Abstract
Currently, there is no animal model known that mimics natural nasal colonization by Staphylococcus aureus in humans. We investigated whether rhesus macaques are natural nasal carriers of S. aureus. Nasal swabs were taken from 731 macaques. S. aureus isolates were typed by pulsed-field gel electrophoresis (PFGE), spa repeat sequencing and multi-locus sequence typing (MLST), and compared with human strains. Furthermore, the isolates were characterized by several PCRs. Thirty-nine percent of 731 macaques were positive for S. aureus. In general, the macaque S. aureus isolates differed from human strains as they formed separate PFGE clusters, 50% of the isolates were untypeable by agr genotyping, 17 new spa types were identified, which all belonged to new sequence types (STs). Furthermore, 66% of macaque isolates were negative for all superantigen genes. To determine S. aureus nasal colonization, three nasal swabs from 48 duo-housed macaques were taken during a 5 month period. In addition, sera were analyzed for immunoglobulin G and A levels directed against 40 staphylococcal proteins using a bead-based flow cytometry technique. Nineteen percent of the animals were negative for S. aureus, and 17% were three times positive. S. aureus strains were easily exchanged between macaques. The antibody response was less pronounced in macaques compared to humans, and nasal carrier status was not associated with differences in serum anti-staphylococcal antibody levels. In conclusion, rhesus macaques are natural hosts of S. aureus, carrying host-specific lineages. Our data indicate that rhesus macaques are useful as an autologous model for studying S. aureus nasal colonization and infection prevention.
Introduction
In the light of the rapid, worldwide emergence of antibiotic resistance in and the lack of an effective long-term elimination strategy against Staphylococcus aureus (S. aureus) nasal carriage, new approaches are needed to prevent staphylococcal carriage and its consequent diseases. Many different S. aureus animal models have been described for studying the pathogenesis of S. aureus colonization and infection. These models have provided insight into the role of bacterial virulence genes and have assisted in the estimation of vaccine efficacy. Models have been set up in various species, such as insects, worms, mice, rats, guinea pigs, hamsters, chickens, rabbits, sheep, dogs, pigs, and cows [1]–[4]. Notably, most of these animals, unlike humans, are not natural nasal carriers of S. aureus, only pigs, sheep, and cows may be naturally colonized by S. aureus. For instance, S. aureus sequence type (ST) 398 strains belong to a biotype associated with pigs and other species of livestock [5], [6]. S. aureus strain RF122 is a member of a bovine mastitis-associated clone that is genetically different from human clones of S. aureus [7], [8]. Humans can acquire these S. aureus strains during intensive short-term exposure to livestock, but in most cases the strain is lost again within 24 hours [9]. However, lack of a natural, human-like animal model of nasal S. aureus carriage is still a problem. Therefore, we investigated whether a non-human primate could provide a natural model for human nasal carriage of S. aureus.
The rhesus macaque (Macaca mulatta) belongs to the old world monkeys and has been used in several studies involving S. aureus. Kuklin et al. used rhesus macaques to study the immunogenicity of IsdB [10]. Protection against lethal SEB aerosol exposure by passive transfer of SEB-specific antibodies was also studied in macaques [11]. In addition to these protection studies, rhesus macaques were also used for safety evaluations. For example, the tolerability and potential toxicity of the thrombolytic agent staphylokinase was investigated in healthy rhesus macaques [12]. To our knowledge, natural nasal S. aureus carriage and the consequences on natural immunity in rhesus macaques has never been studied before.
Using a cross sectional setup, we isolated 287 S. aureus strains from 731 rhesus macaques after nasal sampling. We compared S. aureus strains isolated from rhesus macaques and humans. Furthermore, we followed a group of 48 rhesus macaques in time for studying persistence of nasal carriage of S. aureus. In addition, serum samples from these macaques were analyzed for anti-staphylococcal immunoglobulin G and A (IgG and IgA) levels.
Materials and Methods
Study population
For comparison of S. aureus strains isolated from rhesus macaques with those from humans, 731 rhesus macaques from the breeding colony of the Biomedical Primate Research Centre (Rijswijk, The Netherlands) were studied. These animals were of Indian, Burmese and Chinese origin. These macaques were housed in groups of 2–44 individuals. Furthermore, 48 young rhesus macaques that were recently imported from China were followed in time for studying the persistence of S. aureus nasal carriage as well as their serum anti-staphylococcal antibody levels. These animals were duo-housed in 4 different animal rooms. Physical contact with the macaques in the neighbouring cage was possible. In each room 2 groups of cages were located opposite to each other.
Human S. aureus strains
For reasons of comparison, 56 human isolates of S. aureus were included. These carriage (n = 30) and bacteremia derived (n = 20) MSSA isolates have been described before [13], [14]. Three MSSA isolates from animal care-takers and 3 S. aureus strains for which the genome sequence is known were included as well (N315, Mu50, MRSA252).
Ethics statement
Sampling of the longitudinally screened macaques was approved by the Animal Experiments committee of the Biomedical Primate Research Centre (Dierexperimentencommissie (DEC), which is the ethical committee installed and officially recognised as required by the Dutch Law on Experimental Animals and which is the Dutch analogue for the IACUC). The approval number is: DEC#579, dated October 28, 2008. The study was conducted in compliance with all relevant Dutch laws and in agreement with international and scientific standards and guidelines.
Sampling for bacteriology of the animals in the breeding groups was performed as part of their yearly routine health screening, which is by Dutch law not considered to be an animal experiment. Therefore no permission of the ethical committee was necessary for this part of the study. All samplings were performed under ketamine anaesthesia (which is routine for all health checks of non-human primates) and all efforts were made to minimize stress and suffering of the animals. The housing, care and handling of all animals were according to the Dutch Law on Experimental Animals and the European Directive 86/609/EEC. The research described is in accordance to the Weatherall Report recommendations for good welfare.
Human samples were described before [13], [14]. For carriage isolates and serum samples, volunteers provided written informed consent and the local Medical Ethics Committee of the Erasmus Medical Centre Rotterdam approved the study (MEC-2007-106). For bacteremia derived isolates, S. aureus strains were collected for routine culture. The Medical Ethics Committee of the Erasmus Medical Centre Rotterdam approved the study (MEC-2007-106, addendum 2).
Sampling procedures
A total of 731 macaques were sampled once for nasal carriage of S. aureus, while another 48 macaques were screened three times during a 5 month period. Nasal cultures were taken by streaking both anterior nares using a sterile cotton swab (Swab Transystem, Greiner Bio One, Alphen aan de Rijn, The Netherlands) during regular animal medical check-up. All swabs were processed within 24 hours. Nasal swabs were plated on a Columbia sheep blood agar plate-medium (bioTRADING, Mijdrecht, The Netherlands). Plates were read after one and two days of incubation at 35°C. Identification of S. aureus was based on colony morphology and coagulase plasma test (Becton Dickinson, Breda, The Netherlands) and confirmed by API Staph (bioMérieux, Boxtel, The Netherlands).
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) of SmaI digested chromosomal DNA from all S. aureus strains from rhesus macaques and 56 strains from humans was performed as described previously [15]. Relatedness among the PFGE profiles was evaluated with Bionumerics software (version 3.0; Applied Maths, Ghent, Belgium). A dendrogram was produced using the Dice coefficient and an unweighted-pair group method using arithmetic averages (UPGMA). Band tolerance was set at 2.0%.
Anti-staphylococcal antibodies
IgG and IgA antibody levels in serum directed against the following antigens were semi-quantified: S. aureus proteins clumping factor A and B (ClfA and ClfB); surface protein G (SasG); iron-responsive surface determinants A and H (IsdA and IsdH); fibronectin-binding proteins A and B (FnbpA and FnbpB); serine-aspartate dipeptide repeat protein D and E (SdrD and SdrE); staphylococcal enterotoxins A-E, G-J, M-O, Q, and R (SEA - SEE, SEG - SEJ, SEM - SEO, SEQ, SER); toxic shock syndrome toxin 1 (TSST-1); chemotaxis inhibitory protein of S. aureus (CHIPS); staphylococcal complement inhibitor (SCIN); extracellular fibrinogen-binding protein (Efb); exfoliative toxin A and B (ETA and ETB); alpha toxin; γ hemolysin B (HlgB); leukocidin (Luk) S-PV, LukF-PV, LukD-PV, and LukE-PV; and staphylococcal superantigen-like proteins 1, 3, 5, 9, and 11 (SSL1, SSL3, SSL5, SSL9, and SSL11). Antibodies were semi-quantified simultaneously in a single multiplex assay using a bead-based flow cytometry technique (xMap; Luminex Corporation). Methods have been described elsewhere [14], [16], [17], with the exception that a 1∶50 dilution of R-phycoerythrin (RPE)-conjugated AffiniPure goat anti-human secundairy IgA was used. Tests were performed in independent duplicates, and the median fluorescence intensity (MFI) values, reflecting semiquantitative antibody levels, were averaged. In each experiment, control beads (no protein coupled) were included to determine nonspecific binding. In the event of nonspecific binding, the nonspecific MFI values were subtracted from the antigen-specific results.
