Abstract
Background and Aims Eleusine
(Poaceae) is a small genus of the subfamily Chloridoideae exhibiting considerable morphological and ecological diversity in East Africa and the Americas. The interspecific phylogenetic relationships of Eleusine are investigated in order to identify its allotetraploid origin, and a chronogram is estimated to infer temporal relationships between palaeoenvironment changes and divergence of Eleusine in East Africa.
Methods
Two low-copy nuclear (LCN) markers, Pepc4 and EF-1α, were analysed using parsimony, likelihood and Bayesian approaches. A chronogram of Eleusine was inferred from a combined data set of six plastid DNA markers (ndhA intron, ndhF, rps16-trnK, rps16 intron, rps3, and rpl32-trnL) using the Bayesian dating method.
Key Results
The monophyly of Eleusine is strongly supported by sequence data from two LCN markers. In the cpDNA phylogeny, three tetraploid species (E. africana, E. coracana and E. kigeziensis) share a common ancestor with the E. indica–E. tristachya clade, which is considered a source of maternal parents for allotetraploids. Two homoeologous loci are isolated from three tetraploid species in the Pepc4 phylogeny, and the maternal parents receive further support. The A-type EF-1α sequences possess three characters, i.e. a large number of variations of intron 2; clade E-A distantly diverged from clade E-B and other diploid species; and seven deletions in intron 2, implying a possible derivation through a gene duplication event. The crown age of Eleusine and the allotetraploid lineage are 3·89 million years ago (mya) and 1·40 mya, respectively.
Conclusions
The molecular data support independent allotetraploid origins for E. kigeziensis and the E. africana–E. coracana clade. Both events may have involved diploids E. indica and E. tristachya as the maternal parents, but the paternal parents remain unidentified. The habitat-specific hypothesis is proposed to explain the divergence of Eleusine and its allotetraploid lineage.
Keywords: Allotetraploid origin, chloroplast markers, East Africa, Eleusine, low-copy nuclear markers, phylogeny, Poaceae
INTRODUCTION
Hybridization resulting in the formation of allopolyploids is a frequent mode of plant speciation and evolution (Ellstrand et al., 1996; Seehausen, 2004; Paun et al., 2009), and 21·8 % of grass species (Poaceae) are estimated to have arisen as a result of hybridization events (Knobloch, 1968, 1972). Following advances in molecular phylogeny in recent decades, the evolutionary consequences of merging parental genomes into a polyploid nucleus have attracted much attention (Ge et al., 1999; Petersen et al., 2006; Mason-Gamer et al., 2010). Allopolyploid species present substantial challenges to molecular phylogenetic reconstruction. For example, nuclear gene data are more expensive and time-consuming to obtain than plastid DNA (cpDNA) data, and they are difficult to interpret in allopolyploid taxa where genomes have experienced gene recombination, homogenization or copy loss (Wendel, 2000; Petersen et al., 2006). However, low-copy nuclear (LCN) markers have already been successfully used to elucidate the allopolyploid origins of some plants. Recently, LCN markers have provided useful information (e.g. Fortune et al., 2007; Jakob and Blattner, 2010). Here we apply phylogenetic analyses based on two LCN markers and plastid data of Eleusine Gaertn., a genus of the sub-tribe Eleusininae in the C4 grass subfamily Chloridoideae (Peterson et al., 2010).
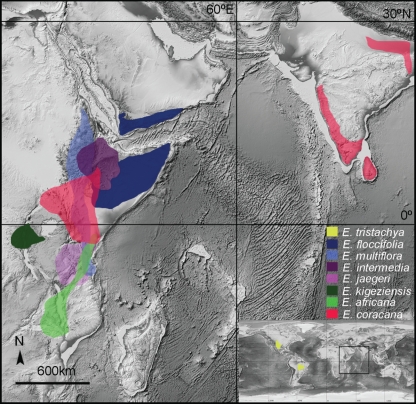
Eleusine is a genus of six diploid (2n = 16, 18 and 20) and three tetraploid taxa (2n = 36 or 38; Phillips, 1995) exhibiting considerable morphological and ecological diversity in East Africa and the Americas, except that the cytologically unknown E. semisterilis S. M. Phillips, known only from the holotype specimen from Kenya (Phillips, 1972), is not included in this study (for the chromosome number and the genome composition see Supplementary Data Table S1, available online). The genus is characterized by having digitate or sub-digitate inflorescences with sub-sessile spikelets arranged along the main axis and 1−16 primary long branches, three-nerved awnless lemmas, and caryopses that have simple verrucate or compound reticulate sculpturing (Liu et al., 2005, 2007; Jiang et al., 2011). The genus is distributed in East Africa and the Americas, and has been introduced to Asia (Fig. 1). Six species are distributed along the eastern branch rift, E. kigeziensis S. M. Phillips is endemic to the western branch rift of East Africa (for definition of two branch rifts see Lærdal and Talbot, 2002), E. tristachya (Lam.) Lam. is endemic to the Americas, and E. indica (L.) Gaertn. has a cosmopolitan distribution (Phillips, 1995; Liu and Peterson, 2010). Over 2 Mt per annum of E. coracana (L.) Gaertn. (finger millet) is produced in Africa, and >4·5 Mt per annum is produced worldwide (National Research Council, 1996). In East Africa, finger millet adapts well to highlands of 1000–2000 m elevation and produces high quality caryopses that are able to withstand frequent drought ((Oduori, 2005). All parts of the finger millet plant provide the raw materials for antiphlogistic medicines used to treat leprosy, liver disease, measles, pleurisy, pneumonia, and smallpox (Duke and Wain, 1981).
Fig. 1.
Natural distribution of Eleusine (unmarked E. indica widespread on four continents except for Antarctica). The background map was downloaded from http://www.ngdc.noaa.gov/mgg/global/global.html (Amante C, Eakins BW. ETOPO1 1 arc-minute global relief model: procedures, data sources and analysis. NOAA Technical Memorandum NESDIS NGDC-24, March 2009).
The taxonomic difficulty of Eleusine is reflected in the controversy of the taxonomic status of E. africana Kenn.-O'Byrne sensu stricto (s.s.), E. coracana s.s., and E. indica. Phillips (1974) recognized two species, E. coracana and E. indica, with ‘africana’ being considered a subspecies of E. indica due to its long, easily shattered spikelets. Phillips (1995) subsequently treated the three taxa as separate species due to the presumed genetic isolation between E. indica and two tetraploid species. This also is the opinion of several other authors (Hiremath and Salimath, 1991; Bisht and Mukai, 2002). More recently, Neves et al. (2005) recognized two species, E. coracana and E. indica, and relegated ‘africana’ as a subspecies of E. coracana, owing to the presumed wild progenitor status of ‘africana’. Therefore, phylogenetic analyses of these taxa are necessary if we are to understand their status and genetic isolation more clearly.
