempted in saturated regions, though it is still possible and sometimes necessary in non-saturated regions. In addition, 3D reconstruction cannot be carried out accurately when saturated a regions is included within the region being used for the mask for the reconstruction. The adjustment of exposure time, binning and F/stop for image acquisition as well as modification of the imaging region or animal position can control the signal obtained and allow quantitative analysis of data. Thus, in many experiments it is helpful to consistently image animals under multiple exposure conditions to ensure that at least one set of images will be within the required limits to allow quantitative analysis.
It is important to remember that expression of any reporter gene can affect virulence, making it necessary to carry out pilot studies to evaluate potential impacts on fitness in recombinants strains that are to be used for imaging. This is one of the limitations of bioluminescent imaging, since it requires expression of foreign genes in the bacteria. This may often be overcome through the use of optimized constructs that use the appropriate codon usage for the bacterial species to be studied, titration of expression levels for reporter genes to reduce impact on metabolism or examination of multiple luminescent reporter systems that may differ in their impact on fitness.
Another primary variable for imaging studies with infectious agents is the level of signal produced by the agent being imaged. Since there are differences in the threshold of detection for each agent in animals as compared to in vitro, it is recommended that pilot studies using multiple infectious doses to optimize agent numbers prior to carrying out more extensive experiments. The bioluminescent reporters themselves should also be optimized for transcription, translation and low toxicity, to ensure that the level of signal produced is as high as possible. Optimization of reporters can be accomplished in most cases in vitro, but the wavelength of light emission will also impact the ability light produced by optimal reporters to penetrate mammalian tissue, making it important to also evaluate different reporters directly in animals. Although each animal can be followed individually, which controls for a great deal of the variability between animals, it is still necessary to include sufficient animals to allow statistical significance between groups to be determined. Usually, animal numbers should be 4 - 6 per group, allowing differences as low as two-fold between groups to be observed in many cases. Lastly, the inoculation route and accuracy of delivery to the appropriate tissues and of precise agent numbers will help to ensure consistent results by reducing overall variability between animals.
The spectral properties of the bioluminescence produced is also an important factor affecting the ability to image infections in vivo. In mammalian tissues, light of wavelengths less than 600 nm are largely absorbed by hemoglobin, a major component of mammalian tissue. The depth of penetration for bioluminescence wavelengths drastically increases for wavelengths longer than 600 nm8,12. Both CBRluc and FFluc produce bioluminescence with a broad wavelength, including over 600 nm, making these luciferases nearly ideal reporters for in vivo imaging, even in deep tissues including lungs and liver7-8.
Disclosures
Animal studies were performed in accordance with the guidelines and regulations set forth by the Institutional Animal Care and Use Commitee of Texas A&M University.
Acknowledgments
The authors thank the Cirillo laboratory members for valuable discussions and assistance throughout this study. We thank Dr. Joshua Hill and the laboratory of Dr. James Samuel for assistance with during the filming of this protocol. This work was funded by grant 48523 from the Bill & Melinda Gates Foundation and grant AI47866 from the National Institutes of Health.
References
- Wilson T, Hastings JW. Bioluminescence. Annu Rev Cell Dev Biol. 1998;14:197–230. doi: 10.1146/annurev.cellbio.14.1.197. [DOI] [PubMed] [Google Scholar]
- Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- Hutchens M, Luker GD. Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol. 2007;9:2315–2322. doi: 10.1111/j.1462-5822.2007.00995.x. [DOI] [PubMed] [Google Scholar]
- Doyle TC, Burns SM, Contag CH. In vivo bioluminescence imaging for integrated studies of infection. Cell Microbiol. 2004;6:303–317. doi: 10.1111/j.1462-5822.2004.00378.x. [DOI] [PubMed] [Google Scholar]
- Wood KV, Lam YA, Seliger HH, McElroy WD. Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science. 1989;244:700–702. doi: 10.1126/science.2655091. [DOI] [PubMed] [Google Scholar]
- Wet JRde, Wood KV, Helinski DR, DeLuca M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings JW. Chemistries and colors of bioluminescent reactions: a review. Gene. 1996;173:5–11. doi: 10.1016/0378-1119(95)00676-1. [DOI] [PubMed] [Google Scholar]
- Zhao H. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10:41210–41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- Rice BW, Cable MD, Nelson MB. In vivo imaging of light-emitting probes. J Biomed Opt. 2001;6:432–440. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- Contag CH. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- Kuo C, Coquoz O, Troy TL, Xu H, Rice BW. Three-dimensional reconstruction of in vivo bioluminescent sources based on multispectral imaging. J Biomed Opt. 2007;12:024007–024007. doi: 10.1117/1.2717898. [DOI] [PubMed] [Google Scholar]
- Weissleder R. A clearer vision for in vivo imaging. Nat Biotechnol. 2001;19:316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]


