Abstract
Background
In animals, prolyl 4-hydroxylases (P4Hs) are regarded as oxygen sensors under hypoxia stress, but little is known about their role in the response to waterlogging in maize.
Methods
A comprehensive genome-wide analysis of P4H genes of maize (zmP4H genes) was carried out, including gene structures, phylogeny, protein motifs, chromosomal locations and expression patterns under waterlogging.
Key Results
Nine zmP4H genes were identified in maize, of which five were alternatively spliced into at least 19 transcripts. Different alternative splicing (AS) events were revealed in different inbred lines, even for the same gene, possibly because of organ and developmental specificities or different stresses. The signal strength of splice sites was strongly correlated with selection of donor and receptor sites, and ambiguous junction sites due to small direct repeats at the exon/intron junction frequently resulted in the selection of unconventional splicing sites. Eleven out of 14 transcripts resulting from AS harboured a premature termination codon, rendering them potential candidates for nonsense-mediated RNA degradation. Reverse transcription–PCR (RT–PCR) indicated that zmP4H genes displayed different expression patterns under waterlogging. The diverse transcripts generated from AS were expressed at different levels, suggesting that zmP4H genes were under specific control by post-transcriptional regulation under waterlogging stress in the line HZ32.
Conclusions
Our results provide a framework for future dissection of the function of the emerging zmP4H family and suggest that AS might have an important role in the regulation of the expression profile of this gene family under waterlogging stress.
Keywords: Maize, Zea mays, prolyl 4-hydroxylase, zmP4H, alternative splicing, AS, waterlogging, flooding stress
INTRODUCTION
Depletion of oxygen is a major feature of waterlogging in the roots of plant species (Armstrong, 1980). To elucidate how plants respond to hypoxia, it is important to identify the sensor of oxygen change and the signal cascades involved in waterlogging stress. Despite many studies, the basis for sensing and signalling of hypoxia stress remains unresolved in plants (Bailey-Serres and Chang, 2005).
Previous studies on prolyl 4-hydroxylases (P4Hs; EC 1·14·11·2) under hypoxia in mammals provided clues to the sensing of hypoxia. P4Hs are a large family of oxidoreductases acting on paired donors, with O2 as the oxidant and involving incorporation or reduction of oxygen (Bruick and McKnight, 2001; Epstein et al., 2001). P4H is upregulated in response to hypoxia. It is oxygen dependent and controls a transcription factor [hypoxia-induced factor (HIF)], which is considered to be a global regulator of hypoxia in various organisms, through proline hydroxylation. Proline hydroxylation targets HIF for rapid ubiquitination and proteosomal degradation when oxygen is available (Ivan et al., 2001; Jaakkola et al., 2001). Three HIF-P4H isozymes have been cloned in mammals. Regulation of the response to hypoxia is governed by this family of HIF-P4H enzymes (Bruick and McKnight, 2001; Epstein et al., 2001).
Furthermore, available data suggest that, for HIF-P4H genes, AS is involved in the response to a hypoxic environment, which represents a novel molecular mechanism of gene regulation (Hirsila et al., 2003; Cervera et al., 2006). Changes in the splicing pattern can therefore be expected to influence the amounts of the active enzymes produced (Hirsila et al., 2003). AS is an important post-transcriptional regulatory mechanism that can increase protein diversity and affect mRNA stability. Eighty per cent of human genes have been demonstrated to undergo AS (Modrek and Lee, 2002; Leipzig et al., 2004). Over 20 % of rice genes undergo AS (Campbell et al., 2006; Wang and Brendel, 2006), and about 42 % of transcripts of arabidopsis are alternatively spliced (Filichkin et al., 2010), suggesting that AS is also a common post-transcriptional regulatory mechanism in plant species. Emerging reports suggested that AS events were affected by tissue-specific expression and responded to various stresses (Palusa et al., 2007; Tanabe et al., 2007), suggesting that AS events had a biological role and were evolved to achieve quantitative post-transcriptional regulation (Maquat, 2004).
Recently, the P4H genes have been cloned and functionally characterized in tobacco (Yuasa et al., 2005), tomato (Bucher et al., 1997) and arabidopsis (Hieta and Myllyharju, 2002). Specifically, 13 members of the arabidopsis P4H family were cloned, and some of the members were induced by hypoxia, indicating that P4H genes might be involved in the early response to hypoxia (Vlad et al., 2007). Over-expression of AtP4H1 in arabidopsis led to a phenotype similar to the hypoxia response, including upregulation of several anaerobic genes, which indicated a direct role for AtP4H1 in hypoxia stress (Asif et al., 2009).
There is little information available concerning maize HIF-P4H genes. In this study, a genome-wide analysis of the P4H family was performed in maize. Nine P4H genes were identified. Genes encoding P4H proteins were named according to their location on the chromosomes. Phylogenetic analysis revealed that the maize P4H family could be divided into three subfamilies. For five of the P4H genes, various transcripts were generated by AS. We found that ambiguous exon/intron junction sites resulted from small direct nucleotide repeats, leading to weaker signal strength of splice sites for unconventional sites compared with those for conventional sites. Reverse transcription–PCR (RT–PCR) indicated that different variants had different expression profiles under waterlogging, suggestive of regulation by AS under waterlogging in maize for the first time. Our results provide a framework for future studies of the function of this emerging gene family, and suggest that post-transcriptional regulation by AS might have an important role in the regulation of zmP4H genes under waterlogging stress in plants.
MATERIALS AND METHODS
Database query and identification of sequences encoding P4H proteins
To obtain maize P4H genes, ‘prolyl 4-hydroxylase’ was used as a keyword to query the maize sequence database (www.maizesequence.org). Thirteen P4H sequences reported in arabidopsis (Vlad et al., 2007) were subjected to tblastn searching against the database of expressed sequence tags (ESTs) in PlantGDB (www.plantgdb.org), PUT (PlantGDB-assembled Unique Transcripts) and GSS (Genome survey sequences) for maize, using default parameters. Redundant sequences were removed from the data set by aligning the sequences using ClustalW2 (www.ebi.ac.uk/Tools/clustalw2/). This procedure was repeated with each newly identified set of genes until no further sequences with significant similarity were identified. For each gene, the longest EST was translated using the ORF Finder tool (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), and only those carrying the entire coding sequence (CDS) were chosen for subsequent analyses.
Multiple sequence alignment and phylogenetic analysis of P4Hs
Multiple sequence alignments with default parameters were performed on the obtained sequences of predicted P4H proteins of maize, arabidopsis and humans using ClustalX (version 1·83). Phylogenetic trees were constructed using the Neighbor–Joining (NJ) method in MEGA 4·0. Bootstrap analysis was performed using 1000 replicates to evaluate the reliability of different phylogenetic groups. The trees thus obtained were viewed using TREEVIEW software.
Amplification of zmP4H genes in HZ32
Seeds of HZ32 (a waterlogging-tolerant inbred maize line) were germinated for 3 d and then the seedlings were individually transplanted into sand chambers. Plants were grown under 30 °C/22 °C (light/ dark, 16/8 h) until they initiated three leaves in total, with two leaves expanded. Uniform seedlings were selected and divided into two groups: one group was cultured with a normal water supply as the control and the other was submerged in water with all leaves in the air as the treatment. Roots treated for 1 h were immediately harvested and stored at –70 °C. Roots of the controls were also harvested and treated in the same way.
