Abstract
Recent reports highlight the severity and the morbidity of disease caused by the long neglected malaria parasite Plasmodium vivax. Due to inherent difficulties in the laboratory-propagation of P. vivax, the biology of this parasite has not been adequately explored. While the proteome of P. falciparum, the causative agent of cerebral malaria, has been extensively explored from several sources, there is limited information on the proteome of P. vivax. We have, for the first time, examined the proteome of P. vivax isolated directly from patients without adaptation to laboratory conditions. We have identified 153 proteins from clinical P. vivax, majority of which do not show homology to any previously known gene products. We also report 29 new proteins that were found to be expressed in P. vivax for the first time. In addition, several proteins previously implicated as anti-malarial targets, were also found in our analysis. Most importantly, we found several unique proteins expressed by P. vivax.This study is an important step in providing insight into physiology of the parasite under clinical settings.
Introduction
Plasmodium vivax has been long neglected as a major malaria parasite since it is the causative agent of benign malaria, in favor of Plasmodium falciparum that causes fatal cerebral malaria. However, P. vivax is the second leading cause of malaria outside Africa inflicting about 40% of the world population. In Asia, P. vivax accounts for about 50% of malaria cases [1]. P. vivax is known to cause severe morbidity in individuals infected with this parasite leading to paroxysmic fever, severe anemia and thrombocytopenia [1]. Although drug-resistant species of both P. vivax as well as P. falciparum have been reported, P. vivax vaccine and drug target discovery operate only in the wake of P. falciparum vaccine and drug target development [2].
Evidences from recent studies have suggested significant differences in the gene families employed by these parasites. Genome analysis of P. vivax isolated from malaria patients have shown that in spite of its resemblance with other malaria parasites in its gene content and metabolic processes, P. vivax possesses novel gene families and alternative invasion pathways [3]. These differences may lead to distinctive clinical features of P. vivax. For example, unlike P. falciparum which infects red blood cells, P. vivax preferentially infects reticulocytes expressing Duffy blood group antigens (Fya and Fyb), which act as receptors for proteins PvRBP1 and PvRBP2 expressed at the apical pole of P. vivax merozoites. Additionally, the presence of hypnozoite stage and the absence of cytoadherence in P.vivax make its biology different from that of P. falciparum. Due to these differences in the biology as well as clinical properties of P. vivax and P. falciparum, our knowledge of P. falciparum cannot be extrapolated to P. vivax.
One of the major hurdles in the study of P. vivax-malaria is the absence of a long-term in vitro culture system, unlike P. falciparum [4]. As a result, our knowledge about P. vivax is limited to a few studies that have been carried out using short-term cultures of P. vivax and clinically isolated parasites. Our current understanding about the P. vivax life cycle is restricted only to the information provided by the global transcriptome analyses of this parasite from clinical samples and limited proteomics analysis of only schizont stage P. vivax parasites that have been culture adapted [5], [6], [7]. Although transcriptome analysis provides useful understanding at the level of gene expression, they do not reflect the active protein component of a cell. Further, parasites such as Plasmodium, interact with and respond to host environmental cues which can often be revealed by proteomic analyses of the parasite as it is present within the host. However unlike P. falciparum, the proteome of this parasite has not yet been explored either from short-term culture adapted parasites or parasites directly isolated from malaria patients. Dearth of any available information regarding the active protein profile employed by this parasite in order to invade and establish infection in humans makes it imperative to carry out proteomic analysis of the parasite directly isolated from patients who suffer from vivax-malaria. This will indeed facilitate deeper understanding of the pathology of this disease. The proteomic analysis of P. vivax, however, is hampered by limited parasitemia of usually <0.5%, seen in case of P. vivax infections as it infects only reticulocytes, which comprise 1–3% of the total RBCs [8]. This is in contrast to P. falciparum which is able to infect RBCs of all ages, resulting in a relatively higher average parasitemia of >5%.