The allotetraploid origin of finger millet has not been satisfactorily explained. Genomic in situ hybridization (GISH) studies identified E. indica (AA genome) and E. floccifolia (Forssk.) Spreng. (BB genome) as candidate progenitors for two tetraploids, E. africana and E. coracana (Bisht and Mukai, 2001a, b). However, phylogenetic analyses of nuclear ribosomal DNA (nrDNA) internal transcribed spacer (ITS) and plastid trnT-trnF sequences contradicted this hypothesis (Neves et al., 2005). In the ITS locus A clade (see Neves et al., 2005), E. indica was confirmed to be closely related to three allotetraploids (E. africana, E. coracana and E. kigeziensis), and this group, in turn, was sister to E. tristachya. However, no diploid was detected as the second genome donor for the B locus. The GISH results provided valuable information with respect to the genetic similarity of chromosomes, but were not useful in elucidating interspecific phylogenetic relationships. Bisht and Mukai (2002) reported similar hybridization signals between E. coracana and two hybrid pairs, E. indica–E. floccifolia and E. tristachya–E. floccifolia, but, so far, the degree of relationship between E. indica and E. tristachya remains unresolved. Plastid sequence similarity between E. indica and E. tristachya suggests that E. tristachya became separated from its ancestor very recently (Neves et al., 2005). Therefore, phylogenetic analyses using additional DNA markers are necessary in order to determine the maternal parents.
Accurate estimates of phylogeny and lineage ages are of critical importance in describing the causative reasons for divergence of genera and allotetraploid lineages. In East Africa, the eastern branch rift became uplifted in the early Miocene, the western branch rift subsequently uplifted in the late Miocene, and these processes continued for both branches throughout the middle Pleistocene (Pallister, 1971; Lærdal and Talbot, 2002). It was proposed that the island-like habitats formed by geological lifting movements facilitated the diversification of plants in East Africa (Hedberg, 1969), such as the mountain-dwelling giant senecios along the eastern branch rift (Knob and Palmer, 1995). Palaeoclimatic studies of East Africa have shown that glacial–interglacial oscillations were characterized by cooler and drier climates during the Pliocene–Pleistocene interval (DeMenocal, 1995; Cerling et al., 1997), and such extreme variations in palaeoclimates might explain the observed divergence in allopolyploid lineages (Stebbins, 1980). However, the paucity of information relating to plant divergence times has impeded our understanding of whether generic diversification was catalysed by palaeotectonic movements. More importantly, the temporal relationships that exist between variation in the palaeoclimate and the divergence of allopolyploid lineages requires investigation.
In this study, we sampled nine extant species of Eleusine within its native distribution range and employed two LCN markers and six plastid markers to explore their evolutionary history. Our study aims to: (1) investigate interspecific phylogenetic relationships within Eleusine; (2) identify the maternal parents and suggest reasons for the equivocal paternal parents of three allotetraploids; and (3) estimate divergence times for Eleusine and its tetraploid lineages, and determine their temporal relationships with palaeoclimate variation in East Africa.
MATERIALS AND METHODS
Taxon sampling
Nineteen accessions were sampled to cover the native distribution range of Eleusine in East Africa and the Americas. Four species of Astrebla F. Muell., Coelachyrum Hochst. & Nees and Vaseyochloa Hitchc. were chosen as outgroups, based on a recent phylogeny of the Chloridoideae (Peterson et al., 2010). Seed germination and seedling cultivation followed the method of Peterson and Annable (1991). Voucher specimens were deposited at four herbaria: IBSC, K, RSA-POM and US (see Supplementary Data Table S1).
DNA extraction, amplification and sequencing
Two LCN markers, phosphoenolpyruvate carboxylase 4 (Pepc4) and eukaryotic elongation factor 1-alpha (EF-1α), were chosen for this study. The Pepc4 gene product catalyses carboxylation of phosphoenolpyruvate to form oxaloacetate and inorganic phosphate during the C4 photosynthetic pathway (Lepiniec et al., 1994), whereas the EF-1α gene product plays a crucial role in the channelling and compartmentalization of protein synthesis in the translational apparatus of higher eukaryotic cells (Negrutskii and El'skaya, 1998). Both of these loci are considered good markers for phylogeny reconstruction. Pepc4 has been successfully used to reconstruct the phylogeny of Poaceae (Christin et al., 2008), and a preliminary screen of both regions indicated no gene homogenization (Q. Liu et al., unpubl. res.). The two LCN markers also appear to represent independent phylogenetic estimates due to their locations on different chromosomes. The Pepc4 gene is mapped on Oryza group 1 homologous chromosomes, whereas the EF-1α gene is mapped on Oryza group 3 homologous chromosomes (Sasaki et al., 2002; The Rice Chromosome 3 Sequencing Consortium, 2005).
The primer combination Pepc4-8F (5′-ACAACCCTGCGTGCCATC-3') and Pepc4-10R (5'-GGAAGTTCTTGATGTCCTTGTCG-3') was designed for Pepc4 based on sequences derived from Cynodon dactylon (L.) Pers. and Tragus racemosus (L.) All. Primers EF-1a-2F (5'-CATTGACTCCACCACTGGT-3') and EF-1a-3R (5'-TAACGGGCCTTGGAGTACTT-3') were designed for EF-1α based on sequences from Hordeum vulgare L., Oryza sativa L. and Saccharum officinarum L.
All procedures were performed in the Laboratory of Analytical Biology (LAB) at the Smithsonian Institution. Total genomic DNA was extracted from silica-dried leaves using DNeasy Plant Mini Kits (Qiagen). PCR amplifications were performed using 2 µL of genomic DNA (at 10 ng μL−1), 0·6 µL of each primer (at 10 pmol), 2·0 µL of dNTPs (at 10 mm), 1 µL of dimethylsulfoxide, 1 µL of MgCl2 (at 2·5 mm), 1 U of Taq polymerase (Bioline), 2·5 µL of 10× Bioline Taq polymerase buffer and 0·5 µL of bovine serum albumin (at 10 mg mL −1) in a volume of 25 µL under the following conditions: 95 °C/3 min, 35× (94 °C/30 s, 53 °C/30 s, 72 °C/80 s), 72 °C/10 min, ending with 4 °C holding.
Amplified PCR products were purified using the PEG method (Hiraishi et al., 1995). The cycle sequencing reactions were conducted in 10 µL volumes containing 0·25 µL of BigDye 3·1, 0·5 µL of primers, 2·0 µL of purified PCR products and 1·75 µL of sequencing buffer. The sequencing reactions were run on an ABI PRISM 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Direct sequencing revealed two homoeologues for four accessions of the three tetraploids. Therefore, their purified PCR products were cloned into pCR®4-TOPO vectors and transformed into Escherichia coli TOP10 competent cells in accordance with the protocol for the TOPO TA cloning kit (Invitrogen). Fragments obtained from white clones were amplified using the original PCR primers. Accessions with unique substitutions were repeatedly sequenced to avoid amplification error. GenBank accession numbers are presented in Table S1.