Total RNA was isolated using TRIzol (Invitrogen, USA) following the manufacturer's recommendations. Total RNA was treated with RNase-free DNase. Reverse transcription of total RNA (5 µg) was performed with an M-MLV RTase cDNA Synthesis Kit (Takara, Japan), following the manufacturer's instructions. Based on the full-length cDNA sequences of putative alternative P4H gene sequences in NCBI, specific primers were designed using PRIMER3 software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and the pair of primers was located in the first and the last exons to cover all the conjunction sites of the genes (Supplementary Data Table S1, available online). For each gene amplified, 1 µL of cDNA was used for the detection of amplified products. The PCR amplification conditions were as follows: 94 °C for 2 min, then 38 cycles of 95 °C for 10 s, 60 °C for 30 s and 72 °C for 90 s, and a final extension period of 72 °C for 10 min to complete the reaction. All PCRs were performed using Takara LA Taq polymerase with GC buffer I (Takara, Japan). The PCR products were purified using an AxyPrep DNA Gel Extraction Kit (Axygen, USA) and cloned into pGEM-T Easy vector (Promega, USA). After transformation, 50 positive clones were picked up and all the clones of different sizes for each gene were sequenced (Invitrogen, USA). Exons and introns in the zmP4H mRNA sequence were analysed using Blast2 at NCBI comparing them with the DNA sequence of B73 (http://www.ncbi.nlm.nih.gov/blast). To confirm the existence of the alternatively spliced transcripts in an independent assay, two biological replications of RNA were used to amplify zmP4H genes in HZ32 under control and treatment conditons, respectively.
RT–PCR analysis in HZ32 under waterlogging
Three biological replications of total RNA were used for RT–PCR. A 1 µL aliquot of first-strand cDNA was used for PCR amplification in a total reaction volume of 20 µL. For each gene, preliminary PCRs were performed using cDNA at different cycles (20, 25, 30 and 35) to determine the linear range of amplification (data not shown). Optimal conditions for the PCR were established in pilot experiments so that linear reaction rates were obtained for each gene. The PCR amplification conditions were as follows: 94 °C for 2 min, then optimal cycles of 95 °C for 10 s, 60 °C for 30 s and 72 °C for 60 s. Gene-specific primers for RT–PCR are listed in Supplementary Data Table S2. Actin and γ-tubulin were used as internal controls. All PCRs were performed using Takara LA Taq polymerase with GC buffer I (Takara, Japan). The amplified PCR products were checked on a 2 % agarose gel containing ethidium bromide. To the expression levels of each variant, primer pairs specific for each transcript bridging the deleted parts of the exons were designed for those alternative transcripts which were visible on the gel.
Quantitative real-time PCR analysis
Three biological replications with two technique replications of total RNA were used for quantitative real-time PCR analysis. First-strand cDNA was synthesized from total RNA with an M-MLV RTase cDNA Synthesis Kit (Takara, Japan). The reactions were carried out using a CFX96 Real-Time System C1000 Thermal Cycler (Bio-RAD, USA) using SYBRGreen PCR Master Mix (Takara, Japan). Primers were designed using PRIMER3 software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and are listed in Supplementary Data Table S3. The expression of actin was used as a control. PCR amplification conditions were as follows: 95 °C for 2 min, then 38 cycles of 95 °C for 10 s, 60 °C for 30 s and 72 °C for 15 s. Relative expression levels were calculated by the comparative CT method. For each gene, expression values of the treatment were normalized against the control.
RESULTS
Identification and analysis of genes encoding P4H in maize
By querying DNA databases and performing bioinformatic analyses, nine non-redundant putative maize P4H genes were obtained and were named relative to their orders on chromosomes. The proteins encoded by the diverse transcripts contained 298–308 amino acids and highly conserved domains, such as the two histidine and aspartate (His-X-Asp) motifs that bind the Fe2+ atom at the catalytic site, and a histidine residue that binds the C-5 carboxyl group of the 2-oxoglutarate (Fig. 1). Interestingly, there were fewer P4H genes in maize than in arabidopsis (Vlad et al., 2007), despite the fact that the size of the maize genome is 2·3 Gb (Schnable et al., 2009), which is much larger than that of arabidopsis.
Fig. 1.
Multiple alignments of zmP4H proteins obtained with ClustalX. Gaps were introduced for optimal alignment. The three Fe2+-binding amino acids, two histidines and one aspartate, the lysine that binds the C-5 carboxyl group of 2-oxglutarate and the serine and arginine in the +2 and +8 positions from the lysine are indicated in bold and boxed.
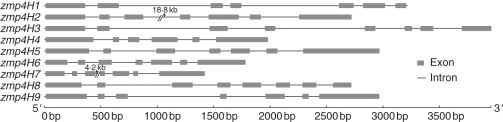
The predicted exon/intron structures were determined by comparing the coding regions of all the zmP4H genes with the B73 genomic sequence. All the coding sequences of the zmP4H genes were disrupted by introns, with the number varying from four to seven (Fig. 2). The smallest intron was only 45 bp and, surprisingly, the largest intron, which appeared to be inserted by a transposon, was 18·8 kb long in zmP4H2. The lengths of the DNA sequences of these genes ranged from 2 to 20·6 kb. The large intron explained the length of the longest gene, zmP4H2.
Fig. 2.
Gene structures of zmP4H genes. Introns and exons are as indicated. The numbers above the introns indicate the length of the introns.
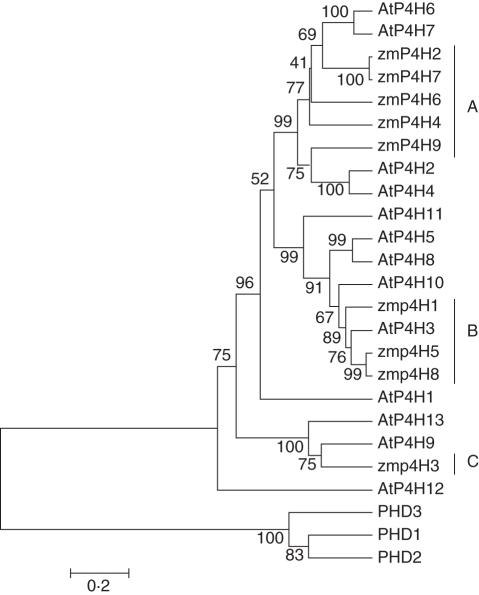
The zmP4H polypeptides share 67–100 % identity, indicating high conservation. This contrasted with the highly divergent proteins, ranging from 18 to 82 % identity, in arabidopsis (Vlad et al., 2007). To evaluate the evolutionary relationships among the genes, phylogenetic trees were constructed with the deduced amino acid sequences of the zmP4H proteins (Fig. 3). Based on phylogenetic analysis, the orthologous relationships can be grouped into three classes, A, B and C, comprising five, three and one member, respectively. The mammalian HIF-P4Hs, as expected, were clustered in a distinct group compared with the plant P4Hs.
Fig. 3.