Our previous study [9] gave the first ever insight into the proteome of malarial parasites at the clinical level. In the case of P. falciparum a reference proteome from lab cultures existed prior to the analysis of the clinical proteome [10], [11]. However, knowledge of P. vivax proteome is extremely limited [7]. In this study, we report the first ever in-depth proteomic analysis of P. vivax isolated directly from malaria patients. We were able to identify 153 proteins from the blood stages of P. vivax. Interestingly, more than 36% of the parasite proteome comprised of hypothetical proteins. We were also able to identify 16 proteins for which there are no homologs in P. falciparum. Overall, our analysis reinforces the belief that P. vivax biology needs to be examined independent of P. falciparum and has several novel features unique to this organism that can be exploited for therapeutic intervention against this major parasite.
Materials and Methods
Ethics Statement
The present study has been approved by the Institutional Scientific Advisory Committee of National Institute of Malaria Research (ICMR), New Delhi, India. A written informed consent was obtained from each patient participated in this study.
Collection of Parasites from Patients
5 ml of blood was collected in heparin coated tubes from patients diagnosed with malaria (with written informed consent). Microscopic examination of peripheral blood smear was done for the presence of malarial parasites. Additionally, Falcivax rapid diagnostic test based on PfHRP-2 and P. vivax specific LDH was used for the differentiation of P. falciparum and P. vivax malaria. Samples positive only for P. vivax were used for this study. Here we must mention that RDT may sometimes detect mixed infections of P. vivax with P. falciparum. In order to avoid this, more specific tests such as nested PCR can be performed. However, since samples used in our study were directly taken from patients for proteomics-processing, we have ensured greater stringency during data analyses as detailed below.
Sample Preparation
Parasitized whole blood cells were centrifuged at 1,500 g for 15 min for the removal of plasma and buffy coat. Blood pellet thus obtained was diluted with PBS and was layered on Histopaque (Sigma Aldrich) for the removal of white blood cells. P. vivax infected cells was purified from uninfected cells by layering the cell suspension on a 45, 50 and 65% Percoll gradient. An enriched fraction of asexual stages of P. vivax was obtained with little contamination of host cells. For extraction of P. vivax proteins, enriched parasitized cells were either boiled directly in SDS sample buffer or lysed with Saponin (Fluka) to obtained soluble and insoluble fractions. Saponin insoluble pellets were further processed as described earlier [12]. Briefly, the saponin pellet was lysed first in Triton X containing buffer, Buffer A (10 mM Tris HCl, pH 7.4; 5 mM EDTA; 1% Triton X 100) followed by lysis in urea containing buffer, Buffer B (10 Mm Tris HCl, pH 7.4; 5 mM EDTA; 1% SDS; 6 M Urea). Soluble fractions obtained at each step were acetone precipitated and the final insoluble pellet was directly boiled in SDS loading buffer. Purified parasites or saponin pellet was maintained at 4°C throughout the lysis procedure. All theses samples were separated on a 10% SDS-PAGE followed by in gel digestion as described by Wilm et al. 1996 with slight modification [13]. In brief, each lane was cut into pieces of width 2 mm. These pieces were further chopped into small pieces, which were reduced with 10 mM dithiotreitol followed by alkylation using 55 mM iodoacetamide. Alkylated gel pieces were incubated with 400 ng of trypsin (20 µl of trypsin from the solution of 20 ng/µl trypsin (Trypsin Gold, Promega), in 25 mM NH4OAc for 14–16 hr at 37°C. Trypsin digested peptides were eluted from the gel pieces using 60% acetonitrile (ACN) and 5% formic acid and were subjected to LC-MS/MS.