Serum samples from 47 out of the 48 rhesus macaques from China were analyzed. Anti-staphylococcal antibody levels in these sera were compared to those in sera from 20 Dutch healthy human volunteers [14]. Pooled serum from all rhesus macaques included in the present study or from humans was used as a standard.
DNA isolation
DNA isolation of S. aureus was performed for representative isolates from each PFGE cluster. Total DNA of S. aureus was isolated with the QIAamp DNA Mini Kit (QIAGEN, Venlo, The Netherlands) according to the manufacturer's instructions, or with MagNA Pure LC DNA isolation kit III (bacteria, fungi) using the MagNA Pure LC instrument (Roche Diagnostics, Almere, The Netherlands) [18].
spa genotyping
PCR for amplification of the S. aureus protein A (spa) repeat region was performed according to the published protocol [19]. PCR products were purified with the QIAquick PCR Purification Kit (QIAGEN, Venlo, The Netherlands) and sequenced using two amplification primers from a commercial supplier (SeqLab, Goettingen, Germany). The forward and reverse sequence chromatograms were analyzed with the Ridom StaphType software (Ridom GmbH, Würzburg, Germany).
Multi-locus sequence typing
For one representative S. aureus isolate of each spa type, the sequence type (ST) was determined using multi-locus sequence typing (MLST) [20]. goeBURST [21], as implement in Phyloviz software (http://www.phyloviz.net/wiki/), and which used the same priority rules for linking STs as eBURST but with a global optimization, was used to infer relatedness between STs. goeBURST was run on the whole S. aureus MLST database (http://saureus.mlst.net/; interrogated July 2011) supplemented with STs of rhesus macaque isolates not present in the S. aureus MLST database at the time of interrogation. Clonal complexes (CCs) are defined as STs that are linked through single locus variants (SLVs) and are named on the basis of the predicted founder ST, which is the ST having the most SLVs.
Staphylococcal antigen PCRs
Methicillin resistance was detected with mecA-specific primers [22]. Six sets of multiplex PCRs were established to amplify 19 superantigen genes (sea to see, seg to ser, seu, tst), eta, etd and agr-1 to -4, as described previously [23]. The presence of the scn, chp, clfA, clfB, efb, fnbA, fnbB and tst genes was determined by PCR as well [13], [24]–[26].
Statistical analysis
Statistical analyses were performed with SPSS software, version 15.0 (SPSS). The Mann-Whitney U test was used to compare median differences in anti-staphylococcal antibody levels. Differences were considered statistically significant when 2-sided P-values were <0.05.
Results
Characterization of S. aureus isolates in rhesus macaques
A total of 731 macaques was screened once for presence of S. aureus in their nasal cavity. Thirty-nine percent (287/731) of these cultures were positive for S. aureus, clearly indicating that rhesus macaques can be S. aureus nasal carriers.
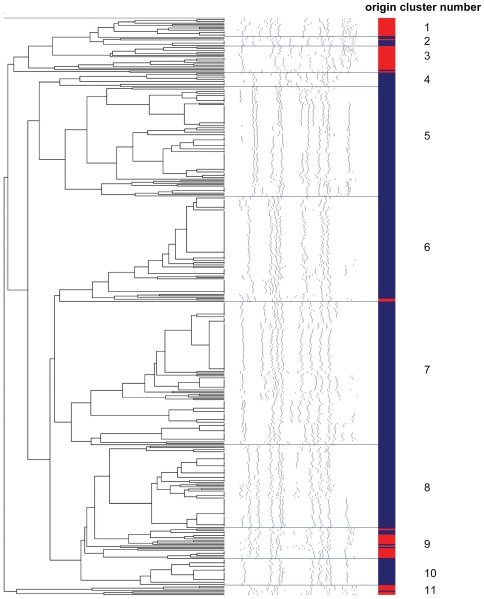
S. aureus isolates from the 287 culture positive macaques as well as all collected isolates from the 48 macaques that were sampled three times (a total of 355 isolates) were typed using molecular methods including PFGE. This revealed 11 major clusters (Figure 1). Five of these consisted of only rhesus macaque isolates, one comprised only human isolates, while five clusters included isolates from both macaques and humans.
Figure 1. Dendrogram of the PFGE data of S. aureus strains isolated from rhesus macaques and humans.
Blue color indicates S. aureus strains isolated from rhesus macaques, red color indicates S. aureus strains isolated from humans. Five clusters consisted of only rhesus macaque isolates, one comprised only human isolates, and five clusters included isolates from both macaques and humans.
For further characterization of the S. aureus isolates from rhesus macaques, spa genotyping was performed on representative isolates from each PFGE cluster (n = 108) (Table 1). These isolates were all spa-positive, re-confirming that these were indeed S. aureus strains. This revealed 22 different spa types, of which five have already been described before. Consequently, we identified 17 new spa types not included in the Ridom SpaServer database at the time of analysis. These new spa types comprised 59% of the isolates. The most prominent clone was t516, which comprised 21 isolates (19%). Spa types t4168, t729 and t4167 were represented by 18 (17%), 15 (14%) and 11 (10%) isolates, respectively. Nine spa types were present as single isolates. Each major PFGE cluster comprised 1 to 8 different spa types, though the repeat succession of different spa types within a cluster were clearly related in some cases (Table 2).
Table 1. Different spa types found in 108 representative S. aureus strains isolated from rhesus macaques.
| spa type | No. of isolates | Percentage of isolates | |
| t189 | 1 | 0.9 | |
| t516 | 21 | 19.4 | |
| t729 | 15 | 13.9 | |
| t786 | 4 | 3.7 | |
| t1814 | 3 | 2.8 | |
| t4167 | new | 11 | 10.2 |
| t4168 | new | 18 | 16.7 |
| t4980 | new | 2 | 1.9 |
| t4981 | new | 1 | 0.9 |
| t5010 | new | 2 | 1.9 |
| t5011 | new | 1 | 0.9 |
| t5012 | new | 1 | 0.9 |
| t5013 | new | 6 | 5.6 |
| t5014 | new | 1 | 0.9 |
| t5015 | new | 4 | 3.7 |
| t5016 | new | 1 | 0.9 |
| t5017 | new | 5 | 4.6 |
| t5018 | new | 5 | 4.6 |
| t5135 | new | 3 | 2.8 |
| t5136 | new | 1 | 0.9 |
| t5137 | new | 1 | 0.9 |
| t5458 | new | 1 | 0.9 |
Table 2. Relatedness of spa types within each PFGE cluster.