Data analyses
Sequencher 4·5 (Gene Codes Corporation, 2005) was used to evaluate chromatograms for base confirmation and to edit contiguous sequences. Sequences were initially aligned with Muscle (Edgar, 2004), followed by manual adjustments using Se-Al v. 2·0a11 (Rambaut, 2007). Several chimeric sequences, identified by the repeated sequencing of the same individual and closer inspection of alignments prior to analysis, were excluded from phylogenetic analyses (Cronn et al., 2003).
Maximum parsimony (MP) searches were performed with 1000 random addition sequence replicates, followed by tree bisection–reconnection (TBR) branch swapping, MulTrees option in effect and character state changes weighted equally in PAUP* v. 4·0b10 (Swofford, 2003). The gaps of the sequence matrix were treated as missing data. The bootstrap percentages for support (PB) of internal nodes were obtained with 1000 replicates. In each replicate, we performed ten random sequence addition replicates followed by the TBR swapping algorithm and saved all trees in each replicate (Felsenstein, 1985). Maximum likelihood (ML) analysis was implemented in GARLI v. 0·951 (Zwickl, 2006), starting from random trees and using 10 000 000 generations per search. The ML bootstrap (LB) support was estimated from 1000 bootstrap replicates in GARLI. The output file containing the best trees for bootstrap reweighted data was then read into PAUP*, where the majority-rule consensus tree was constructed and bootstrap support values were calculated. For ML analysis, the substitution model was identified under the Akaike Information Criterion (AIC) implemented in Modeltest v. 3·7 (Posada and Crandall, 1998). Bootstrap values of 90–100 were interpreted as strong support, 70–89 as moderate and 50–69 as weak (Mason-Gamer and Kellogg, 1996). Bayesian posterior probabilities (PP) were estimated using MrBayes v. 3·1·2 (Ronquist and Huelsenbeck, 2003). Two independent runs of Bayesian Markov Chain Monte Carlo (MCMC) analyses were conducted simultaneously, each run having one cold chain and three incrementally heated chains, all starting from a random tree and sampling one out of 1000 generations. Tracer v. 1·5 was used to check the chain stationarity (Rambaut and Drummond, 2007). The first 10 % of trees were discarded as burn-in, and the remaining trees were used to calculate the PP. The sequence and tree statistics of two LCN markers are presented in Table 1.
Table 1.
Sequence and tree statistics of two LCN markers used in the study
| LCN marker | Aligned full sequence length (bp) | GC% | Coding region length (bp) | Proportion of coding region (%) | No. of variable sites | No. of informative characters | Proportion of informative characters (%) | Ti/Tv | Tree length | CI | RI | Model selected by AIC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pepc4 | 1047 | 54·3 | 793 | 75·7 | 271 | 171 | 16·3 | 1·37 | 350 | 0·85 | 0·96 | TVM + G |
| EF-1α | 1143 (remainder 954) | 37·0 | 101 | 10·6 | 561 | 534 | 56·0 | 0·97 | 1189 | 0·75 | 0·92 | HKY + I + G |
We rooted the Pepc4 tree using species of Astrebla and Coelachyrum as outgroups and rooted the EF-1α tree using species of Vaseyochloa as outgroups because we could not get clean EF-1α sequences for Astrebla and Coelachyrum in the laboratory. The appropriate choice of outgroups was confirmed by phylogenetic proximity (the monophyletic ingroup being supported), genetic proximity (short branch length being observed) and base compositional similarity (ingroup-like GC%, Table 1) (Rota-Stabelli and Telford, 2008).
Divergence time estimates
Because divergence time estimates using cpDNA data are able to avoid potential errors caused by heterogeneity of substitution rates or the existence of paralogous copies of nuclear markers for allopolyploid lineages (Renner, 2005), six plastid markers (ndhA intron, ndhF, rps16-trnK, rps16 intron, rps3, and rpl32-trnL) were amplified using published primers (Peterson et al., 2010) for eight accessions representing eight species, except for E. indica (downloaded from GenBank). The six amplified plastid sequences were then added to an alignment matrix derived from 268 species (Peterson et al., 2010) and five species of Centropodieae in the Chloridoideae (Peterson et al., 2011) in order to estimate the divergence times of Eleusine and its tetraploid lineages.
In order to evaluate the molecular clock assumption for our data, likelihood scores for clock and non-clock models were compared using a likelihood ratio (LR) test (Felsenstein, 1981). The LR test was calculated as 2× (ln Lclock – ln Lnonclock) and assumed to follow a χ2 distribution, with the number of degrees of freedom (n) equal to the number of terminals minus two. The assumption of rate constancy was rejected for this study because the constrained and unconstrained analyses differed significantly (LR = 1647·2395, d.f. = 279, P = 0). A Bayesian method, which allows a relaxed evolutionary model to estimate divergence times, was then employed.
The Bayesian analysis was conducted in BEAST v. 1·5·3 (Drummond and Rambaut, 2007), which employed a Bayesian MCMC to co-estimate topology, substitution rates and node ages. BEAUti was used to set criteria for the analysis. The AIC estimated by Modeltest 3·7 (Posada and Crandall, 1998) was used to determine which nucleotide substitution model best fit our data. The oldest C4 lineage in Chloridoideae was dated to be 32·0 million years ago (mya) (Christin et al., 2008; Vicentini et al., 2008), and thus the crown age of the subfamily was set at 32·0 mya since isotopic surveys provide no evidence for an older date of C4 grass origin so far (Osborne and Beerling, 2006). In addition, an uncorrelated lognormal model of rate variation among branches in the tree was assumed and a Yule prior on the birth rate of new lineages employed (Drummond et al., 2006). Posterior distributions of parameters were calculated by two independent MCMC analyses of 20 000 000 generations with 10 % burn-in. Results were checked in Tracer to ensure that plots of both analyses converged on the same area and then were combined using TreeAnnotator v. 1·5·2 (part of the BEAST package). Final trees were checked and edited in FigTree v. 1·3·1 (Rambaut, 2009), and the divergence times were shown as the mean and the 95 % highest posterior density (HPD) in millions of years.