Nano LC-MS/MS analysis of malaria proteins
Digested peptides were analyzed by nano LC- MS/MS as described previously [8]. Briefly, peptide mixtures were dissolved in 25 µl of sample preparation solution (98% water, 2% Acetonitrile and 0.5% Formic acid) and injected into Nano-LC through an autosampler system. Peptides were eluted using nano- reverse phase column (Michrom C18 5 µ 300 Å) which was further connected to the Nano Spray ESI- QTOF system (Qstar Elite, Applied Biosystems). A gradient of water and acetonitrile was set up for 60 minutes with a flow rate of 400 nL/minute. Eluted peptides from the column were ionized using ESI source with ion spray voltage 2250 V and temperature 120°C. Ionized peptides were analyzed by one full MS scan and four consecutive product ion scans of the four most intense peaks, using rolling collision energy. An Information Dependant Acquisition (IDA) experiment was used to specify the criteria for selecting each parent ion for fragmentation which included selection of ions in m/z range: >400 and <1600, of charge state of +2 to +5, exclusion of former target ions for 30 seconds, accumulation time of 1 second for a full scan and 2 seconds for MS/MS. The data generated by the Analyst software was stored in a .wiff format.
Data Analysis
Data was analyzed using ProteinPilot version 4.0 software. The original data files were analyzed using the ProteinPilot version 4.0 software with a combined database (NCBI Human DB 2008 (containing 39125 non-redundant protein entries, 18.8 Mb), PlasmoDB Plasmodium vivax version 7.1 (containing 5432 redundant protein entries, 4.4 Mb), and PlasmoDB Plasmodium falciparum October 2008 (containing 11669 redundant protein entries, 4.8 Mb) with a total of 56226 protein entries using Paragon Algorithm. The number of missed cleavages permitted was two. During the analysis, in the search parameters modification of cysteine by idoacetamide and biological modifications programmed in algorithm were allowed. Mass tolerance for precursor ion and fragment ions were set to 100 ppm and 0.2 Da respectively. In Paragon Algorithm, protein score is calculated on the basis of percentage confidence level of the peptides identified. Protein score of minimum 0.47 (fit incorrect rate is 0%) corresponding to a confidence level greater than 66% were used. Several P. vivax proteins detected had a score of less than 0.47 and these have been included in a separate list in Table S6.
In order to rule out false discoveries, we have carried out False Discovery Rate (FDR) analysis. For this, ProteinPilot 4.0 with Paragon algorithm has been used for data analysis. As part of the Paragon analysis method, a false discovery rate (FDR) analysis of the results has been carried out by the Proteomics Performance Evaluation Pipeline Software (PSPEP). Finally, proteins have been selected on the basis of their critical FDR value i.e. 1%.
In order to avoid identifications based on redundant peptides in our proteome, we have not included proteins that have no unique peptide identifications (Table S4). Proteins which share some peptides as well as have unique peptide-identifications, have been grouped accordingly.
Annotation of hypothetical proteins
About 36% of the P. vivax proteome consisted of hypothetical proteins. We have carried out sequence based domain search using the SMART and Pfam tools and identifying domains present in them. A molecular class was assigned to the hypothetical proteins based on the identified domain.