| PFGE cluster | spa types in PFGE cluster | Repeat succession | ||||||||||||||||||||
| 1 | - | |||||||||||||||||||||
| 2 | t4980 | 4 | - | 16 | - | 21 | - | 12 | - | 12 | - | 17 | - | 13 | - | 39 | - | 13 | ||||
| t5135 | 210 | - | 23 | - | 34 | - | 34 | - | 16 | - | 17 | - | 34 | - | 17 | - | 17 | - | 23 | - | 17 | |
| 3 | - | |||||||||||||||||||||
| 4 | t516 | 8 | - | 16 | - | 2 | - | 25 | - | 51 | - | 68 | - | 2 | - | 24 | - | 2 | - | 24 | ||
| t5014 | 15 | - | 12 | - | 16 | - | 16 | - | 2 | - | 25 | - | 34 | - | 24 | - | 17 | - | 24 | - | 17 | |
| t5015 | 15 | - | 12 | - | 16 | - | 16 | - | 25 | - | 25 | - | 34 | - | 24 | - | 17 | - | 17 | - | 17 | |
| t5017 | 8 | - | 34 | - | 2 | - | 43 | - | 34 | - | 43 | - | 43 | - | 16 | - | 2 | - | 17 | - | 83 | |
| 5 | t516 | 8 | - | 16 | - | 2 | - | 25 | - | 51 | - | 68 | - | 2 | - | 24 | - | 2 | - | 24 | ||
| t5011 | 8 | - | 16 | - | 16 | - | 25 | - | 25 | - | 34 | - | 24 | - | 17 | - | 17 | - | 17 | |||
| t5015 | 15 | - | 12 | - | 16 | - | 16 | - | 25 | - | 25 | - | 34 | - | 24 | - | 17 | - | 17 | - | 17 | |
| 6 | t516 | 8 | - | 16 | - | 2 | - | 25 | - | 51 | - | 68 | - | 2 | - | 24 | - | 2 | - | 24 | ||
| t729 | 7 | - | 12 | - | 21 | - | 17 | - | 13 | - | 34 | - | 34 | - | 34 | - | 33 | - | 34 | |||
| t786 | 7 | - | 12 | - | 21 | - | 17 | - | 13 | - | 34 | - | 34 | - | 33 | - | 34 | |||||
| t1814 | 7 | - | 12 | - | 21 | - | 17 | - | 34 | - | 34 | - | 34 | - | 33 | - | 34 | |||||
| t5012 | 8 | - | 34 | - | 2 | - | 43 | - | 34 | - | 43 | - | 43 | - | 16 | - | 2 | - | 17 | - | 16 | |
| t5136 | 4 | - | 2 | - | 17 | - | 17 | - | 34 | - | 24 | - | 17 | - | 17 | - | 17 | |||||
| t5458 | 210 | - | 23 | - | 34 | - | 17 | - | 34 | - | 22 | - | 34 | - | 17 | - | 23 | - | 34 | |||
| 7 | t4167 | 7 | - | 23 | - | 17 | - | 22 | - | 249 | - | 12 | - | 117 | - | 24 | - | 25 | - | 17 | ||
| t4168 | 8 | - | 23 | - | 21 | - | 17 | - | 13 | - | 13 | - | 13 | - | 24 | - | 23 | |||||
| t4980 | 4 | - | 16 | - | 21 | - | 12 | - | 12 | - | 17 | - | 13 | - | 39 | - | 13 | |||||
| t5013 | 7 | - | 23 | - | 23 | - | 17 | - | 34 | - | 21 | - | 12 | - | 20 | - | 24 | |||||
| t5016 | 8 | - | 34 | - | 2 | - | 43 | - | 34 | - | 43 | - | 17 | - | 83 | |||||||
| t5017 | 8 | - | 34 | - | 2 | - | 43 | - | 34 | - | 43 | - | 43 | - | 16 | - | 2 | - | 17 | - | 83 | |
| t4981 | 8 | - | 16 | - | 2 | - | 43 | - | 17 | - | 34 | |||||||||||
| t5018 | 8 | - | 16 | - | 2 | - | 43 | - | 17 | - | 173 | - | 34 | - | 17 | - | 34 | |||||
| 8 | t4168 | 8 | - | 23 | - | 21 | - | 17 | - | 13 | - | 13 | - | 13 | - | 24 | - | 23 | ||||
| t5010 | 8 | - | 23 | - | 12 | - | 17 | - | 13 | - | 13 | - | 13 | - | 24 | - | 23 | |||||
| 9 | t4167 | 7 | - | 23 | - | 17 | - | 22 | - | 249 | - | 12 | - | 117 | - | 24 | - | 25 | - | 17 | ||
| t4168 | 8 | - | 23 | - | 21 | - | 17 | - | 13 | - | 13 | - | 13 | - | 24 | - | 23 | |||||
| t5015 | 15 | - | 12 | - | 16 | - | 16 | - | 25 | - | 25 | - | 34 | - | 24 | - | 17 | - | 17 | - | 17 | |
| t5137 | 4 | - | 16 | - | 13 | |||||||||||||||||
| 10 | t4168 | 8 | - | 23 | - | 21 | - | 17 | - | 13 | - | 13 | - | 13 | - | 24 | - | 23 | ||||
| 11 | t189 | 7 | - | 23 | - | 12 | - | 21 | - | 17 | - | 34 | ||||||||||
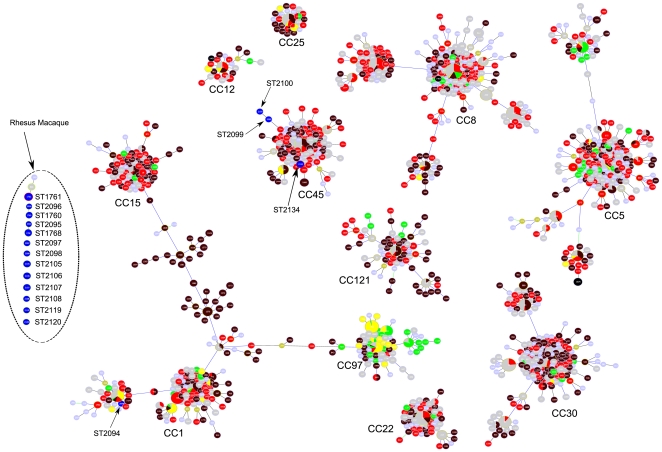
For one S. aureus isolate of each spa type, we performed MLST and compared STs found in macaque strains with all STs available in the S. aureus MLST database at the moment of interrogation (Figure 2). Seventeen different STs were found, which were all not present in the S. aureus MLST database at the time of interrogation. ST2099, ST2100 and ST2134 were part of CC45, which mostly contained STs found in human S. aureus isolates. ST2099 and ST2100 were SLVs. Isolates with these 3 STs were found in different PFGE clusters. ST2094 had relations with STs in CC1, which contained a lot of STs found in human isolates as well. The PFGE pattern of this S. aureus isolate differed from all other macaque isolates, as this isolate was found in PFGE cluster 11, in which this isolate was the only one from macaque origin. All other STs found in the macaque isolates (ST1760, ST1761, ST1768, ST2095, ST2096, ST2097, ST2098, ST2105, ST2106, ST2107, ST2108, ST2119, ST2120) were not part of the larger CCs, but were present as singletons. ST1760 and ST2096 as well as ST1768 and ST2095 were SLVs, and were both found in PFGE clusters 7 and 6, respectively, clusters with mainly isolates from macaques. Isolates with STs present as a singleton were distributed among PFGE clusters 4 to 9, which were clusters with mainly isolates from rhesus macaques.
Figure 2. Partial population snapshot of S. aureus.
This snapshot was created by goeBURST v1.2 software using the dataset downloaded from http://saureus.mlst.net/ that included 2010 STs representing 3887 isolates. A subset of this dataset (1304 STs, representing 2875 isolates) is presentend in this figure including all major Clonal Complexes (CCs) supplemented with all rhesus macaque ST as far as they were not part of the major CCs. The area of each circle in the goeBURST diagram corresponds to the relative abundance of the STs in the input data. The names of major CCs have been indicated. ST colors refer to the source S. aureus was isolated from. STs representing macaque isolates are indicated in blue and by their ST number; brown colors correspond with community-acquired human isolates; red colors correspond with hospital-acquired human isolates; green colors correspond with animal isolates; yellow colors correspond with S. aureus isolates from food; grey, light green, and light blue colors correspond with isolates with unknown source.
Next, we determined the genomic presence of mecA, 19 superantigen genes, and eta and etd by PCR. None of the 108 selected isolates harboured the mecA gene, which indicates that none of these isolates were MRSA. Thirty-two isolates that were representative for each spa type were selected for testing presence of superantigen genes and eta and etd (Table 3). These genes were absent in 66% (21/32) of these isolates. The etd gene was detected in 9% of the isolates (3/32), 16% of the isolates were sei-positive (5/32), 19% were positive for sem, sen, and seo (6/32), and 25% were positive for seg (8/32). Genes encoding SEA to SEE, SEH, SEJ to SEL, SEP to SER, SEU, TSST-1 and ETA were absent in all 32 isolates. Only 6 isolates harboured a complete or incomplete egc locus. Four of these were strains from PFGE clusters comprising only macaque isolates, while 2 isolates belong to clusters including both macaque and human isolates.