Neighbor–Joining phylogenetic tree of zmP4H members with deduced P4H amino acid sequences from arabidopsis and humans. The unrooted tree was generated using the MEGA4·0 program by the Neighbor–Joining method. Bootstrap values are shown at each node. The accession numbers for the P4Hs are AtP4H1, AT2G43080; AtP4H2, AT3G06300; AtP4H3, AT1G20270; AtP4H4, AT5G18900; AtP4H5, AT2G17720; AtP4H6, AT3G28490; AtP4H7, AT3G28480; AtP4H8, AT4G35810; AtP4H9, AT4G33910; AtP4H10, AT5G66060; AtP4H11, AT4G35820; AtP4H12, AT4G25600; AtP4H13, AT2G23096; prolyl hydroxylase 1 (PHD1), CAC42509; PHD2, CAC42510; and PHD3, CAC42511.
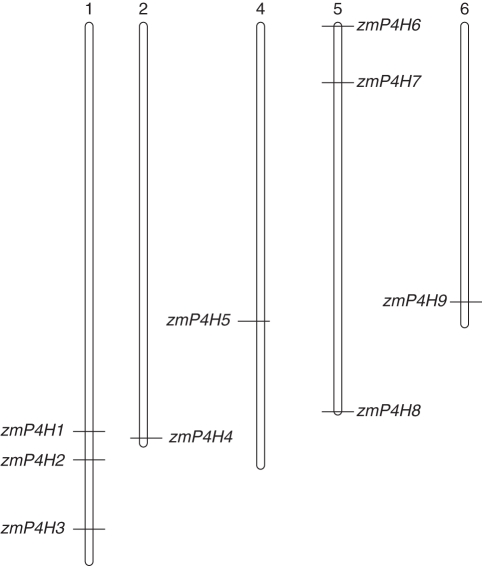
To investigate further the relationship between the genetic divergence within the zmP4H family and gene duplication in maize, the chromosomal location of each zmP4H gene was determined. The results showed that zmP4H genes were located on five chromosomes (Fig. 4), three on chromosome 5 (zmP4H6, zmP4H7 and zmP4H8), three on chromosome 1 (zmP4H1, zmP4H2 and zmP4H3) and one each on chromosomes 2 (zmP4H4), 4 (zmP4H5) and 6 (zmP4H9).
Fig. 4.
Chromosomal distribution of zmP4H genes in maize. The chromosome number is indicated at the top of each chromosome.
AS events in the zmP4H gene family
From the large number of sequences collected from public databases, it was noted that four members of the P4H family showed AS, i.e. zmP4H2, zmP4H4, zmP4H6 and zmP4H8 (Table 1). We then investigated whether AS events exist in the waterlogging-tolerant inbred line HZ32. To confirm the existence of different transcripts derived from a single gene, an RT–PCR experiment was performed with primers located in the first and the last exons, which can indicate the presence of transcripts generated by AS in HZ32. The results showed that some zmP4H genes produced more isoforms than those predicted from EST alignments, suggesting that the limited number of available ESTs/cDNAs did not predict all the AS events for these genes. The abundance of the AS transcripts was very different. Although the AS transcripts were successfully cloned from HZ32, most of them were not visible when the products of RT–PCR were checked on a 2 % agarose gel containing ethidium bromide because of their low abundance. Taking zmP4H8 as an example, one pair of primers was designed based on the results of aligning the transcript of zmP4H8 from GenBank with the genome sequence from B73, and the forward primer was located in the first exon and the reverse primer was located in the last exon. As revealed by the results from the sequencing of clones from PCR products, zmP4H8 and zmP4H8-1 were not obtained from HZ32 and four other variants were identified, i.e. zmP4H8-2, zmP4H8-3, zmP4H8-4 and zmP4H8-5. As shown by the results obtained by checking the RT–PCR products on the agarose gel (Supplementary Data Fig. S1), only zmP4H8-4 (1100 bp) and zmP4H8-5 (1219 bp) were visible, while zmP4H8-2 (600 bp) and zmP4H8-3 (750 bp) were not, although up to 40 cycles of PCR were performed and non-specific bands were obtained.
Table 1.
The sequence resources of zmP4H genes used to analyse the AS event
| Namea | NCBI resourceb | Controlc | Waterloggingd |
|---|---|---|---|
| zmP4H2 | B73 | HZ32 | HZ32 |
| zmP4H2-1 | Hybrid 35A19 | HZ32 | HZ32 |
| zmP4H4 | B73 | HZ32 | HZ32 |
| zmP4H4-1 | B73 | ||
| zmP4H4-2 | B73 | ||
| zmP4H5 | Hybrid 35A19 | HZ32 | HZ32 |
| zmP4H5-1 | HZ32 | HZ32 | |
| zmP4H5-2 | HZ32 | HZ32 | |
| zmP4H6 | Hybrid 35A19 | HZ32 | HZ32 |
| zmP4H6-1 | B73 | ||
| zmP4H6-2 | HZ32 | HZ32 | |
| zmP4H6-3 | HZ32 | HZ32 | |
| zmP4H6-4 | HZ32 | HZ32 | |
| zmP4H8 | B73 | ||
| zmP4H8-1 | Hybrid 35A19 | ||
| zmP4H8-2 | HZ32 | HZ32 | |
| zmP4H8-3 | HZ32 | HZ32 | |
| zmP4H8-4 | HZ32 | HZ32 | |
| zmP4H8-5 | HZ32 | HZ32 |
aNames of genes. bThese sequences were obtained from GenBank and indicate from which inbred line these sequences were produced. c and dThese sequences were confirmed or newly identified in HZ32. cThese sequences could be amplified in the roots of HZ32 under control conditions. dThese sequences could be amplified in the roots of HZ32 under waterlogging.
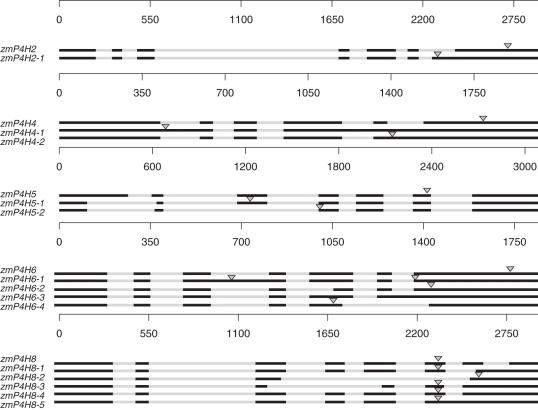
In total, combining the results of amplification in HZ32 and public data from GenBank, we discovered 19 alternatively spliced zmP4H transcripts, representing two, three, three, five and six transcripts for zmP4H2, zmP4H4, zmP4H5, zmP4H6 and zmP4H8, respectively (Fig. 5) and the resource of these transcripts were listed in Table 1. The zmP4H sequences were deposited in GenBank and were assigned the GenBank accession numbers shown in Supplementary Data Table S4. The following analyses of the AS transcripts were based on the data from both GenBank and the results of amplification from HZ32 in this study.
Fig. 5.