Results
Functional classification of proteins expressed by P. vivax
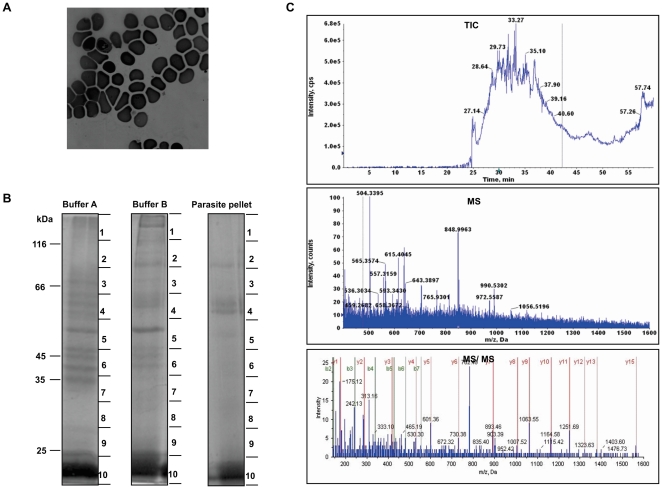
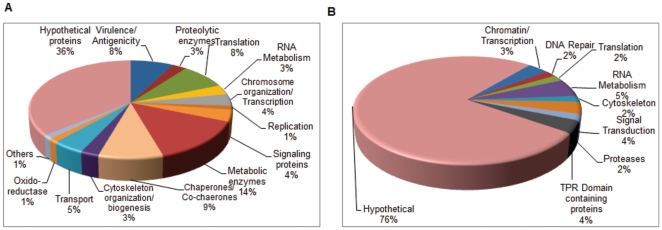
To study the proteome of P. vivax, we collected peripheral blood from P. vivax infected patients attended to the OPD of Wenlock Government Hospital, Mangalore, Karnataka, India and S.P. Medical College and Associated Group of Hospitals, Bikaner, India. P. vivax infected blood was collected before administration of any drugs. To minimize the host proteins contamination parasitized erythrocytes were purified by percoll density gradient as described in “Materials and Methods”. A typical sample consisting of about 5 mL of parasitized blood of 0.1% parasitemia contains 1×107 infected reticulocytes. However, at each step of purification there is a loss of sample which therefore, resulted in a 1×105 infected reticulocytes after percoll purification. Proteins were extracted from enriched fraction either by directly boiling in sample buffer or by sequential lysis using SDS containing buffer or urea containing buffer. After each lysis step proteins were separated on SDS-PAGE (Figure 1B). Each of these lanes were divided into 10 pieces and were subjected to in-gel trypsin digestion. Digested peptides were extracted in 60% ACN and 5% formic acid and were analyzed by LC-MS/MS-QTOF. The representative TIC, MS and MS/MS spectra for one of the sample is given in Figure 1C. We were able to identify 137 proteins from the asexual stages of P.vivax. In our previous study, we have identified 16 P. vivax proteins. Altogether, we here represent 153 proteins expressed by asexual stages of P. vivax (Table S1). P. vivax proteome comprises of proteins consisting of chaperones, glycolytic enzymes, signaling proteins, RNA processing, chromatin and chromosome organization, cytoskeleton organization, transport, virulence, transcription and translational regulatory and hypothetical proteins (Figure 2A). Majority of these proteins belong to the hypothetical protein group followed by proteins involved in metabolic processes, protein folding (chaperones and co-chaperones) and pathogenicity. Based on the presence of domains, we have classified hypothetical proteins into various categories along with other annotated proteins as described below. Of all the hypothetical proteins about 76% did not contain any identifiable domains. From here onwards, the annotated hypothetical proteins are no longer analyzed as hypothetical proteins. Instead, they are categorized as proteins with known putative functions (Figure 2B) (Table S2).
Figure 1. Mass spectrometric analysis of proteins employed by asexual stages of P. vivax.
A. Giemsa-stained image of peripheral blood smear of P. vivax infected patient. B. SDS-PAGE profile of proteins extracted from asexual stages of P. vivax by sequential lysis using SDS buffer (lane 1), urea containing buffer (lane 2) and direct boiling of pellet in Laemmli buffer (lane 3). C. Represents the Total Ion Chromatogram (TIC), MS, and MS/MS spectra for enolase (PVX_095015).
Figure 2. Functional profiles of protein expressed by asexual stages of P. vivax.
A. Proteins identified in asexual stages were plotted as a function of their broad functional classification as defined in PlasmoDB or GO. Only one class was assigned to one protein to avoid any redundancy. Major group in the P. vivax is represented by hypothetical proteins followed by metabolic enzymes, chaperones and proteins involved in virulence. B. Showing functional plot for hypothetical proteins. About 25% of the hypothetical proteins were assigned to different functional classes on domain architecture.