Table 3. Presence of genes encoding superantigens, exfoliative toxins, and agr types in 32 representative S. aureus strains isolated from rhesus macaques.
| Genea | |||||||||||||
| Isolate no. | seg | sei | sem | sen | seo | etd | agr-1 | agr-2 | agr-3 | agr-4 | Gene profile | PFGE cluster | Origin isolates in PFGE clusterb |
| 08-1178 | − | − | − | − | − | − | + | − | − | − | 1 | 5 | RM |
| 08-1192 | − | − | − | − | − | − | + | − | − | − | 1 | 4 | RM |
| 08-1276 | − | − | − | − | − | − | + | − | − | − | 1 | 7 | RM |
| 08-1494 | − | − | − | − | − | − | + | − | − | − | 1 | 7 | RM |
| 08-1495 | − | − | − | − | − | − | + | − | − | − | 1 | 7 | RM |
| 08-1185 | + | + | + | + | + | − | + | − | − | − | 2 | 8 | RM |
| 08-1415 | + | + | + | + | + | − | + | − | − | − | 2 | 8 | RM |
| 08-1197 | + | − | − | − | − | − | − | − | − | + | 3 | 6 | RM+HV |
| 08-1255 | + | − | − | − | − | − | − | − | − | − | 4 | 11 | RM+HV |
| 08-1299 | − | − | − | − | − | − | − | − | − | + | 5 | 5 | RM |
| 08-1514 | − | − | − | − | − | − | − | − | − | + | 5 | 5 | RM |
| 08-1332 | − | − | − | − | − | − | − | + | − | − | 6 | 4 | RM |
| 08-1342 | − | − | − | − | − | − | − | − | + | − | 7 | 6 | RM+HV |
| 08-1404 | − | − | − | − | − | − | − | − | − | − | 8 | 5 | RM |
| 08-1412 | − | − | − | − | − | − | − | − | − | − | 8 | 6 | RM+HV |
| 08-1423 | − | − | − | − | − | − | − | − | − | − | 8 | 7 | RM |
| 08-1430 | − | − | − | − | − | − | − | − | − | − | 8 | 7 | RM |
| 08-1432 | − | − | − | − | − | − | − | − | − | − | 8 | 7 | RM |
| 08-1487 | − | − | − | − | − | − | − | − | − | − | 8 | 6 | RM+HV |
| 08-1504 | − | − | − | − | − | − | − | − | − | − | 8 | 7 | RM |
| 08-1507 | − | − | − | − | − | − | − | − | − | − | 8 | 7 | RM |
| 08-1528 | − | − | − | − | − | − | − | − | − | − | 8 | 7 | RM |
| 08-1737 | − | − | − | − | − | − | − | − | − | − | 8 | 2 | RM+HV |
| 08-1749 | − | − | − | − | − | − | − | − | − | − | 8 | 7 | RM |
| 08-1756 | − | − | − | − | − | − | − | − | − | − | 8 | 7 | RM |
| 08-1419 | − | − | − | − | − | + | − | − | + | − | 9 | 7 | RM |
| 08-1420 | − | − | − | − | − | + | − | − | + | − | 9 | 7 | RM |
| 08-1486 | + | − | + | + | + | − | − | − | − | − | 10 | 7 | RM |
| 08-1583 | + | + | + | + | + | − | − | − | − | − | 11 | 6 | RM+HV |
| 08-1602 | + | + | + | + | + | − | − | − | + | − | 12 | 9 | RM+HV |
| 08-1765 | + | + | + | + | + | − | − | − | + | − | 12 | 7 | RM |
| 08-1630 | − | − | − | − | − | + | − | − | − | − | 13 | 2 | RM+HV |
| No. of positive isolates (%) | 8 (25) | 5 (16) | 6 (19) | 6 (19) | 6 (19) | 3 (9) | 7 (22) | 1 (3) | 5 (16) | 3 (9) | |||
All isolates were negative for sea, seb, sec, sed, see, seh, sej, sek, sel, sep, seq, ser, seu, tst, and eta.
Isolates belong to typically rhesus macaque (RM) PFGE clusters or to mixed clusters including both macaque and healthy human volunteer (HV) strains.
These 32 isolates were also agr genotyped (Table 3). The most common agr type was type I (22%), followed by agr type III (16%), type IV (9%), and type II (3%). Sixteen isolates (50%) did not belong to any of the 4 known agr types, which is intriguing. Isolates with agr type I or type II belonged to PFGE clusters comprising only macaque isolates, while isolates with other agr types belonged to clusters including both macaque and human isolates. Overall, 13 combinations of agr type and toxin genes were found. Two of the agr type I isolates, two of the agr type III isolates and two of the unknown agr type isolates harboured a complete or incomplete egc locus. Five agr type I isolates, the single agr type II isolate, one agr type III isolate, two agr type IV isolates, and 12 isolates with an unknown agr type harboured none of the superantigen and exfoliative toxin genes. S. aureus isolates with the same agr type and toxin genes were distributed among the different PFGE clusters.
S. aureus nasal carriage in rhesus macaques
Forty-eight (4 groups of 12) macaques were screened three times for nasal carriage of S. aureus over a 5 month period. The four groups of twelve macaques were housed in separate rooms. For 9 macaques (18.8%), all nasal culture results were negative. Eighteen macaques (37.5%) had one positive culture, 13 (27.1%) had two positive cultures, and for 8 macaques (16.7%) S. aureus was found in all three cultures.
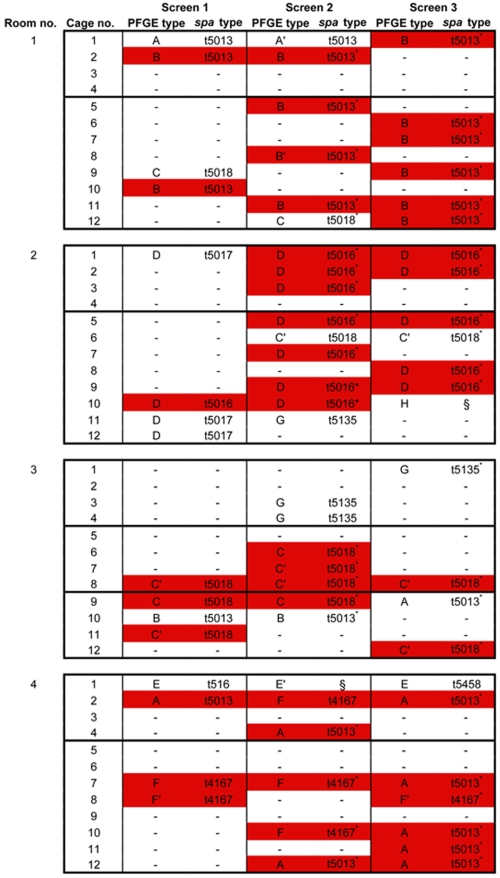
Isolates from these 48 duo-housed macaques were also typed by PFGE, and spa types were determined for representative isolates from each PFGE cluster (Figure 3). In macaques with more than one S. aureus positive nasal culture, a strain replacement occurred in half of the animals.
Figure 3. Persistence of S. aureus nasal carriage in rhesus macaques.
Macaques were duo-housed. Physical contact to the macaques in the neighbouring cage was possible. Each block indicates an animal room, horizontal lines in blocks indicate which macaques were not able to have physical contact. Red color indicates the “epidemic” strain in each room. – indicates a S. aureus negative nasal culture. * indicates that the isolate was not spa genotyped itself, spa type was derived from typed isolates with identical PFGE pattern. § indicates that the isolate was not spa genotyped, and this could not be derived from typed isolates with identical PFGE pattern. S. aureus strains were frequently exchanged between macaques in the same room.
It seemed that there was one most prevalent “epidemic” strain in each animal room. This strain spread from one or a couple of macaques to most others. This phenomenon was clear for rooms 1 (PFGE type B, spa type t5013), 2 (PFGE type D, spa type t5016) and 3 (PFGE type C′, spa type t5018), while for room 4, the epidemic strain was less clear (PFGE type A, spa type t5013, or PFGE type F, spa type t4167). In some cases, a macaque did not acquire the epidemic strain, e.g. the macaque in animal room 2, cage 6 had two nasal cultures positive with a S. aureus isolate with PFGE type C′, while the epidemic isolate had PFGE type D.