RESULTS
Analysis of Pepc4 sequences
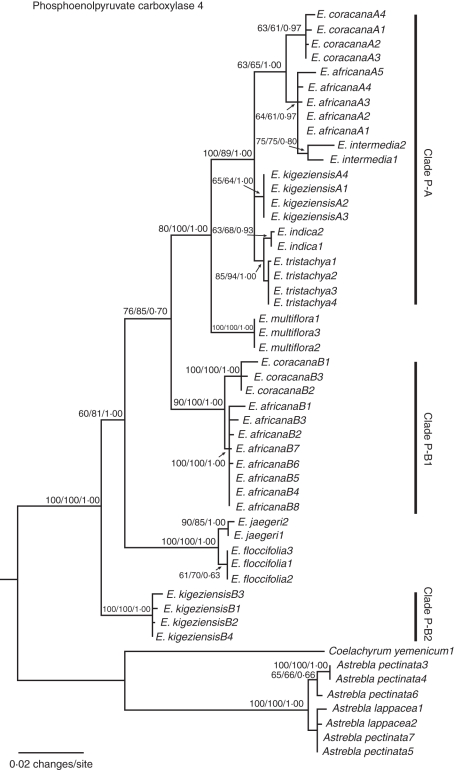
The matrix of Pepc4 comprises 1047 characters, including partial exon 8, complete exon 9, partial exon 10, intron 8 and intron 9 at a length of 71, 531, 181, 131 and 133 bp, respectively. Among the 1047 characters, 171 are parsimony-informative (16·3 %) (Table 1). The log likelihood scores of 56 substitution models range from 3520·6231 to 3599·8337, and Modeltest indicates that the best fit model under AIC is TVM + G. The ML tree has the same topology as the MP strict consensus tree and the Bayesian majority-rule consensus tree. The ML tree is presented in Fig. 2, along with bootstrap support values from MP and ML analyses and posterior probabilities from the Bayesian analysis. The monophyly of Eleusine was strongly supported by the three support indices (PB = 100 %, LB = 100 %, PP = 1·00).
Fig. 2.
Maximum likelihood phylogeny of Eleusine inferred from the nuclear Pepc4 data. Numbers above nodes indicate bootstrap values obtained from parsimony and maximum likelihood analyses, and Bayesian posterior probabilities.
A single sequence type for the Pepc4 locus was identified for all diploid species of Eleusine (Fig. 1). These sequences formed three monophyletic groups: the E. indica–E. tristachya clade (PB = 85 %, LB = 94 %, PP = 1·00); the E. multiflora clade (PB = 100 %, LB = 100 %, PP = 1·00); and the strongly supported E. floccifolia–E. jaegeri clade (PB = 100 %, LB = 100 %, PP = 1·00). Two Pepc4 sequence types (A- and B-) were identified in one accession of E. africana and E. coracana, and in two accessions of E. kigeziensis, consistent with tetraploidy. These sequences fell into three distinct groups. Clade P-A (PB = 100 %, LB = 89 %, PP = 1·00) contains a sub-clade with E. intermedia and the A-type sequences of E. africana and E. coracana (PB = 63 %, LB = 65 %, PP = 1·00), the A-type sequences of E. kigeziensis (PB = 65 %, LB = 64 %, PP = 1·00) and the E. indica–E. tristachya clade (PB = 85 %, LB = 94 %, PP = 1·00). Clade P-B1 contains B-type sequences of E. africana and E. coracana (PB = 90 %, LB = 100 %, PP = 1·00). Clade P-B2 contains a monophyletic clade and contains B-type sequences of E. kigeziensis (PB = 100 %, LB = 100 %, PP = 1·00). Clade P-A is sister to E. multiflora, and this group, in turn, is sister to clade P-B1 (PB = 76 %, LB = 85 %, PP = 0·70). Clade P-A + E. multiflora + clade P-B1 is sister to the E. jaegeri–E. floccifolia clade, while clade P-B2 (E. kigeziensis) is sister to the remaining species of Eleusine (Fig. 2).
Analysis of EF-1α sequences
The matrix of EF-1α comprises 1143 characters, including partial exon 2, partial exon 3 and complete intron 2 at a length of 78, 23 and 1042 bp, respectively. An ambiguous region (positions 235−423) in intron 2 is excluded from phylogenetic analyses. Among the remaining 954 characters, 534 are parsimony-informative (56·0 %) (Table 1). The log likelihood scores of 56 substitution models range from 5882·0400 to 6096·7363, and Modeltest indicates that the best fit model under AIC is HKY + I + G. The ML tree has the same topology as the MP strict consensus tree and the Bayesian majority-rule consensus tree. The ML tree is presented in Fig. 3, along with bootstrap support values from MP and ML analyses and posterior probabilities from Bayesian analysis. The monophyly of Eleusine is strongly supported by the three support indices (PB = 100 %, LB = 100 %, PP = 1·00).
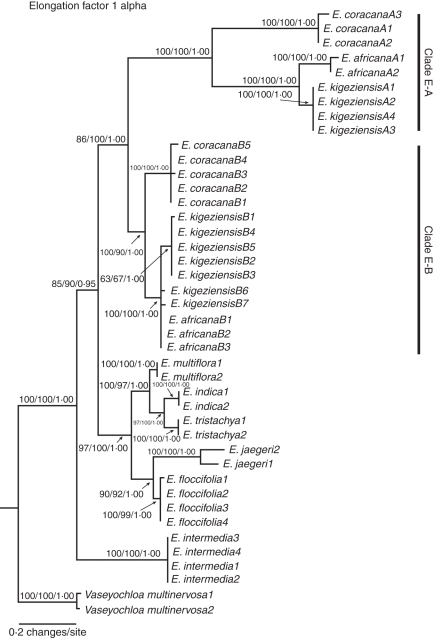
Fig. 3.
Maximum likelihood phylogeny of Eleusine inferred from the nuclear EF-1α data. Numbers above nodes indicate bootstrap values obtained from parsimony and maximum likelihood analyses, and Bayesian posterior probabilities.
A single sequence type for the EF-1α locus was identified for all diploid species of Eleusine. These sequences formed three monophyletic groups: the E. multiflora and E. indica–E. tristachya clade (PB = 100 %, LB = 97 %, PP = 1·00); the strongly supported E. floccifolia–E. jaegeri clade (PB = 90 %, LB = 92 %, PP = 1·00); and the E. intermedia clade (PB = 100 %, LB = 100 %, PP = 1·00). The EF-1α sequences of the three allotetraploids diverged from diploid lineages. As with Pepc4, two sequence types (A- and B-) were recovered from one accession of E. africana and E. coracana, and two accessions of E. kigeziensis. These sequences fell into two distinct groups (Fig. 3). Clade E-A contains the A-type sequences of E. coracana and sister E. africana and E. kigeziensis (PB = 100 %, LB = 100 %, PP = 1·00), while clade E-B contains the B-type sequences of E. coracana, E. africana and E. kigeziensis (PB = 100 %, LB = 90 %, PP = 1·00). Clade E-A is sister to clade E-B (PB = 86 %, LB = 100 %, PP = 1·00), and together they are sister to a clade containing E. multiflora + the E. indica–E. tristachya clade + the E. floccifolia–E. jaegeri clade (PB = 97 %, LB = 100 %, PP = 1·00). Eleusine intermedia is sister to all remaining species of Eleusine.
The A-type sequences of the three allopolyploids possess three characters: a large number of variations regularly distributed along intron 2 (e.g. notice the long branch length in Fig. 3); clade E-A distantly diverged from clade E-B and other diploid species (Fig. 3); and seven deletions (6–12 bp in length) in intron 2, implying the likelihood of gene divergence after speciation.