AS forms of zmP4H and their exon/intron composition. ZmP4H cDNAs are aligned with the genomic zmP4H DNA. Exons and introns are indicated by boxes and lines, respectively. The arrowheads represents the stop codon.
zmP4H2
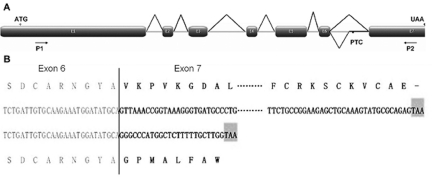
Sequencing of the RT–PCR products revealed two transcripts for zmP4H2 in HZ32, while there was only one transcript in GenBank. Compared with the normal transcript of zmP4H2, zmP4H2-1 retained an extra 135 bp fragment (Fig. 6A). The resulting change in the reading frame inserts a premature stop codon (Fig. 6B) and the deduced protein lacks 95 amino acids, resulting in the loss of the functional domain.
Fig. 6.
Identification of two AS transcripts from the zmP4H2 gene in HZ32. (A) Schematic diagram showing zmP4H2 pre-mRNA and splicing patterns that produce zmP4H2 and zmP4H2-1 transcripts. Also shown are the primer-binding sites for PCR. (B) Comparison of zmP4H2 and zmP4H2-1 nucleotide and amino acid sequences from exons 6 and 7.
zmP4H4
RT–PCR in HZ32 indicated that there was only one mature transcript for zmP4H4, not as many as the three variants identified in B73 (Table 1). The deduced protein sequence from zmP4H4, which was the only mature transcript identified in HZ32, was a normal P4H protein containing a typical domain structure. The primary transcripts of the other two variants were interrupted by unspliced introns. The zmP4H4-2 gene retained an unspliced intron 5. The zmP4H4-1 gene retained three unspliced introns: 1, 4 and 5. Although intron retention lengthened the transcripts of zmP4H4-1 and zmP4H4-2, the transcripts contained premature termination codons producing proteins of 213 and 67 amino acids, respectively.
zmP4H5
There is only one zmP4H5 transcript deposited in GenBank identified from hybrid 35A19; however, two other variants, zmP4H5-1 and zmP4H5-2, were identified in HZ32. Both zmP4H5-1 and zmP4H5-2 have a non-canonical 5′ splice site shifted 256 bp upstream of the normal site in exon 1 and a non-canonical 3′ splice site shifted 24 bp downstream in exon 2. zmP4H5-2 also lacked exon 3 compared with zmP4H5-1. The truncated protein products of zmP4H5-1 and zmP4H5-2 were only 21 amino acids long because of an AS donor site in exon 1.
zmP4H6
There are two variants of zmP4H6 in public databases from different inbred lines, zmP4H6 from hybrid 35A19 and zmP4H6-1 from B73. zmP4H6-1 retained intron 3 compared with zmP4H6. In HZ32, four variants were identified, i.e. zmP4H6 and three other variants: zmP4H6-2, zmP4H6-3 and zmP4H6-4. zmP4H6-2 has a 3′ AS site at intron 4, 101 bp downstream of the normal splice site. Intron 6 was retained in zmP4H6-3. zmP4H6-4 showed a complicated AS phenotype, involving skipping of exon 6, 5′ AS shifted by 45 bp upstream in exon 5 and 3′ AS shifted 53 bp downstream in exon 6. The truncated protein product of zmP4H6-2 resulting from an AS acceptor site lacked 135 amino acids. Although intron retention lengthened the transcripts of zmP4H6-1 and zmP4H6-3, the transcripts contained premature termination codons and produced proteins of 175 and 89 amino acids, respectively. zmP4H6-4 encoded a protein of 146 amino acids, which was shorter by 152 acids compared with zmP4H6.
zmP4H8
Six variants were identified for zmP4H8 based the data from GenBank and the amplification from HZ32. The two sequences from the public databases were identified in hybrid 35A19 (zmP4H8) and in B73 (zmP4H8-1); however, neither of them could be amplified from HZ32. In contrast, four other transcripts were obtained from HZ32 (zmP4H8-2, zmP4H8-3, zmP4H8-4 and zmP4H8-5), which have not been previously deposited in public databases. Compared with zmP4H8, zmP4H8-1 retained intron 7 and had a non-canonical 5′ splice site shifted 256 bp downstream of the normal site in exon 6. zmP4H8-2 was produced by complicated AS, involving skipping of exons 4, 5 and 6, retention of intron 7, 5′ AS 34 bp upstream in exon 3, and 3′ AS 35 bp downstream in exon 7. In zmP4H8-3, exon 4 was skipped, intron 7 was retained, 5′ AS shifted 112 bp upstream in exon 3, and 3′ AS 111 bp downstream in exon 5. Compared with zmP4H8, zmP4H8-4 was produced by retention of intron 7. zmP4H8-5 was produced by retention of introns 6 and 7. The deduced proteins of only three transcripts, zmP4H8-1, zmP4H8-4 and zmP4H8-5, were unaffected by AS: the same amino acid sequences as the reference protein were predicted because the AS was involved with only the 3′-untranslated region of these transcripts. The variants of zmP4H8-2 and zmP4H8-3 were produced by complex AS events, including all four types AS events. The protein sequence deduced from zmP4H8-2 was only 185 amino acids, lacking 123 acids. The transcript of zmP4H8-3 encoded a protein lacking 119 acids.
Splice site strength
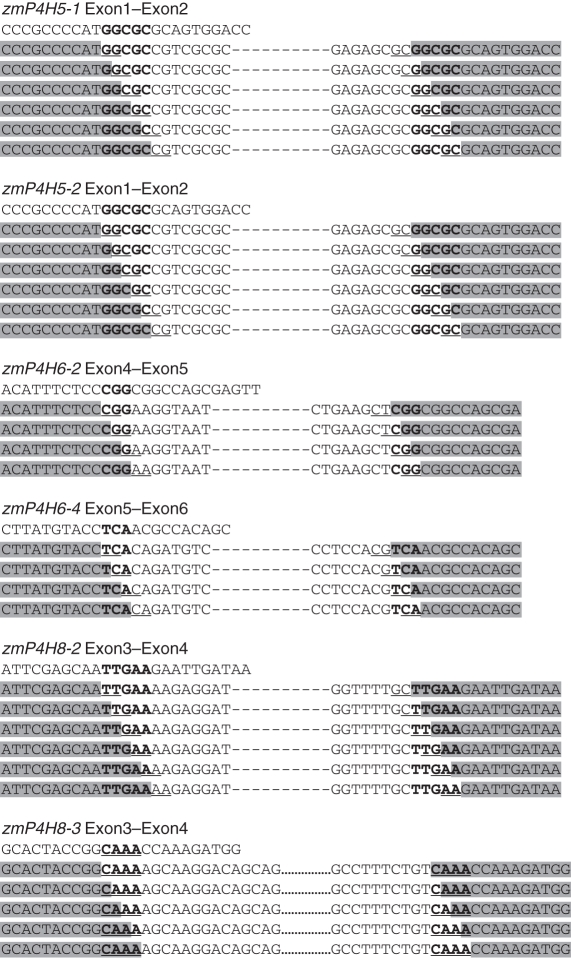
In this study, we analysed sequence elements located at the 5′ and 3′ splice sites in all the transcripts of zmP4H (Supplementary Data Table S4). Among the 122 splice sites analysed, eight splice sites were unconventional sites. The results showed that 106 out of 122 junction sites contained short direct nucleotide repeats (Table 2). The small repeats existed at the junction sites of the normal transcripts, and were all of the GT–AG type (Supplementary Data Fig. S2). However, for six out of the eight unconventional sites, which resulted from splicing in the short direct repeats, their splice sites could not be precisely identified. All possibilities were deduced (Fig. 7).