Anti-staphylococcal antibodies in rhesus macaques
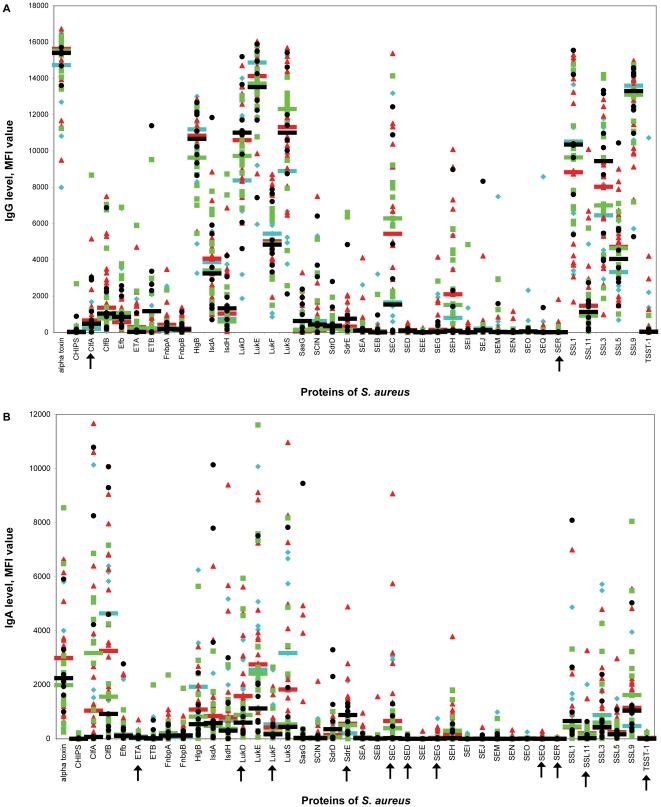
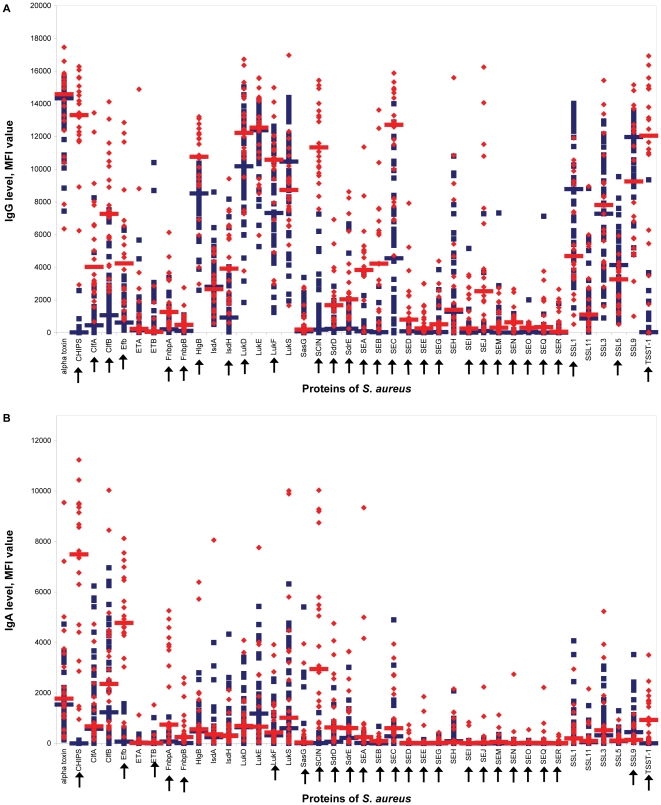
Serum samples from 47 of the 48 macaques that were screened thee times for nasal carriage were analyzed for anti-staphylococcal antibodies directed against 40 different S. aureus proteins. The MFI values reflecting the serum IgG and IgA levels for each macaque and each antibody isotype are shown in Figure 4. An extensive diversity in individual antibody responses was observed, similar to the findings in humans. Individual macaques had high antibody levels against a number of antigens, while anti-staphylococcal antibody levels against other antigens were low.
Figure 4. Relation between S. aureus nasal colonization and level of anti-staphylococcal IgG (A) and IgA (B).
Antibody levels are reflected by Median fluorescence intensity (MFI) value. Each symbol represents a single rhesus macaque. Blue diamonds represent macaques without S. aureus positive culture, red triangles represent macaques with one positive culture, green squares represent macaques with two positive cultures, and black circles represent macaques with three positive cultures. Median values are indicated by horizontal lines. Arrows indicate statistically significant differences in median values (Mann-Whitney U test). More S. aureus positive cultures were not related to high – or low – anti-staphylococcal antibody levels. CHIPS, chemotaxis inhibitory protein of S. aureus; Clf, clumping factor; Efb, extracellular fibrinogen-binding protein; ET, exfoliative toxin; Fnbp, fibronectin-binding protein; HlgB, γ hemolysin B; Isd, iron-responsive surface determinant; Luk, leukocidin; SasG, S. aureus surface protein G; SCIN, staphylococcal complement inhibitor; Sdr, serine-aspartate dipeptide repeat protein; SE, staphylococcal enterotoxin; SSL, staphylococcal superantigen-like protein; TSST-1, toxic shock syndrome toxin 1.
For most antigens, there was no apparent quantitative difference in antibody level between macaques with 0, 1, 2, or 3 positive cultures. However, the median serum levels of IgG directed against ClfA were significantly higher in rhesus macaques with one positive culture than in those without positive culture (MFI value, 657 vs. 191; P<0.05). Additionally, median IgA levels were higher in macaques without a positive culture than in those with three positive cultures for LukF (532 vs. 167; P<0.05). In macaques with three positive cultures, median IgA levels were higher than in macaques without positive culture for SdrE (881 vs. 200; P<0.05). In macaques with one positive culture, median IgA levels were higher than in macaques with three positive cultures for LukD (1580 vs. 599; P<0.005), SEC (658 vs. 23; P<0.05), and SSL11 (204 vs 22; P<0.05). In general, a larger number of positive cultures was not related to elevated or decreased anti-staphylococcal antibody levels. Other comparisons were also made (non-carriers vs. intermittent vs. persistent carriers; carriers of the most prominent isolate in the animal room vs. carriers of another isolate). Again, no association was found between nasal carrier status and anti-staphylococcal antibody levels in serum (data not shown).
Isolates from rhesus macaques differ from those from humans in PFGE type, spa type, agr type, and prevalence of superantigen genes. To determine whether these different S. aureus isolates induce different humoral antibody responses, anti-staphylococcal antibody levels in sera of the 47 rhesus macaques were compared to those in sera of 20 healthy human volunteers (Figure 5). Significant differences in anti-staphylococcal IgG and IgA levels between rhesus macaques and healthy volunteers were observed for 29 and 25 out of 40 antigens, respectively (Table 4). In general, the median antibody serum levels in humans were higher than those in macaques. However, the median serum IgG level directed against SSL1 and SSL5 was significantly higher in macaques than in humans. The median serum IgA level directed against SSL9 was significantly higher in macaques than in humans.
Figure 5. Level of anti-staphylococcal IgG (A) and IgA (B) in rhesus macaques and human volunteers.
Antibody levels are reflected by Median fluorescence intensity (MFI) value. Each symbol represents a single rhesus macaque or human. Blue squares represent healthy rhesus macaques and red diamonds represent healthy human volunteers. Median values are indicated by horizontal lines. Arrows indicate statistically significant differences in median values (Mann-Whitney U test). Anti-staphylcoccal IgG and IgA levels were significantly different between rhesus macaques and humans for 29 and 25 out of 40 antigens, respectively. CHIPS, chemotaxis inhibitory protein of S. aureus; Clf, clumping factor; Efb, extracellular fibrinogen-binding protein; ET, exfoliative toxin; Fnbp, fibronectin-binding protein; HlgB, γ hemolysin B; Isd, iron-responsive surface determinant; Luk, leukocidin; SasG, S. aureus surface protein G; SCIN, staphylococcal complement inhibitor; Sdr, serine-aspartate dipeptide repeat protein; SE, staphylococcal enterotoxin; SSL, staphylococcal superantigen-like protein; TSST-1, toxic shock syndrome toxin 1.
Table 4. Median fluorescence intensity (MFI) values reflecting antigen-specific IgG and IgA levels in rhesus macaques (RM) and healthy human volunteers (HV).