Divergence times
A combined matrix of six plastid markers from 281 accessions comprises 6737 characters, of which 2129 are parsimony-informative (31·6 %). Modeltest indicates that the best fit model under AIC is GTR + G. Divergence times for nodes in the phylogeny are shown in Fig. 4.
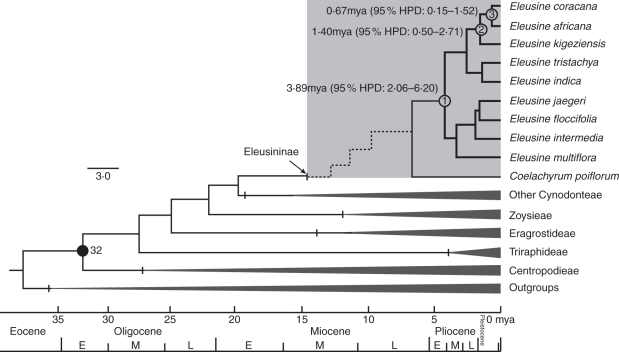
Fig. 4.
Bayesian divergence time estimates of Eleusine (shaded) based on the six combined plastid gene markers (ndhA intron, ndhF, rps16-trnK, rps16 intron, rps3, and rpl32-trnL) of Chloridoideae. Clade constraint is marked by the black circle. An interrupted line indicates the origin of Eleusine from sub-tribe Eleusininae. Node numbers 1, 2 and 3 indicate the crown age of Eleusine, allotetraploid clade and E. coracana, respectively. Numbers in parentheses indicate 95 % highest posterior density (HPD) intervals.
Using the combination of six plastid DNA markers calibrated with the constrained calibration point of the oldest C4 lineage in Chloridoideae, the crown age of Eleusine was determined to be 3·89 (95 % HPD: 2·06–6·20) mya in the Miocene–early Pliocene interval. The crown age of allotetraploid lineage was 1·40 (95 % HPD: 0·50–2·71) mya in the middle Pleistocene. The divergence of E. coracana was estimated to have occurred 0·67 (95 % HPD: 0·15–1·52) mya in the late Pleistocene (Fig. 4). In the BEAST topology, three tetraploids and the diploid E. indica–E. tristachya clade comprised a monophyletic lineage, and then the lineage was sister to a clade that included E. floccifolia, E. jaegeri, E. intermedia and E. multiflora.
DISCUSSION
Interspecific phylogenetic relationships in Eleusine
A sister relationship between E. indica and E. tristachya was strongly supported by Pepc4, EF-1α and cpDNA data sets (Figs 2–4). Biochemical and genetic evidence also supports this relationship: the two species have similar karyotypes (Hiremath and Chennaveeraiah, 1982), sub-equal 2C DNA amounts of 2·9 pg (Hiremath and Salimath, 1991), high Rogers' similarity value of isozyme S = 0·794 (Werth et al., 1994) and a shared intergenic spacer region (IGS) phenotype in ribosomal DNA (rDNA) (Hilu and Johnson, 1992). The close relationship between E. indica and E. tristachya in the chronogram suggests that the arrival of E. tristachya in the Americas was recent (Fig. 4). Therefore, the disjunctive distribution of Eleusine is best explained in terms of long-distance dispersal. The hypothesis that the American E. tristachya had separated from its African ancestor in ancient times is not supported by our data since it appears to be of recent origin (Hilu and Johnson, 1997).
A sister relationship between E. floccifolia and E. jaegeri also received strong support from our data. Similarly, biochemical and genetic characters have been useful for interpreting phylogenetic relationships between the two species, including the possession of isozyme gene Idh-2 duplicated loci (Werth et al., 1993), a sub-equal 2C DNA amount of 3·3 pg (Hiremath and Salimath, 1991) and highly variable IGS phenotypes in rDNA (Hilu and Johnson, 1992). Apparent morphological differences between the two species could be due to elevation-related phenotypic plasticity. Eleusine floccifolia is unique in forming less robust tufts with slender culms and leaf blades with scattered tufts of soft white hairs; the species is distributed in seasonal climate highlands at moderate elevations (about 1800 m) in Ethiopia, Kenya, Somalia and Yemen (Phillips, 1972; Lovett, 1993). Eleusine jaegeri is easily recognized since it forms a very robust tussock with fasciculate-branched culms and has glabrous leaf blades (Phillips, 1972, 1995); the species is restricted to extremely arid regions at moderate to high elevations (1800–3200 m) in Tanzania and Kenya (Fig. 1). The phylogenetic lineage thus exemplifies the modern synthetic evolution theory: change is based on a combination of chance and natural selection that affects phenotype and genetic characters unequally (Stebbins, 1986).
The taxonomic separation of E. africana s.s., E. coracana s.s. and E. indica is supported by our study. Eleusine indica was reported to be a diploid species (Moinuddin et al., 1994), and only one type of Pepc4 and EF-1α sequence was detected from the sampled accessions. Thus, it appears that E. indica is genetically distant from the two allotetraploids (Chennaveeraiah and Hiremath, 1974). Two types of Pepc4 and EF-1α sequences were identified from both E. africana and E. coracana accessions. In the Pepc4 phylogeny, A-type sequences of E. coracana grouped with A-type sequences of E. africana and E. kigeziensis, E. intermedia, E. indica and E. tristachya (i.e. clade P-A), while B-type sequences of E. coracana grouped with B-type sequences of E. africana in clade P-B1 (Fig. 2). In the EF-1α phylogeny, both A- and B-type sequences of E. coracana grouped with the A- and B-type sequences of E. africana and E. kigeziensis, respectively (Fig. 3). The relationships among allotetraploids were incongruent between two nuclear gene trees. A possible interpretation of the conflicting pattern was that a hybridization event was followed by speciation at the polyploid level, producing two species of the allopolyploid clade (E. africana and E. coracana). Furthermore, the recurrent gene flow might have occurred between E. kigeziensis and the lineage of E. africana and E. coracana (Kellogg et al., 1996; Emshwiller and Doyle, 1998). Contrasting phylogenetic signals in the IGS region (rDNA) occur between E. coracana and E. africana, and it is evident that finger millet is domesticated from a limited number of wild populations due to a less variable IGS region (Hilu and Johnson, 1992). This finding indicates that finger millet had a more narrow genetic base than E. africana, while the unique simple verrucate sculpturing of finger millet caryopses highlighted a rapid process aided by domestication (Paterson et al., 2004; Jiang et al., 2011). Therefore, recognition of these three taxa at the species level as opposed to placing E. africana as a subspecies either of E. indica or of E. coracana is compatible with our results (Phillips, 1995).