Table 2.
Small nucleotide repeats at exon/intron junctions
| Gene | Exon 1–2* | Intron 1† | Exon 2–3 | Intron 2 | Exon 3–4 | Intron 3 | Exon 4–5 | Intron 4 | Exon 5–6 | Intron 5 | Exon 6–7 | Intron 6 | Exon 7–8 | Intron 7 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| zmP4H1 | 1 | GT … AG | 0 | GT … AG | 5 | GT … AG | 2 | GT … AG | 0 | GT … AG | 4 | GT … AG | ||
| zmP4H2 | 3 | GT … AG | 2 | GT … AG | 4 | GT … AG | 3 | GT … AG | 3 | GT … AG | 5 | GT … AG | ||
| zmP4H2-1 | 3 | GT … AG | 2 | GT … AG | 4 | GT … AG | 3 | GT … AG | 3 | GT … AG | 3 | GT … AG | ||
| zmP4H3 | 4 | GT … AG | 3 | GT … AG | 2 | GT … AG | 2 | GT … AG | 3 | GT … AG | 6 | GT … AG | 1 | GT … AG |
| zmP4H4 | 3 | GT … AG | 2 | GT … AG | 4 | GT … AG | 3 | GT … AG | 4 | GT … AG | ||||
| zmP4H4-1 | 2 | GT … AG | 4 | GT … AG | ||||||||||
| zmP4H4-2 | 3 | GT … AG | 2 | GT … AG | 4 | GT … AG | 3 | GT … AG | ||||||
| zmP4H5 | 3 | GT … AG | 1 | GT … AG | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | 2 | GT … AG | ||
| zmP4H5-1 | 5 | AS site | 1 | GT … AG | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | 2 | GT … AG | ||
| zmP4H5-2 | 5 | AS site | 1 | GT … AG | 0 | GT … AG | 2 | GT … AG | 2 | GT … AG | ||||
| zmP4H6 | 4 | GT … AG | 2 | GT … AG | 3 | GT … AG | 2 | GT … AG | 3 | GT … AG | 4 | GT … AG | ||
| zmP4H6-1 | 4 | GT … AG | 2 | GT … AG | 2 | GT … AG | 3 | GT … AG | 4 | GT … AG | ||||
| zmP4H6-2 | 4 | GT … AG | 2 | GT … AG | 3 | GT … AG | 3 | AS site | 4 | GT … AG | ||||
| zmP4H6-3 | 4 | GT … AG | 2 | GT … AG | 3 | GT … AG | 2 | GT … AG | 3 | GT … AG | ||||
| zmP4H6-4 | 4 | GT … AG | 2 | GT … AG | 3 | GT … AG | 2 | GT … AG | 3 | AS site | ||||
| zmP4H7 | 3 | GT … AG | 2 | GT … AG | 7 | GT … AG | 3 | GT … AG | 3 | GT … AG | 5 | GT … AG | ||
| zmP4H8 | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | 3 | GT … AG | 0 | GT … AG |
| zmP4H8-1 | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | 2 | GT … AG | ||
| zmP4H8-2 | 2 | GT … AG | 0 | GT … AG | 5 | AS site | ||||||||
| zmP4H8-3 | 2 | GT … AG | 0 | GT … AG | 4 | AS site | 2 | GT … AG | 3 | GT … AG | ||||
| zmP4H8-4 | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | 3 | GT … AG | ||
| zmP4H8-5 | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | 0 | GT … AG | 2 | GT … AG | ||||
| zmP4H9 | 5 | GT … AG | 1 | GT … AG | 4 | GT … AG | 2 | GT … AG | 3 | GT … AG | 3 | GT … AG |
* The numbers of nucleotidesfor the small direct repeats.
† The sequences at the ends of the introns. Bold indicates non-canonical splicing sites and AS site indicates that this splicing site has a small direct repeat and it cannot be precisely identified.
Fig. 7.
All the possibilities of conjunction sites deduced at the non-canonical splice sites. Highlighted sequences are located in the 5′ exon and 3′ exon. The sequences in black and underlined are the small direct nucleotide repeat.
Changes in mRNA levels of zmP4H under waterlogging
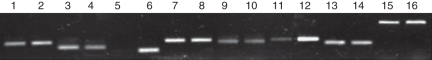
We first examined the effect of waterlogging on the expression of those genes without AS. Only one transcript could be amplified from zmP4H1, zmP4H3, zmP4H4, zmP4H7 and zmP4H9 in HZ32 whether under control or waterlogging conditions in this study. As indicated by RT–PCR (Fig. 8), there was significant induction in the transcript level of a marker gene for waterlogging, Adh1, indicating that our experimental conditions were valid. The differences in expression of zmP4H genes between the control and waterlogging conditions were not significant, except for zmP4H4. Very low expression was detected for zmP4H4 under control conditions, and zmP4H4 was significantly induced by waterlogging (Fig. 8). This analysis revealed that zmP4H1, zmP4H3, zmP4H7 and zmP4H9 did not alter their expression level in response to waterlogging (Fig. 8). To verify the result of RT–PCR of these five zmP4H genes, real-time PCR was carried out in an independent assay. actin-1 was used as a housekeeping gene for the normalization of the expression levels of zmP4H genes. Real-time PCR confirmed the RT–PCR expression pattern for all of the five zmP4H genes (Table 3).
Fig. 8.
The expression pattern of zmP4H genes in HZ32. RT–PCR analysis was performed with control and treated roots of three-leaf seedlings. actin-1 and γ-tubulin were used as an internal control for all the following RT–PCR experiments. Adh1 was used as the marker gene for waterlogging. Lanes 1, 3, 5, 7, 9, 11, 13 and 15 are the amplification results of zmP4H1, zmP4H3, zmP4H4, zmP4H7, zmP4H9, Adh1, actin-1 and γ-tubulin in the control, and 2, 4, 6, 8, 10, 12, 14 and 16 are those of each gene under waterlogging stress. PCR cycles for zmP4H1, zmP4H3, zmP4H4, zmP4H7, zmP4H9, Adh1, actin-1 and γ-tubulin were set at 30, 32, 32, 30, 32, 28, 30 and 32, respectively.
Table 3.
Relative expression profiles of zmP4H genes analysed by real-time PCR in the roots of seedlings of HZ32
| Gene | Ratio |
|---|---|
| zmP4H1 | 1·21 ± 0·05 |
| zmP4H3 | 0·84 ± 0·10 |
| zmP4H4 | 47·53 ± 0·05 |
| zmP4H7 | 1·09 ± 0·07 |
| zmP4H9 | 1·03 ± 0·07 |
| Adh1 | 11·09 ± 0·03 |
The transcript levels of each gene in the roots of seedlings of HZ32 were plotted as the relative expression (fold) of the control seedlings exposed to waterlogging. actin-1 was used as an internal control. Values are means ± s.e.