| MFI value, median (range) | MFI value, median (range) | ||||||
| S. aureus protein (antibody isotype) | RM | HV | P a | S. aureus protein (antibody isotype) | RM | HV | P a |
| alpha toxin (IgG) | 14337 (7438–15574) | 14587 (6355–17465) | 0.285 | SEA (IgG) | 88 (17–3565) | 3839 (29–11365) | <0.001 |
| alpha toxin (IgA) | 1540 (139–4729 | 1775 (215–9549) | 0.154 | SEA (IgA) | 16 (1–449) | 247 (5–9345) | <0.001 |
| CHIPS (IgG) | 20 (0–2580) | 13310 (2933–16281) | <0.001 | SEB (IgG) | 34 (0–2836) | 4223 (190–13622) | <0.001 |
| CHIPS (IgA) | 1 (0–130) | 7494 (957–11238) | <0.001 | SEB (IgA) | 2 (0–673) | 90 (2–398) | <0.001 |
| ClfA (IgG) | 454 (16–8250) | 4029 (670–13449) | <0.001 | SEC (IgG) | 4551 (1–14006) | 12720 (4713–15884) | <0.001 |
| ClfA (IgA) | 580 (4–6237) | 673 (145–4730) | 0.373 | SEC (IgA) | 280 (0–4896) | 600 (63–3943) | 0.004 |
| ClfB (IgG) | 1066 (65–7518) | 7272 (2499–14127) | <0.001 | SED (IgG) | 84 (3–533) | 800 (33–7924) | <0.001 |
| ClfB (IgA) | 1232 (0–6962) | 2357 (255–10038) | 0.125 | SED (IgA) | 1 (0–85) | 19 (0–736) | <0.001 |
| Efb (IgG) | 622 (162–6325) | 4238 (770–12855) | <0.001 | SEE (IgG) | 3 (0–229) | 258 (7–3017) | <0.001 |
| Efb (IgA) | 69 (0–1575) | 4775 (257–8125) | <0.001 | SEE (IgA) | 0 (0–151) | 12 (0–1856) | <0.001 |
| ETA (IgG) | 156 (10–5670) | 234 (16–14891) | 0.217 | SEG (IgG) | 37 (0–3861) | 517 (5–4385) | 0.002 |
| ETA (IgA) | 30 (1–366) | 19 (6–1127) | 0.742 | SEG (IgA) | 5 (0–353) | 22 (0–432) | 0.035 |
| ETB (IgG) | 72 (0–10405) | 55 (1–2855) | 0.515 | SEH (IgG) | 1282 (0–10804) | 1400 (9–15602) | 0.468 |
| ETB (IgA) | 3 (0–1023) | 20 (1–1529) | 0.026 | SEH (IgA) | 88 (0–2083) | 44 (0–2158) | 0.821 |
| FnbpA (IgG) | 208 (20–3374) | 1269 (222–6132) | <0.001 | SEI (IgG) | 4 (0–5154) | 257 (0–3544) | <0.001 |
| FnbpA (IgA) | 73 (1–1047) | 744 (26–5261) | <0.001 | SEI (IgA) | 0 (0–111) | 8 (0–840) | <0.001 |
| FnbpB (IgG) | 111 (9–1089) | 478 (54–2758) | <0.001 | SEJ (IgG) | 105 (5–7274) | 2538 (95–16246) | <0.001 |
| FnbpB (IgA) | 15 (2–253) | 253 (19–2603) | <0.001 | SEJ (IgA) | 1 (0–217) | 25 (0–2236) | <0.001 |
| HlgB (IgG) | 8519 (2595–10365) | 10761 (2979–13212) | 0.001 | SEM (IgG) | 12 (0–7325) | 307 (8–2738) | <0.001 |
| HlgB (IgA) | 474 (10–2799) | 556 (44–6394) | 0.233 | SEM (IgA) | 1 (0–354) | 56 (0–1128) | <0.001 |
| IsdA (IgG) | 2811 (508–8602) | 2665 (576–5183) | 0.732 | SEN (IgG) | 7 (0–1166) | 631 (0–2672) | <0.001 |
| IsdA (IgA) | 248 (13–3997) | 357 (9–8058) | 0.940 | SEN (IgA) | 0 (0–155) | 7 (0–2743) | <0.001 |
| IsdH (IgG) | 929 (44–8170) | 3921 (605–9420) | <0.001 | SEO (IgG) | 40 (7–2009) | 306 (147–653) | <0.001 |
| IsdH (IgA) | 311 (3–4328) | 294 (43–2117) | 0.661 | SEO (IgA) | 3 (0–117) | 20 (4–544) | <0.001 |
| LukD (IgG) | 1082 (1852–15031) | 12226 (3781–16731) | 0.011 | SEQ (IgG) | 5 (0–7125) | 341 (0–3763) | <0.001 |
| LukD (IgA) | 641 (0–3418) | 694 (106–4088) | 0.753 | SEQ (IgA) | 0 (0–132) | 19 (0–2219) | <0.001 |
| LukE (IgG) | 12386 (5270–14191) | 12541 (5955–15590) | 0.732 | SER (IgG) | 3 (0–1797) | 63 (0–2654) | <0.001 |
| LukE (IgA) | 1181 (28–5431) | 664 (38–7766) | 0.524 | SER (IgA) | 0 (0–40) | 9 (0–357) | 0.001 |
| LukF (IgG) | 7321 (1240–12631) | 10579 (2984–14996) | 0.007 | SSL1 (IgG) | 8783 (1520–14022) | 4683 (518–11914) | 0.005 |
| LukF (IgA) | 321 (0–2554) | 433 (38–3916) | 0.041 | SSL1 (IgA) | 198 (0–4066) | 195 (6–2947) | 0.956 |
| LukS (IgG) | 10470 (1939–14385) | 8734 (1662–16976) | 0.135 | SSL11 (IgG) | 867 (43–8824) | 1107 (122–8955) | 0.311 |
| LukS (IgA) | 602 (0–6321) | 1015 (13–10014) | 0.524 | SSL11 (IgA) | 46 (3–1495) | 96 (8–2162) | 0.215 |
| SasG (IgG) | 89 (3–3376) | 197 (3–2791) | 0.093 | SSL3 (IgG) | 7280 (898–12949) | 7813 (1618–15432) | 0.403 |
| SasG (IgA) | 8 (0–5408) | 33 (4–3950) | 0.009 | SSL3 (IgA) | 321 (5–3062) | 519 (24–5234) | 0.244 |
| SCIN (IgG) | 187 (5–7259) | 11333 (7572–15438) | <0.001 | SSL5 (IgG) | 4138 (621–9547) | 3267 (511–5928) | 0.021 |
| SCIN (IgA) | 14 (0–1220) | 2945 (211–10036) | <0.001 | SSL5 (IgA) | 97 (6–1293) | 115 (27–933) | 0.264 |
| SdrD (IgG) | 233 (17–2844) | 1683 (400–6924) | <0.001 | SSL9 (IgG) | 11968 (4788–13605) | 9255 (1005–15161) | 0.150 |
| SdrD (IgA) | 64 (3–2205) | 634 (48–4758) | <0.001 | SSL9 (IgA) | 444 (35–3523) | 138 (20–1158) | 0.001 |
| SdrE (IgG) | 250 (28–6655) | 2045 (251–8626) | <0.001 | TSST-1 (IgG) | 32 (4–9350) | 12051 (0–16927) | <0.001 |
| SdrE (IgA) | 218 (0–3017) | 604 (28–3650) | 0.015 | TSST-1 (IgA) | 4 (0–125) | 932 (10–3504) | <0.001 |
Differences in antigen-specific MFI values between groups were considered to be statistically significant at P<0.05 (Mann-Whitney U test).
Association of anti-staphylococcal antibody levels with presence of genes
To study whether the low antibody levels in rhesus macaques were due to absence of the genes encoding the antigens, 10 isolates from the macaques that were screened three times, were selected for determination of the presence of genes encoding antigens against which a low MFI value was found in macaques, while MFI values in humans were high (SCIN, CHIPS, ClfA, ClfB, Efb, TSST-1, FnbpA, and FnbpB). Results are shown in Table 5. None of the isolates from rhesus macaques were scn-, tst-, or fnbB-positive, which explains why antibody levels were at background level. On the other hand, all 10 isolates were clfB- and efb-positive, and the macaques that carry these isolates had high antibody levels against these antigens. Nine out of 10 S. aureus isolates did not possess the gene encoding for CHIPS, which explains why the MFI values were at background level. The macaque from which the chp-positive strain was isolated also had MFI values at background level. Eight isolates were clfA-positive, with corresponding high MFI values for 6 macaques. Interestingly, the two macaques from which the clfA-negative strains were isolated, still had high MFI values. Nine isolates possessed the gene encoding FnbpA, which corresponds with MFI values just above background level (range 22–616). However, the macaque with the fnbA-negative S. aureus strain had MFI values above background level as well. These differences are probably due to a previous exposure to an other S. aureus strain.