Allotetraploid origin
As inferred from the results of two sets of molecular markers (plastid and Pepc4), three tetraploids are of allopolyploid origin. In the plastid phylogeny, all three allotetraploids share a common ancestor with the E. indica–E. tristachya clade, which represents a source for the maternal parents (Fig. 4). In the Pepc4 phylogeny, two homoeologous loci were isolated from each of the allotetraploids, providing strong evidence for the presence of two divergent genomes in each allotetraploid. The maternal lineage identified by plastid phylogeny is also confirmed by the A homoeologues of three allotetraploids in Fig. 2 (clade P-A). However, the EF-1α tree shows that the collected gene copies may be derived through a gene duplication event, but the sets of gene copies being derived from the allopolyploidization event (suggested by the Pepc4 tree) may be degraded or not collected at all. With the exception of E. intermedia, the diploid species formed a single lineage separating from the remaining species in the EF-1α phylogeny, thus providing indirect evidence that hardly supports the species as a potential source for the maternal parents.
The Pepc4 data support independent allotetraploid origins for E. kigeziensis and the E. africana and E. coracana clade. Based on cpDNA evidence, both events may have involved species from the E. indica–E. tristachya clade as the maternal parents, but the paternal parents involved in the original hybridization events remain unknown. We propose three hypotheses to explain why the paternal parents remain unidentified: (1) the paternal parents of E. kigeziensis may come from outside of Eleusine, and thus they remain unidentified due to restricted sampling at the intergeneric level; (2) genomes derived from paternal parents may have undergone a rapid evolutionary divergence (or degradation) after allotetraploid speciation and are therefore untraceable; or (3) the paternal parents are extinct in the wild.
For the E. africana and E. coracana clade, the Pepc4 and nrDNA ITS data supported its location nested within Eleusine, suggesting that their paternal parents should be members of the genus, but the earliest diverging clade P-B2 in Pepc4 phylogeny indicated the possibility of B homoeologues of E. kigeziensis derived from intergeneric hybridization (hypothesis 1). It would be worthwhile to test the hypothesis with targeted sampling among allied genera (Peterson et al., 2010). The second proposal provides evolutionary scenarios to reconcile conflicting patterns between two LCN trees. The clade P-B1 is sister to clade P-A + E. multiflora in the Pepc4 tree; the similar branching pattern is shared by the Waxy tree (Q. Liu et al., unpubl. res.) for the lineage of E. africana and E. coracana, and so its paternal parents must be members of the genus. However, the long branch of clade E-B in the EF-1α tree implies its derivation from a gene duplication event. The paternal gene copies derived from the allopolyploidization event (suggested by the Pepc4 tree) are diverged so greatly that they are too rare to find even after extensive cloning of the EF-1α gene (Wendel, 2000; Petersen et al., 2006). In constrast to EF-1α phylogeny, sequence variation of clade P-B2 reflects complex evolutionary scenarios for the genome origin following the allopolyploidization. Furthermore, the third hypothesis is probably reasonable because the B homoeologues of E. kigeziensis, E. africana and E. coracana do not correspond to any diploid taxon sampled for chloroplast, LCN or a previous nrDNA ITS phylogeny (Neves et al., 2005). Since the genus has been thoroughly studied throughout its natural range in East Africa and the Americas, the only unsampled E. semisterilis in this study seems an unlikely candidate for paternal parents due to its unusual morphology of laxly arranged spikelets that disarticulate as a unit (vs. between the florets for all other species in the genus; Phillips, 1972). The natural distribution of E. coracana also excludes the possibility of a genetic contribution by extra-African diploid taxa. Therefore, the extinction hypothesis is reasonable to explain the intriguing parental origins for the two hybridization events.
Palaeoenvironmental hypothesis for divergence of genus and allotetraploid lineage
The habitat-specific hypothesis might apply to the current species distribution pattern within Eleusine. This premise could account for the divergence of species within Eleusine since phenotypic traits, such as racemes terminating in a fertile spikelet and hypervariable sculpturing of the caryopsis surface, are apparently adaptations to seasonal rainfall in East Africa (Stebbins, 1986; Liu and Peterson, 2010; Jiang et al., 2011). East African summer rainfall has been highly variable due to rain shadow effects, which were shaped by the topographic barrier of the eastern branch rift to moist, maritime air from the Indian Ocean (Sepulchre et al., 2006). The Kenyan and Ethiopian highlands capture moisture, and summer monsoonal run-off favours the rise and prosperity of C4 plants (e.g. Eleusine) in habitat-specific mountain savannas. The same premise (habitat-specific hypothesis) might account for the divergence of the allotetraploid lineages (e.g. E. africana, E. coracana and E. kigeziensis). The geological records support the claim that East African climates had changed from warmer and wetter at the Miocene–early Pliocene interval to cooler and drier during the glacial–interglacial oscillations of the Pliocene–Pleistocene interval (DeMenocal, 1995; Cerling et al., 1997). The divergence time of the allotetraploid lineage at 1·40 (95 % HPD: 0·50–2·70) mya falls into the interval of 1·6–1·8 mya, when increasing amplitudes of palaeoclimate variability induced plant speciation, adaptation and evolution (Vicentini et al., 2008; Edwards et al., 2010). Widespread interspecific gene exchange might have occurred during hybridization during the allopolyploidization process (Wakeley and Hey, 1997; Emshwiller and Doyle, 1998). The periodic increases in drier conditions in East Africa during the periods of 1·6–1·8 and 0·8–1·2 mya might have established discrete opportunities for ecological fragmentation and genetic isolation, leading to rapid diversification of allotetraploids adapted to mountain savannas at elevations <3200 m (DeMenocal, 1995).
Conclusions
Our molecular results based on plastid and LCN markers support the premise that E. africana, E. coracana and E. kigeziensis (tetraploid species) are of allopolyploid origin. Our results support two separate allopolyploidization origins for E. kigeziensis and for the E. africana–E. coracana clade. Both events may have involved the diploid E. indica–E. tristachya clade as the maternal parent, but the paternal parents are unknown. The most likely explanation for the difficulty in determining the paternal parents of three tetraploids is that they no longer exist. Our study has identified two promising sister lineages for further study, i.e. the E. indica–E. tristachya clade and the E. floccifolia–E. jaegeri clade. The habitat-specific hypothesis proposes that the divergences of the allotetraploid lineage within Eleusine in East Africa may be associated with habitat-specific mountain savannas formed by topographic uplift and increasing amplitudes of palaeoclimate variability at crucial geological periods. In the future, it may be informative to sample extra-generic relationships, and this can be achieved with a broader sampling of chloridoid genera.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank USDA-Beltsville National Germplasm System, ILRI-Addis Ababa and J. Travis Columbus for help with caryopsis and leaf collection; Lee Weigt, Jeffery Hunt, Rong Li and Xinwei Xu for help in the laboratory; Robert J. Soreng, Dianxiang Zhang, Gabriel Johnson and Tieyao Tu for helpful discussions; and six anonymous reviewers for their constructive comments that improved the manuscript. This work was supported by the National Geographic Society Committee for Research and Exploration (grant no. 8087-06), the Smithsonian Institution's Small Grants Program, National Natural Science Foundation of China (no. 30700043), the China Scholarship Council Awards (no. 2008491004), the Key Laboratory of Plant Resources Conservation and Sustainable Utilization of Chinese Academy of Sciences (no. 200922) and the Knowledge Innovation Program of the Chinese Academy of Sciences (no. KSCX2-EW-J-28).