RT–PCR of AS transcripts of the zmP4H genes was complicated. Most of the AS transcripts were expressed at a very low level under both control and waterlogging conditions, which makes the transcripts difficult to visualize when the products of RT–PCR were checked on the agarose gel with ethidium bromide, even though they could be cloned. A better experimental approach would be real-time PCR to analyse the change in the genes involved with AS; however, there were two impediments to this: (1) the homology between the members of this gene family was very high, sharing 67–100 % identity; and (2) more than one transcript was generated from AS, and these were the same in most regions in the CDS as the normal transcripts, and it is thus difficult to design suitable primers specific to each transcript for real-time PCR. Hence, we performed RT–PCR only for those transcripts for which it was possible.
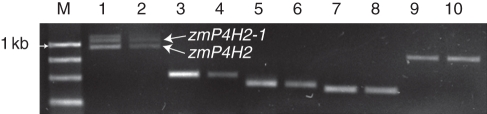
We first examined the effect of waterlogging stress on the expression of zmP4H2 in HZ32 (Fig. 9). The first set of primers, P2-NM-RT-1 and -2, was designed to be specific for zmP4H2 because P2-NM-RT-2 bridged exon 6 and exon 7. It was found that zmP4H2 was decreased under waterlogging. The second set of primers P2-AS-RT-1 and -2 could only amplify zmP4H2-1. The results indicated that zmP4H2-1 was decreased under waterlogging. Furthermore, another pair of primers, P2-5 and -3, located in exon 1 and exon 7, were designed to amplify both zmP4H2 and zmP4H2-1. Both the transcripts were detectable under control and waterlogging conditions. Compared with the control, the zmP4H2/zmP4H2-1 ratio was reduced under waterlogging stress conditions.
Fig. 9.
The expression patterns of variants of zmP4H2 genes in HZ32. RT–PCR analysis was performed with control and treated roots of three-leaf seedlings. Lanes 1 and 2: fragments amplified by primers for both zmP4H2 and zmP4H2-1 for 34 cycles. Lanes 3 and 4: fragments amplified by primers only for zmP4H2 for 32 cycles. Lanes and 5 and 6: fragments amplified by primers only for zmP4H2-1 for 32 cycles. Lanes 7 and 8: actin-1 for 30 cycles. Lanes 9 and 10: γ-tubulin for 32 cycles. Lanes 1, 3, 5, 7 and 9 are the amplification results in the control, and 2, 4, 6, 8 and 10 are those of each gene under waterlogging stress.
zmP4H5 was expressed at a very low level, and the result of real-time PCR amplified for all the variants of zmP4H5 revealed that the abundance of zmP4H5 was estimated to be approx. 25-fold less compared with that of actin under both control and waterlogging conditions. It was not suitable to perform RT–PCR for zmP4H5 to study the expression pattern under waterlogging because of the low expression level. It was also difficult to perform real-time PCR for this gene because there were no suitable primers to distinguish different variants from each other and we were also not able to design primers specific to the functional form only.
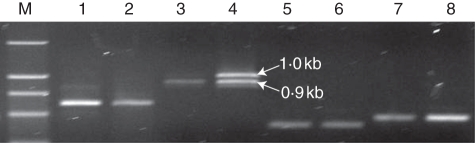
Only one transcript, zmP4H6 (600 bp), was visible in the control using primers designed to amplify all four variants of zmP4H6 (Fig. 10). zmP4H6 was reduced under waterlogging (Fig. 10). Under control conditions, there was only one visible band representing zmP4H8-4 (approx. 900 bp), while under waterlogging, as shown in Fig. 10, zmP4H8-4 was induced and another band could been clearly seen, representing zmP4H8-5 (approx. 1·0 kb), which indicated that zmP4H8-5 was induced in response to waterlogging.
Fig. 10.
The expression pattern of zmP4H6 and zmP4H8 genes in HZ32. RT–PCR analysis was performed with control and treated roots of three-leaf seedlings. actin-1 was used as an internal control. Adh1 was used as the marker gene for waterlogging. M: DL 2000 (Takara, Japan). Lanes 1, 3, 5 and 7 are the amplification results of zmP4H6, zmP4H8, actin-1 and Adh1 in the control, and 2, 4, 6 and 8 are those of each gene under waterlogging stress. PCR cycles for zmP4H6, zmP4H8, actin-1 and Adh1 were set at 33, 33, 28 and 28, respectively.
DISCUSSION
The ability to sense and respond to changes in oxygen concentration represents a fundamental property of all metazoan cells. For over a decade, little progress has been made in determining the genetic and molecular basis of sensing and signalling in response to hypoxia in plants. Research in animals revealed that the transcription factor HIF-1 can be hydroxylated by P4H in response to oxygen limitation (Semenza, 2004). Previous studies revealed that three oxygen-dependent prolyl hydroxylase enzymes [PHD1 (prolyl hydroxylase domain 1), PHD2 and PHD3] control the abundance of HIF in mammals. Two of the three HIF-P4H genes, PHD2 and PHD3, were found to be subject to AS (Hirsila et al., 2003). Furthermore, in the eukaryotic parasitic protist, Perkinsus olseni, transcripts resulting from AS of the Perkinsus PHD2 gene (Leite et al., 2008) were identified. Thus, it was revealed that variants of P4H genes were generated from AS in some animal species. Although P4H genes were isolated from some plant species, they have never been reported to undergo AS in plants. Here, we performed a genome-wide analysis of the P4H gene family in maize. Our phylogenetic analysis of the P4H gene family provides a basis for future functional genomic studies in maize. Interestingly, data from public databases and the results of amplification from HZ32, a waterlogging-tolerant inbred line, suggested that zmP4H genes were alternatively spliced under waterlogging stress.
Unconventional splicing sites were caused by short direct repeats at intron/exon junctions
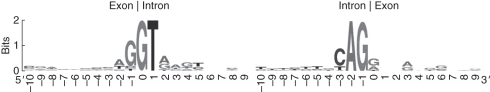
In our study, the assay of 5′ and 3′ sequences flanking conventional splicing sites, as well as unconventional sites, revealed that most of the splicing sites contained short direct nucleotide repeats, and the frequency of the short direct repeats was not biased between conventional and unconventional sites. The existence of small repeats flanking the constitutive splice sites can ensure that the spliceosome accurately recognizes the GT–AG splice signal (Supplementary Data Table S3). The analysis of all splice signals of the constitutively spliced junction sites revealed that there was a conserved consensus sequence at the 5′ conventional splicing site at the exon/intron boundary, TAG/guragu (where Y stands for pyrimidines, R for purines and the slash indicates the exon/intron boundary) (Fig. 11). There was also a YAG consensus sequence at the intron/exon junction at the 3′ splice site. Generally, it is widely assumed that the sequences of exon/intron boundaries (Black, 2003) are conserved in different transcripts, although some junctions are involved in exon skipping or intron retention. On the other hand, the results of the analysis of the regions around the exon/intron junction revealed that, for six out of the eight unconventional sites, the splice sites could not be precisely identified because of short direct repeats. All the possibilities were deduced (Fig. 7) and none of the possible combinations fitted a consensus derived for either U2-type introns, characterized by GT–AG junctions, or U12-type introns, characterized by an NNATCCTN sequence (Patel and Steitz, 2003). It was difficult to identify the precise structure at the exon/intron boundary.