Table 5. Association of anti-staphylococcal antibody levels with presence of genes in 10 S. aureus strains isolated from rhesus macaques.
| SCIN | CHIPS | ClfA | ClfB | |||||||||
| Isolate no. | Gene | MFI value, IgG | MFI value, IgA | Gene | MFI value, IgG | MFI value, IgA | Gene | MFI value, IgG | MFI value, IgA | Gene | MFI value, IgG | MFI value, IgA |
| 1 | − | 356 | 2 | − | 6 | 0 | − | 664 | 82 | + | 291 | 45 |
| 2 | − | 6404 | 77 | − | 23 | 1 | + | 2893 | 8252 | + | 6871 | 10067 |
| 3 | − | 5 | 3 | − | 15 | 1 | + | 3033 | 10785 | + | 997 | 4604 |
| 4 | − | 305 | 2 | − | 8 | 0 | + | 142 | 660 | + | 816 | 238 |
| 5 | − | 759 | 44 | − | 65 | 0 | + | 275 | 5213 | + | 1382 | 805 |
| 6 | − | 3702 | 26 | + | 173 | 3 | − | 282 | 4223 | + | 2423 | 9292 |
| 7 | − | 3067 | 127 | − | 30 | 2 | + | 26 | 7 | + | 866 | 462 |
| 8 | − | 492 | 11 | − | 886 | 0 | + | 27 | 37 | + | 2232 | 310 |
| 9 | − | 155 | 9 | − | 338 | 11 | + | 664 | 3549 | + | 462 | 549 |
| 10 | − | 27 | 76 | − | 8 | 27 | + | 17 | 1574 | + | 192 | 1574 |
Discussion
Nasal carriage of S. aureus plays a key role in the epidemiology and pathogenesis of staphylococcal infections [27], [28]. Eradication of S. aureus from the nose has proven to be effective in the reduction of staphylococcal infections [29]–[32]. This indicates that the anterior nasal region is a primary ecological reservoir of S. aureus [33], [34], although throat and perineum are important reservoirs as well [35]–[37]. However, nasal re-colonization may occur within weeks to months in those who have successfully been decolonized [38], [39]. In order to develop new strategies in prevention of staphylococcal disease, acquiring additional knowledge about the underlying mechanisms of S. aureus nasal carriage is important. Artificial inoculation of human volunteers offers opportunities to study these mechanisms [40]. While such semi-clinical studies in humans remain the most informative, animal models of S. aureus colonization sometimes enable a more detailed investigation of the processes involved in pathogenesis by allowing for more risky interventions.
In the present study, we describe for the first time natural nasal S. aureus carriage in rhesus macaques. The nasal cavity of these macaques appears to be an important reservoir of S. aureus, as is also observed in humans. In a single screening of their noses, 39% of the 731 rhesus macaques had a positive culture, which is comparable to the human situation with ∼20% persistent carriers and ∼30% intermittent carriers [28], [41], [42]. Most of these S. aureus isolates were different from human S. aureus isolates: rhesus macaque isolates formed separate PFGE clusters, 59% had one of the 17 spa types that were not yet described, the previously described spa types t189, t516, t729, t786, and t1814 were rare as well (Ridom SpaServer), in most macaque isolates genes encoding superantigens were not present, and half of the isolates could not be agr typed. Moreover, in the majority of macaque isolates new STs were found which were not related to described STs. Three isolates were part of CC45, and one was part of CC1. These macaque isolates were therefore more comparable to human isolates than the other ones. Differences between macaque and human S. aureus isolates are underlined by major differences in both anti-staphylococcal antibody levels and gene content of the S. aureus isolates between these two species. This indicates that new host-specific lineages of S. aureus have now been found in rhesus macaques.
In contrast to the macaque S. aureus isolates, most human strains harbour at least one gene encoding superantigens, on average 5 or 6 genes, among which those encoding the egc superantigens SEG, SEI, SEM, SEN, and SEO are most prevalent [23], [43], [44]. These genes were also most prevalent in the macaque isolates. The relatively low number of isolates containing superantigen genes could also be due to the fact that the primers used in this study were designed for S. aureus strains isolated from humans. Therefore, it can not be concluded that most macaque S. aureus isolates do not carry superantigen genes at all, but that, at least, if they are present, these genes differ from those found in human S. aureus isolates.
In addition, half of the macaque isolates could not be typed with the current agr multiplex PCR system. Of those that could be typed, most belonged to agr types I or III, which is in contrast to human S. aureus isolates. These agr types are also the largest groups (agr type I 35% and type III 38%), but 25% of the isolates contains agr type II [45]. In contrast, agr type IV is hardly found among human isolates (2%), while 19% of the typeable macaque isolates harboured this type. The agr untypeable S. aureus isolates probably have a deletion of the agr locus or extensive sequence variation at the primer sites and could therefore not be typed. This further emphasizes that S. aureus isolates in rhesus macaques differ from those in humans.
Next, we studied the persistence of nasal S. aureus carriage in 48 rhesus macaques. PFGE and spa typing of these isolates, as well as the absence of certain antigen genes while serum anti-staphylococcal antibody levels against these antigens were high, suggest that persistent nasal S. aureus carriage as observed in humans is not present among these rhesus macaques. Human persistent carriers are usually colonized by the same S. aureus strain over a long time period [46], while half of the studied rhesus macaques carried different S. aureus isolates, even over a 5 month period. Frequent transmission of S. aureus isolates rather than persistent nasal carriage was suggested by the presence of one or two epidemic strains in each animal room.
The absence of persistent nasal carriage was underlined by the absence of an association between nasal carrier status and anti-staphylococcal antibody levels in serum of rhesus macaques, as was observed in humans. Differences in IgG levels between human persistent carriers and non-carriers were reported for alpha hemolysin, major autolysin, IsdA and IsdH, immunodominant secretory antigen A (IsaA), major histocompatibility complex class II analogue protein w (Map-w), ClfB, TSST-1, and SEA. IgA levels were different between persistent carriers and non-carriers for TSST-1, SEA, ClfA, and ClfB [14], [47], [48].
In addition, the differences in anti-staphylococcal antibody levels in rhesus macaques and humans, independent of nasal carrier status, again underlined the differences between S. aureus isolates in humans and those in macaques. The major immunogenic proteins in human S. aureus strains do not seem to elicit a humoral immune response in rhesus macaques.
In conclusion, the present study demonstrates that rhesus macaques are natural hosts of S. aureus, and that S. aureus isolates from rhesus macaques versus those from humans differ in many aspects. Therefore, rhesus macaques are not suitable for studying S. aureus persistent carriage as is seen in humans. However, the nasal cavity is an important reservoir in both species, which implies that the rhesus macaque provides an interesting model for studying short term nasal colonization, and in particular bacterial factors involved in adherence. Furthermore, rhesus macaque provide an autologous system in which transmission of S. aureus strains between individuals can be studied, and are therefore a useful model for studying infection prevention. Whether data from these studies can be directly extrapolated to humans is unclear. However, possibilities for extrapolation from rhesus macaque to human appear reasonable because of their close evolutionary relatedness.
Acknowledgments
The authors wish to thank Deborah Horst-Kreft for her technical assistance. We thank Timothy Foster, Gerard Lina, Silva Holtfreter, Dorothee Grumann, Suzan Rooijakkers, Jan-Ingmar Flock, and John Fraser for kindly supplying the S. aureus proteins.