LITERATURE CITED
- Bisht MS, Mukai Y. Genomic in situ hybridization identifies genome donor of finger millet (Eleusine coracana) Theoretical and Applied Genetics. 2001a;102:825–832. [Google Scholar]
- Bisht MS, Mukai Y. Identification of genome donors to the wild species of finger millet, Eleusine africana by genomic in situ hybridization. Breeding Science. 2001b;51:263–269. [Google Scholar]
- Bisht MS, Mukai Y. Genome organization and polyploid evolution in the genus Eleusine (Poaceae) Plant Systematics and Evolution. 2002;233:243–258. [Google Scholar]
- Chennaveeraiah MS, Hiremath SC. Genome analysis of Eleusine coracana (L.) Gaertn. Euphytica. 1974;23:489–495. [Google Scholar]
- Christin PA, Besnard G, Samaritani E, et al. Oligocene CO2 decline promoted C4 photosynthesis in grasses. Current Biology. 2008;18:37–43. doi: 10.1016/j.cub.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Cerling TE. Development of grasslands, savannas in East Africa during the Neogene. Palaeogeography, Palaeoclimatology, Palaeoecology. 1992;97:241–247. [Google Scholar]
- Cerling TE, Harris JM, Macfadden BJ, et al. Global vegetation change through the Miocene/Pliocene boundary. Nature. 1997;389:153–158. [Google Scholar]
- Cronn RC, Small RL, Haselkorn T, Wendel JF. Cryptic repeated genomic recombination during speciation in Gossypium gossypioides. Evolution. 2003;57:2475–2489. doi: 10.1111/j.0014-3820.2003.tb01493.x. [DOI] [PubMed] [Google Scholar]
- DeMenocal PB. Plio-Pleistocene African climate. Science. 1995;270:53–59. doi: 10.1126/science.270.5233.53. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. doi:10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. doi:10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke JA, Wain KK. Medicinal plants of the world. Computer index with more than 85000 entries. London: Longman Group Limited; 1981. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, Osborne CP, Strömberg CAE, et al. The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science. 2010;328:587–591. doi: 10.1126/science.1177216. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Whitkus R, Rieseberg LH. Distribution of spontaneous plant hybrids. Proceedings of the National Academy of Sciences, USA. 1996;93:5090–5093. doi: 10.1073/pnas.93.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emshwiller E, Doyle JJ. Origins of domestication and polyploidy in oca (Oxalis tuberosa: Oxalidaceae): nrITS data. American Journal of Botany. 1998;85:975–985. [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fortune PM, Schierenbeck KA, Ainouche AK, Jaquemin J, Wendel JF, Ainouche ML. Evolutionary dynamics of Waxy and the origin of hexaploid Spartina species (Poaceae) Molecular Phylogenetics and Evolution. 2007;43:1040–1055. doi: 10.1016/j.ympev.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Ge S, Sang T, Lu BR, Hong DY. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proceedings of the National Academy of Sciences, USA. 1999;96:14400–14405. doi: 10.1073/pnas.96.25.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Codes Corporation. Sequencher 4·5. Michigan: Gene Codes Corporation; 2005. [Google Scholar]
- Hedberg O. Evolution and speciation in a tropical high mountain flora. Biological Journal of the Linnean Society. 1969;1:135–148. [Google Scholar]
- Hilu KW, Johnson JL. Ribosomal DNA variation in finger millet and wild species of Eleusine (Poaceae) Theoretical and Applied Genetics. 1992;83:895–902. doi: 10.1007/BF00226713. [DOI] [PubMed] [Google Scholar]
- Hilu KW, Johnson JL. Systematics of Eleusine Gaertn. (Poaceae, Chloridoideae): chloroplast DNA and total evidence. Annals of the Missouri Botanical Garden. 1997;84:841–847. [Google Scholar]
- Hiraishi A, Kamagata Y, Nakamura K. Polymerase chain reaction amplification and restriction fragment length polymorphism analysis of 16S rRNA genes from methanogens. Journal of Fermentation and Bioengineering. 1995;79:523–529. [Google Scholar]
- Hiremath SC, Chennaveeraiah MS. Cytogenetical studies in wild and cultivated species of Eleusine (Gramineae) Caryologia. 1982;35:57–69. [Google Scholar]
- Hiremath SC, Salimath SS. The quantitative nuclear DNA changes in Eleusine (Gramineae) Plant Systematics and Evolution. 1991;178:225–233. [Google Scholar]
- Jakob SS, Blattner FR. Two extinct diploid progenitors were involved in allopolyploid formation in the Hordeum murinum (Poaceae: Triticeae) taxon complex. Molecular Phylogenetics and Evolution. 2010;55:650–659. doi: 10.1016/j.ympev.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Jiang B, Peterson PM, Liu Q. Caryopsis micromorphology of Eleusine Gaertn. (Poaceae) and its systematic implications. Journal of Tropical and Subtropical Botany. 2011;19:195–204. [in Chinese]. [Google Scholar]
- Kellogg EA, Appels R, Mason-Gamer RJ. When gene trees tell different stories: the diploid genera of Triticeae. Systematic Botany. 1996;21:321–347. [Google Scholar]
- Knob EB, Palmer JD. Chloroplast DNA variation and the recent radiation of the giant senecios (Asteraceae) on the tall mountains of eastern Africa. Proceedings of the National Academy of Sciences, USA. 1995;92:10349–10353. doi: 10.1073/pnas.92.22.10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch IW. A check list of crosses in the Gramineae. New York: Stechert-Hafner Service Agency; 1968. [Google Scholar]
- Knobloch IW. Intergeneric hybridization in flowering plants. Taxon. 1972;21:97–103. [Google Scholar]
- Lærdal T, Talbot MR. Basin neotectonics of Lakes Edward and George, East African Rift. Palaeogeography, Palaeoclimatology, Palaeoecology. 2002;187:213–232. [Google Scholar]
- Lepiniec L, Vidal J, Chollet R, Gadal P, Crétin C. Phosphoenolpyruvate carboxylase: structure, regulation and evolution. Plant Science. 1994;99:111–124. [Google Scholar]
- Liu Q, Peterson PM. Advances in systematics of adaptively radiated Eleusine Gaertn. (Poaceae) Journal of Tropical and Subtropical Botany. 2010;18:219–226. [in Chinese] [Google Scholar]
- Liu Q, Peterson PM, Columbus JT, Zhang DX, Hao G, Zhang DX. Inflorescence diversification in ‘finger millet clade’ (Chloridoideae, Poaceae): a comparison of molecular phylogeny and developmental morphology. American Journal of Botany. 2007;94:1230–1247. doi: 10.3732/ajb.94.7.1230. [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhao NX, Hao G. Inflorescence structures and evolution in the subfamily Chloridoideae (Gramineae) Plant Systematics and Evolution. 2005;251:183–198. [Google Scholar]
- Lovett JC. Climatic history and forest distribution in eastern Africa. In: Lovett JC, Wasser SK, editors. Biogeography and ecology of the rain forests of eastern Africa. Cambridge: Cambridge University Press; 1993. pp. 23–29. [Google Scholar]