Fig. 11.
Splice site motifs for canonical splicing sites. Numbers on the x-axis represent the sequence positions at exon/intron conjunction sites. The y-axis represents the information content measured in bits. The sequence logos were derived using WebLogo.
When a pre-mRNA is spliced, the spliceosome recognizes splicing signals and catalyses the removal of intronic sequences to ensure accurate gene expression. Distinct transcripts are generated from the same pre-mRNA when different splice sites are chosen (Graveley, 2001). It has been reported that splicing of introns is directed by three main splicing signals: the 5′ splice site (5′ ss) at the 5′ end of the intron, the polypyrimdine tract/3′ splice site (PPT-3′ ss) at the 3′ end of the intron, and a branch site (BS) upstream of the PPT-3′ ss. Alternative splice sites generally show a lower level of sequence conservation in comparison with consensus splice sites (Stamm et al., 2000; Zavolan et al., 2003). Our results revealed that the signal strengths of splice sites participating in unconventional sites in our study were certainly weaker than those of constitutive splice sites for the ambiguous conjunction sites due to the small repeats. It may be one of the underlying mechanisms of AS involved in these sites, because the spliceosome did not recognize the typical sites. This raises another question regarding how these unconventional sites are selected, which requires further study.
AS of P4H genes in maize
In general, AS can be identified by aligning ESTs to genomic DNA and comparing transcripts originating from the same genomic region to identify alternative events. The availability of sequenced genomes and large collections of transcript sequences provides a rich source for identifying AS events by computational methods. The sequences from GenBank representing P4H genes in maize indicated that AS operated in this gene family, which prompted us to investigate whether AS occurs for the zmP4H gene family in HZ32, a waterlogging-tolerant inbred line. The analysis of transcripts collected from public databases and those amplified from HZ32 indicated there were at least five zmP4H genes that were spliced to generate 19 transcripts. This is the first report of AS of P4H genes in maize, or even in plants. AS events from four zmP4H genes were validated, and, in particular, several new transcripts of these genes were isolated in HZ32. These AS events were involved with exon skipping, intron retention and the use of alternative donor or acceptor sites.
To some extent, the transcripts expressed in HZ32 were different from those isolated from other inbred lines (Table 1). The alternatively spliced transcripts detected for zmP4H2, zmP4H5, zmP4H6 and zmP4H8 in HZ32 were not present in the public databases. In contrast, three transcripts generated from zmP4H4 of B73 were deposited in the public database, while only one can be detected in HZ32. Additionally, we found that different inbred lines could generate different transcripts from the same gene, such as for zmP4H6 and zmP4H8. The phenomenon may be partially explained by differences in AS in development stages and stress conditions of the studied materials. The sequences from public databases were identified from various developmental stages, tissues and stress conditions, while the sequences in our study were only obtained from roots of maize seedlings of HZ32 subjected to waterlogging. Numerous studies reported that AS events can be specifically influenced by development and stress (Iida et al., 2004; Morere-Le Paven et al., 2007; Reddy, 2007; Tanabe et al., 2007). Our experiments were designed to focus on the relationship between P4H genes and waterlogging in a tolerant inbred line HZ32, which makes the sequences that we amplified particularly specific for, and restricted to, the material and stress condition studied. Waterlogging is a serious abiotic stress, but relatively few waterlogging-responsive ESTs are deposited in public databases. Thus, we have the chance to detect new transcripts of zmP4H and to enlarge the maize EST database. Our results also indicate that AS of zmP4H is involved in the response to waterlogging. In addition, the depth of the sequence data sets is another factor that impacts on the discovery of AS (Barbazuk et al., 2008). In our study, the limited number of fragments that were sequenced might underestimate the number and kinds of AS transcripts of zmP4H genes.
The response to waterlogging of zmP4H genes in roots of HZ32 seedlings
Gene expression patterns can provide important clues for gene function, and growing evidence indicates that P4H genes are involved in the response to hypoxia in plants. We have previously reported that a P4H gene was induced at the late stage of waterlogging in HZ32 (Zou et al., 2010), so we were interested in analysis of this gene family under waterlogging in HZ32. In this study, when we analysed the EST homology to P4H genes identified from a suppression subtractive hybridization (SSH) library, it was found that these ESTs were located at a homology segment between zmP4H5 and zmP4H8, which aligned at a level of 86 % identity along the entire coding region. Furthermore, these two genes had more than one variant in HZ32 under waterlogging. It was impossible to tell which one was identified from SSH, zmP4H5 or zmP4H8. Consequently, at first, we examined the expression levels of each zmP4H gene in response to waterlogging, which were not involved with AS in HZ32 under waterlogging, including zmP4H1, zmP4H3, zmP4H7 and zmP4H9.
The results showed that zmP4H1, zmP4H3, zmP4H7 and zmP4H9 were constitutively expressed under waterlogging (Fig. 8), implying that they may not be involved in the regulation of the response to waterlogging in HZ32. zmP4H4 was significantly induced under waterlogging, while the expression of zmP4H4 was very low under control conditions, and the abundance of zmP4H4 was almost 30 times lower compared with that of actin1 under control conditions in the roots (data not shown), and was not even visible by RT–PCR checked by agarose gel (Fig. 8). AtP4H7 was induced during the first 3 h under 1·5 % O2 and 5 % O2 (Vlad et al., 2007). In our study, the P4H genes from arabidopsis and maize could be divided into three groups based on the amino acid sequences, and zmP4H4 was clustered with AtP4H7. It is interesting that only zmP4H4 was significantly induced, while the other four zmP4H transcripts were not changed, which may suggest potential roles for zmP4H4 in the waterlogging response in maize.
Variants of zmP4H genes and nonsense-mediated mRNA decay
In this study, four zmP4H genes in HZ32 were involved in AS events, and 11 out of 13 transcripts contained a PTC. A process known as nonsense-mediated RNA degradation (NMD) presumably functions to recognize these mRNAs and degrades them to prevent accumulation of truncated and potentially harmful proteins. All variants containing PTCs were potential targets for NMD. In mammals, transcripts with a PTC located >50 nucleotides upstream of an exon–exon junction are degraded by NMD (Maquat, 2004; Chang et al., 2007). Application of the >50 nucleotide NMD rule to sequenced zmP4H splice variants revealed that only two transcripts contain such a PTC: zmP4H5-1 and zmP4H5-2. However, all the variants including zmP4H5-1 and zmP4H5-2 could be amplified and cloned under both control and waterlogging conditions in this study, indicating that NMD does not remove the PTC-containing transcripts entirely. Other transcripts (zmP4H6-2, zmP4H6-3, zmP4H6-4, zmP4H8-2 and zmP4H8-3), which were presumed to encode truncated proteins, but were not the predicted target of NMD, were detected at a very low level, even not visible on the agarose gel. Interestingly, zmP4H2-1 was detectable at a reasonable abundance in both the control and treatment, and it was decreased under waterlogging. Variants of zmP4H genes containing a PTC or not showed different abundance in HZ32 under control or waterlogging conditions. However, the abundance of a transcript detected in our experiment is the net result of transcription, AS and degradation by NMD; therefore, it was difficult to deduce whether this variant underwent NMD or whether NMD did not remove the PTC-containing transcripts entirely. We cannot exclude the possibility that the NMD system is more complicated and elegant than previously thought, involving unknown mechanisms in addition to the PTC rule.