Footnotes
Competing Interests: The authors have read the journals' policy and have the following conflicts: S.v.d.B. and A.v.B. were in part supported by the Top Institute Pharma project T4-213. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: S.v.d.B. and A.v.B. were in part supported financially by the Top Institute Pharma project T4-213. B.O. and I.K. were in part supported financially by EUPRIM-NET2, grant agreement no. 262443. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1.Needham AJ, Kibart M, Crossley H, Ingham PW, Foster SJ. Drosophila melanogaster as a model host for Staphylococcus aureus infection. Microbiology. 2004;150:2347–2355. doi: 10.1099/mic.0.27116-0. [DOI] [PubMed] [Google Scholar]
- 2.Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun. 2003;71:2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarkowski A, Collins LV, Gjertsson I, Hultgren OH, Jonsson IM, et al. Model systems: modeling human staphylococcal arthritis and sepsis in the mouse. Trends Microbiol. 2001;9:321–326. doi: 10.1016/s0966-842x(01)02078-9. [DOI] [PubMed] [Google Scholar]
- 4.Collins LV, Tarkowski A. Animal models of experimental Staphylococcus aureus infection. In: Fischetti VA, Novick RP, Ferreti JJ, Portnoy DA, Rood JI, editors. Gram-positive pathogens. Washington: ASM Press; 2006. pp. 535–543. [Google Scholar]
- 5.van Belkum A, Melles DC, Peeters JK, van Leeuwen WB, van Duijkeren E, et al. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg Infect Dis. 2008;14:479–483. doi: 10.3201/eid1403.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wulf M, Voss A. MRSA in livestock animals-an epidemic waiting to happen? Clin Microbiol Infect. 2008;14:519–521. doi: 10.1111/j.1469-0691.2008.01970.x. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald JR, Monday SR, Foster TJ, Bohach GA, Hartigan PJ, et al. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J Bacteriol. 2001;183:63–70. doi: 10.1128/JB.183.1.63-70.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herron LL, Chakravarty R, Dwan C, Fitzgerald JR, Musser JM, et al. Genome sequence survey identifies unique sequences and key virulence genes with unusual rates of amino acid substitution in bovine Staphylococcus aureus. Infect Immun. 2002;70:3978–3981. doi: 10.1128/IAI.70.7.3978-3981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Cleef BA, Graveland H, Haenen AP, van de Giessen AW, Heederik D, et al. Persistence of livestock-associated methicillin-resistant Staphylococcus aureus in field workers after short-term occupational exposure to pigs and veal calves. J Clin Microbiol. 2011;49:1030–1033. doi: 10.1128/JCM.00493-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, et al. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun. 2006;74:2215–2223. doi: 10.1128/IAI.74.4.2215-2223.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeClaire RD, Hunt RE, Bavari S. Protection against bacterial superantigen staphylococcal enterotoxin B by passive vaccination. Infect Immun. 2002;70:2278–2281. doi: 10.1128/IAI.70.5.2278-2281.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Q, Li Y, Wen L, Guo S, Chen Y, et al. Safety evaluation of recombinant staphylokinase in rhesus monkeys. Toxicol Pathol. 2003;31:14–21. doi: 10.1080/01926230390173815. [DOI] [PubMed] [Google Scholar]
- 13.Verkaik NJ, Boelens HA, de Vogel CP, Tavakol M, Bode LG, et al. Heterogeneity of the humoral immune response following Staphylococcus aureus bacteremia. Eur J Clin Microbiol Infect Dis. 2010;29:509–518. doi: 10.1007/s10096-010-0888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verkaik NJ, de Vogel CP, Boelens HA, Grumann D, Hoogenboezem T, et al. Anti-staphylococcal humoral immune response in persistent nasal carriers and noncarriers of Staphylococcus aureus. J Infect Dis. 2009;199:625–632. doi: 10.1086/596743. [DOI] [PubMed] [Google Scholar]
- 15.Koning S, van Belkum A, Snijders S, van Leeuwen W, Verbrugh H, et al. Severity of nonbullous Staphylococcus aureus impetigo in children is associated with strains harboring genetic markers for exfoliative toxin B, Panton-Valentine leukocidin, and the multidrug resistance plasmid pSK41. J Clin Microbiol. 2003;41:3017–3021. doi: 10.1128/JCM.41.7.3017-3021.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins TB, Augustine NH, Hill HR. Development of a multiplexed fluorescent immunoassay for the quantitation of antibody responses to group A streptococci. J Immunol Methods. 2006;316:97–106. doi: 10.1016/j.jim.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Verkaik N, Brouwer E, Hooijkaas H, van Belkum A, van Wamel W. Comparison of carboxylated and Penta-His microspheres for semi-quantitative measurement of antibody responses to His-tagged proteins. J Immunol Methods. 2008;335:121–125. doi: 10.1016/j.jim.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Melles DC, Gorkink RF, Boelens HA, Snijders SV, Peeters JK, et al. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J Clin Invest. 2004;114:1732–1740. doi: 10.1172/JCI23083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aires-de-Sousa M, Boye K, de Lencastre H, Deplano A, Enright MC, et al. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J Clin Microbiol. 2006;44:619–621. doi: 10.1128/JCM.44.2.619-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francisco AP, Bugalho M, Ramirez M, Carrico JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics. 2009;10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, et al. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holtfreter S, Grumann D, Schmudde M, Nguyen HT, Eichler P, et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J Clin Microbiol. 2007;45:2669–2680. doi: 10.1128/JCM.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tristan A, Ying L, Bes M, Etienne J, Vandenesch F, et al. Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J Clin Microbiol. 2003;41:4465–4467. doi: 10.1128/JCM.41.9.4465-4467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J Bacteriol. 2006;188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, et al. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med. 2004;10:243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 28.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 29.Kalmeijer MD, Coertjens H, van Nieuwland-Bollen PM, Bogaers-Hofman D, de Baere GA, et al. Surgical site infections in orthopedic surgery: the effect of mupirocin nasal ointment in a double-blind, randomized, placebo-controlled study. Clin Infect Dis. 2002;35:353–358. doi: 10.1086/341025. [DOI] [PubMed] [Google Scholar]
- 30.Perl TM, Cullen JJ, Wenzel RP, Zimmerman MB, Pfaller MA, et al. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N Engl J Med. 2002;346:1871–1877. doi: 10.1056/NEJMoa003069. [DOI] [PubMed] [Google Scholar]
- 31.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 32.Bode LG, Kluytmans JA, Wertheim HF, Bogaers D, Vandenbroucke-Grauls CM, et al. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med. 2010;362:9–17. doi: 10.1056/NEJMoa0808939. [DOI] [PubMed] [Google Scholar]
- 33.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10:505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams RE. Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol Rev. 1963;27:56–71. doi: 10.1128/br.27.1.56-71.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wertheim HF, Verveer J, Boelens HA, van Belkum A, Verbrugh HA, et al. Effect of mupirocin treatment on nasal, pharyngeal, and perineal carriage of Staphylococcus aureus in healthy adults. Antimicrob Agents Chemother. 2005;49:1465–1467. doi: 10.1128/AAC.49.4.1465-1467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acton DS, Plat-Sinnige MJ, van Wamel W, de Groot N, van Belkum A. Intestinal carriage of Staphylococcus aureus: how does its frequency compare with that of nasal carriage and what is its clinical impact? Eur J Clin Microbiol Infect Dis. 2009;28:115–127. doi: 10.1007/s10096-008-0602-7. [DOI] [PubMed] [Google Scholar]
- 37.Mertz D, Frei R, Jaussi B, Tietz A, Stebler C, et al. Throat swabs are necessary to reliably detect carriers of Staphylococcus aureus. Clin Infect Dis. 2007;45:475–477. doi: 10.1086/520016. [DOI] [PubMed] [Google Scholar]
- 38.Leigh DA, Joy G. Treatment of familial staphylococcal infection–comparison of mupirocin nasal ointment and chlorhexidine/neomycin (Naseptin) cream in eradication of nasal carriage. J Antimicrob Chemother. 1993;31:909–917. doi: 10.1093/jac/31.6.909. [DOI] [PubMed] [Google Scholar]
- 39.Watanakunakorn C, Axelson C, Bota B, Stahl C. Mupirocin ointment with and without chlorhexidine baths in the eradication of Staphylococcus aureus nasal carriage in nursing home residents. Am J Infect Control. 1995;23:306–309. doi: 10.1016/0196-6553(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 40.Wertheim HF, Walsh E, Choudhurry R, Melles DC, Boelens HA, et al. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med. 2008;5:e17. doi: 10.1371/journal.pmed.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eriksen NH, Espersen F, Rosdahl VT, Jensen K. Carriage of Staphylococcus aureus among 104 healthy persons during a 19-month period. Epidemiol Infect. 1995;115:51–60. doi: 10.1017/s0950268800058118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu L, Umeda A, Kondo S, Amako K. Typing of Staphylococcus aureus colonising human nasal carriers by pulsed-field gel electrophoresis. J Med Microbiol. 1995;42:127–132. doi: 10.1099/00222615-42-2-127. [DOI] [PubMed] [Google Scholar]
- 43.Thomas D, Chou S, Dauwalder O, Lina G. Diversity in Staphylococcus aureus enterotoxins. Chem Immunol Allergy. 2007;93:24–41. doi: 10.1159/000100856. [DOI] [PubMed] [Google Scholar]
- 44.Becker K, Friedrich AW, Lubritz G, Weilert M, Peters G, et al. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J Clin Microbiol. 2003;41:1434–1439. doi: 10.1128/JCM.41.4.1434-1439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collery MM, Smyth DS, Twohig JM, Shore AC, Coleman DC, et al. Molecular typing of nasal carriage isolates of Staphylococcus aureus from an Irish university student population based on toxin gene PCR, agr locus types and multiple locus, variable number tandem repeat analysis. J Med Microbiol. 2008;57:348–358. doi: 10.1099/jmm.0.47734-0. [DOI] [PubMed] [Google Scholar]
- 46.Noble WC, Williams RE, Jevons MP, Shooter RA. Some aspects of nasal carriage of staphylococci. J Clin Pathol. 1964;17:79–83. doi: 10.1136/jcp.17.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clarke SR, Brummell KJ, Horsburgh MJ, McDowell PW, Mohamad SA, et al. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J Infect Dis. 2006;193:1098–1108. doi: 10.1086/501471. [DOI] [PubMed] [Google Scholar]
- 48.Dryla A, Prustomersky S, Gelbmann D, Hanner M, Bettinger E, et al. Comparison of antibody repertoires against Staphylococcus aureus in healthy individuals and in acutely infected patients. Clin Diagn Lab Immunol. 2005;12:387–398. doi: 10.1128/CDLI.12.3.387-398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]