- Mason-Gamer RJ, Kellogg EA. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae) Systematic Biology. 1996;45:524–545. [Google Scholar]
- Mason-Gamer RJ, Naum M, Burns MM. Phylogenetic relationships and reticulation among Asian Elymus (Poaceae) allotetraploids: analyses of three nuclear gene trees. Molecular Phylogenetics and Evolution. 2010;54:10–22. doi: 10.1016/j.ympev.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Moinuddin M, Vahidy AA, Ali SI. Chromosome counts in Arundinoideae, Chloridoideae, and Pooideae (Poaceae) from Pakistan. Annals of the Missouri Botanical Garden. 1994;81:784–791. [Google Scholar]
- National Research Council. Lost crops of Africa. volume 1. Grains. Washington, DC: National Academy Press; 1996. [Google Scholar]
- Negrutskii BS, El'skaya AV. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Progress in Nucleic Acid Research and Molecular Biology. 1998;60:47–78. doi: 10.1016/s0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- Neves SS, Swire-Clark G, Hilu KW, Baird WV. Phylogeny of Eleusine (Poaceae: Chloridoideae) based on nuclear ITS and plastid trnT-trnF sequences. Molecular Phylogenetics and Evolution. 2005;35:395–419. doi: 10.1016/j.ympev.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Oduori COA. The importance and research status of finger millet in Africa. Nairobi: The McKnight Foundation Collaborative Crop Research; 2005. [Google Scholar]
- Osborne CP, Beerling DJ. Nature's green revolution: the remarkable evolutionary rise of C4 plants. Philosophical Transactions of the Royal Society B: Biological Scinces. 2006;361:173–194. doi: 10.1098/rstb.2005.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallister JW. The tectonics of East Africa. In: Choubert G, Faure-Muret A, editors. Tectonics of Africa. Paris: United Nations Educational, Scientific and Cultural Organization; 1971. pp. 511–542. [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proceedings of the National Academy of Sciences, USA. 2004;26:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O, Forest F, Fay MF, Chase MW. Hybrid speciation in angiosperms: parental divergence drives ploidy. New Phytologist. 2009;182:507–518. doi: 10.1111/j.1469-8137.2009.02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G, Seberg O, Yde M, Berthelsen K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum) Molecular Phylogenetics and Evolution. 2006;39:70–82. doi: 10.1016/j.ympev.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Peterson PM, Annable CR. Systematics of the annual species of Muhlenbergia (Poaceae-Eragrostideae) Systematic Botany Monographs. 1991;31:1–109. [Google Scholar]
- Peterson PM, Romaschenko K, Johnson G. A classification of the Chloridoideae (Poaceae) based on multi-gene phylogenetic trees. Molecular Phylogenetics and Evolution. 2010;55:580–598. doi: 10.1016/j.ympev.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Peterson PM, Romaschenko K, Barker NP, Linder HP. Centropodieae and Ellisochloa, a new tribe and genus in the Chloridoideae (Poaceae) Taxon. 2011;60:1113–1122. [Google Scholar]
- Phillips SM. A survey of the genus Eleusine Gaertn. (Gramineae) in Africa. Kew Bulletin. 1972;27:251–270. [Google Scholar]
- Phillips SM. Eleusine Gaertn. In: Polhill RM, editor. Flora of tropical East Africa. London: Crown Agents for Overseas Governments and Administrations; 1974. pp. 260–267. [Google Scholar]
- Phillips SM. Eleusine Gaertn. In: Hedberg I, Edwards S, editors. Flora of Ethiopia and Eritrea. vol. 7. Addis: Addis Ababa University and Sweden: Uppsala University; 1995. pp. 138–142. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rambaut A. Se-Al sequence alignment editor. 2007 Available from: http://tree.bio.ed.ac.uk/software/seal/ (accessed 20 April 2008). [Google Scholar]
- Rambaut A. FigTree version 1·3·1. 2009 Available from: http://tree.bio.ed.ac.uk/software/figtree/ (accessed 30 November 2009) [Google Scholar]
- Rambaut A, Drummond AJ. Tracer version 1·5. 2007 Available from: http://tree.bio.ed.ac.uk/software/tracer/ (accessed 10 March 2010) [Google Scholar]
- Renner SS. Relaxed molecular clocks for dating historical plant dispersal events. Trends in Plant Science. 2005;10:550–558. doi: 10.1016/j.tplants.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, Telford MJ. A multi criterion approach for the selection of optimal outgroups in phylogeny: recovering some support for Mandibulata over Myriochelata using mitogenomics. Molecular Phylogenetics and Evolution. 2008;48:103–111. doi: 10.1016/j.ympev.2008.03.033. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K. The genome sequence and structure of rice chromosome 1. Nature. 2002;420:312–316. doi: 10.1038/nature01184. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends in Ecology and Evolution. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Sepulchre P, Ramstein G, Fluteau F, Schuster M, Tiercelin JJ, Brunet M. Tectonic uplift and East Africa aridification. Science. 2006;313:1419–1423. doi: 10.1126/science.1129158. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Polyploidy in plants: unsolved problems and prospects. In: Lewis W, editor. Polyploidy: biological relevance. New York: Plenum Press; 1980. pp. 495–520. [Google Scholar]
- Stebbins GL. Grass systematics and evolution: past, present and future. In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME, editors. Grass systematics and evolution. Washington, DC: Smithsonian Institution Press; 1986. pp. 359–367. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (* and other methods) version 4·0b10. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- The Rice Chromosome 3 Sequencing Consortium. Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Research. 2005;15:1284–1291. doi: 10.1101/gr.3869505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini A, Barber JC, Aliscioni SS, Giussani LM, Kellogg EA. The age of the grasses and clusters of origins of C4 photosynthesis. Global Change Biology. 2008;14:2963–2977. [Google Scholar]
- Wakeley J, Hey J. Estimating ancestral population parameters. Genetics. 1997;145:847–855. doi: 10.1093/genetics/145.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
- Werth CR, Hilu KW, Langner CA, Baird WV. Duplicate gene expression for isocitrate dehydrogenase and 6-phosphogluconate dehydrogenase in diploid species of Eleusine (Gramineae) American Journal of Botany. 1993;80:705–710. [Google Scholar]
- Werth CR, Hilu KW, Langner CA. Isozyme of Eleusine (Gramineae) and the origin of finger millet. American Journal of Botany. 1994;81:1186–1197. [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. 2006 PhD thesis, University of Texas at Austin, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.