zmP4H genes are under regulation of AS in the response to waterlogging in HZ32
In our study, although variants generated from AS could be isolated from both the control and treated roots of HZ32, the variants derived from AS including zmP4H2, zmP4H5 and zmP4H8, showed different expression patterns under control and waterlogging conditions in HZ32, indicating that these transcripts were specifically controlled during waterlogging stress. Both zmP4H2 and zmP4H2-1 were downregulated under waterlogging, but the zmP4H2/zmP4H2-1 ratio was reduced slightly under waterlogging stress conditions. For zmP4H8, both zmP4H8-4 and zmP4H8-5 were upregulated in the response to waterlogging, especially zmP4H8-5. AtP4H3 was induced during the first 3 h under 1·5 % O2 and 5 % O2 (Vlad et al., 2007), and zmP4H8 was clustered with AtP4H3. Furthermore, the ratio of variants of zmP4H8 was apparently changed in response to waterlogging. In arabidopsis, it is reported that a disease resistance gene, RPS4, produces multiple transcripts via AS, and the transcript isoform ratios are adjusted to achieve dynamic changes during the resistance response to fine-tune resistance gene activity (Zhang and Gassmann, 2007). Similarly, the relative abundance of the two splicing forms of the arabidopsis SOS4 (salt overly sensitive 4) gene is regulated by salt stress (Shi et al., 2002). In our case, we put forward the hypothesis that the zmP4H2/zmP4H2-1 and zmP4H8-4/zmP4H8-5 ratio might intervene in the control of the response to waterlogging, and AS in zmP4H genes might be useful for acquiring certain adaptive benefits that are important for the response under waterlogging in maize seedlings.
It is clear that the regulation of AS occurs at zmP4H under waterlogging, but its biological roles remain to be elucidated. Our study is only the first step towards understanding the function of zmP4H genes. Further experiments are required to obtain more detailed expression information, especially regarding the regulation of AS during waterlogging. This analysis provides a good starting point for future functional studies.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We sincerely thank Bin Tang, Lei Liu and Fazhan Qiu for their helpful suggestions on writing this paper. This work is supported by the 111 Project (B07041), the Beijing Agricultural Innovative Platform - Beijing Natural Science Fund Program (D08070500690802), National Science Foundation of China (31071428) and National Science Foundation of China (31071429). We also thank two anonymous reviewers for constructive suggestions to improve the manuscript.
LITERATURE CITED
- Armstrong W. Aeration in higher plants. In: Woolhouse HW, editor. Advances in botanical research. New York: Academic Press; 1980. pp. 225–232. [Google Scholar]
- Asif MH, Trivedi PK, Misra P, Nath P. Prolyl-4-hydroxylase (AtP4H1) mediates and mimics low oxygen response in Arabidopsis thaliana. Functional and Integrative Genomics. 2009;9:525–35. doi: 10.1007/s10142-009-0118-y. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Chang R. Sensing and signalling in response to oxygen deprivation in plants and other organisms. Annals of Botany. 2005;96:507–518. doi: 10.1093/aob/mci206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Research. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annual Review of Biochemistry. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Bucher M, Schroeer B, Willmitzer L, Riesmeier JW. Two genes encoding extension-like proteins are predominantly expressed in tomato root hair cells. Plant Molecular Biology. 1997;35:497–508. doi: 10.1023/a:1005869717158. [DOI] [PubMed] [Google Scholar]
- Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. doi:10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera AM, Apostolova N, Luna-Crespo F, Sanjuan-Pla A, Garcia-Bou R, McCreath KJ. An alternatively spliced transcript of the PHD3 gene retains prolyl hydroxylase activity. Cancer Letters. 2006;233:131–138. doi: 10.1016/j.canlet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annual Review of Biochemistry. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Research. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends in Genetics. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Hieta R, Myllyharju J. Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Journal of Biological Chemistry. 2002;277:23965–23971. doi: 10.1074/jbc.M201865200. [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. Journal of Biological Chemistry. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, et al. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Research. 2004;32:5096–5103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Leipzig J, Pevzner P, Heber S. The alternative splicing gallery (ASG): bridging the gap between genome and transcriptome. Nucleic Acids Research. 2004;32:3977–3983. doi: 10.1093/nar/gkh731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite RB, Brito AB, Cancela ML. An oxygen molecular sensor, the HIF prolyl 4-hydroxylase, in the marine protist Perkinsus olseni. Protist. 2008;159:355–368. doi: 10.1016/j.protis.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nature Reviews Molecular Cell Biology. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee C. A genomic view of alternative splicing. Nature Genetics. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Morere-Le Paven MC, Anzala F, Recton A, Limami AM. Differential transcription initiation and alternative RNA splicing of Knox7, a class 2 homeobox gene of maize. Gene. 2007;401:71–79. doi: 10.1016/j.gene.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Palusa SG, Ali GS, Reddy AS. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. The Plant Journal. 2007;49:1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x. [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nature Reviews Molecular Cell Biology. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Reddy AS. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annual Review of Plant Biology. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- Shi H, Xiong L, Stevenson B, Lu T, Zhu JK. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. The Plant Cell. 2002;14:575–588. doi: 10.1105/tpc.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Zhu J, Nakai K, Stoilov P, Stoss O, Zhang MQ. An alternative-exon database and its statistical analysis. DNA Cell Biology. 2000;19:739–756. doi: 10.1089/104454900750058107. [DOI] [PubMed] [Google Scholar]
- Tanabe N, Yoshimura K, Kimura A, Yabuta Y, Shigeoka S. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant and Cell Physiology. 2007;48:1036–1049. doi: 10.1093/pcp/pcm069. [DOI] [PubMed] [Google Scholar]
- Vlad F, Spano T, Vlad D, Daher FB, Ouelhadj A, Kalaitzis P. Arabidopsis prolyl 4-hydroxylases are differentially expressed in response to hypoxia, anoxia and mechanical wounding. Physiologia Plantarum. 2007;130:471–483. [Google Scholar]
- Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proceedings of the National Academy of Sciences, USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa K, Toyooka K, Fukuda H, Matsuoka K. Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. The Plant Journal. 2005;41:81–94. doi: 10.1111/j.1365-313X.2004.02279.x. [DOI] [PubMed] [Google Scholar]
- Zavolan M, Kondo S, Schonbach C, et al. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Research. 2003;13:1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Gassmann W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiology. 2007;145:1577–1587. doi: 10.1104/pp.107.108720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Jiang Y, Liu L, Zhang Z, Zheng Y. Identification of transcriptome induced in roots of maize seedlings at the late stage of waterlogging. BMC Plant Biology. 2010;10:189. doi: 10.1186/1471-2229-10-189. doi:10.1186/1471-2229-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.