Additionally, we have identified 7 proteins which were entirely specific to P. vivax (do not contain homologs in any Plasmodium species) and 16 proteins for which there were no homologs in P. falciparum (Table 1). Of the 16 proteins that did not have homologs in P. falciparum, 2 were Vir proteins and 8 were P-fam proteins, known to be involved in virulence/antigenicity of P. vivax.
Table 1. List of uniquely expressed proteins in P. vivax isolated from patients.
| Serial No. | Accession No. | Protein | Mw/pI | Probable function |
| 1 | PVX_074690* | Variable surface protein Vir4-related | 48.4/9.29 | Virulence/Antigenicity |
| 2 | PVX_096985* | Variable surface protein Vir, putative | 44.6/5.88 | Virulence/Antigenicity |
| 3 | PVX_101520* | Pv-fam-d protein | 49.5/9.7 | Virulence/Antigenicity |
| 4 | PVX_003525* | Hypothetical protein | 211.6/4.49 | Not Known |
| 5 | PVX_089835* | RAD protein | 398.77/10.01 | Virulence/Antigenicity |
| 6 | PVX_092995* | Tryptophan-rich antigen (Pv-fam-a) | 429.99./10.1 | Virulence/Antigenicity |
| 7 | PVX_118695* | Pv-fam-d protein | 103.2/9.85 | Not Known |
| 8 | PVX_003545 | Hypothetical protein, conserved | 34.4/6.31 | Not Known |
| 9 | PVX_003555 | Hypothetical protein, conserved | 119.2/3.82 | Not Known |
| 10 | PVX_083555 | Hypothetical protein | 16.5/4 | Not Known |
| 11 | PVX_090265 | Tryptophan-rich antigen (Pv-fam-a) | 40.1/6.57 | Virulence/Antigenicity |
| 12 | PVX_096950 | Tryptophan-rich antigen (Pv-fam-a) | 39.8/9.92 | Virulence/Antigenicity |
| 13 | PVX_092990 | Tryptophan-rich antigen (Pv-fam-a) | 157.5/3.36 | Virulence/Antigenicity |
| 14 | PVX_112670 | Tryptophan-rich antigen (Pv-fam-a) | 38.5/6.11 | Virulence/Antigenicity |
| 15 | PVX_090230 | Early transcribed membrane protein (ETRAMP) | 15.8/10.78 | Virulence/Antigenicity |
| 16 | PVX_097880 | Hypothetical protein | 145/10.33 | Not Known |
‘*’Refers to proteins only present in P. vivax. Remaining proteins are present in any one species of Plasmodium except P. falciparum.
Drug targets and vaccine candidates in P. vivax
Several proteins that are well known drug targets and vaccine candidates in P. falciparum are also expressed in P. vivax (Table 2). Plasmepsin IV, m1-family aminopeptidaese, spermidine synthase, fructose 1, 6- bisphosphate aldolase, pyridoxal kinase, triosephosphate isomerase, L-lactate dehydrogenase, Hsp90 and Hsp70 are all well known drug targets in P. falciparum against which inhibitors are known [14]–[28]. Some vaccine candidates such as the duffy receptor precursor, which is unique to P. vivax, were also detected [3], [29], [30]. Earlier, sequencing of P. vivax revealed several novel gene families which are located in the subtelomeric regions [3]. Most notably, we found at least five members of Pv-fam-a (tryptophan rich antigen) in patients suffering from vivax malaria (Table 2). There are at least 36 members of pv-fam-a family of proteins, one of which has previously been reported to elicit humoral immune response and hence a probable vaccine candidates [31]. Table 2 contains a list of potential drug targets/vaccine candidates in P. vivax whose efficacy can be revealed only upon further research.
Table 2. Putative vaccine candidates and drug targets identified in P. vivax isolated from patients.
| Potential Drug/Vaccine Targets | Putative Function | Drug Inhibitors (If known) | Ref. | |
| Protein ID | Description | |||
| PVX_092990 PVX_092995 PVX_112670 PVX_096950 PVX_090265 | tryptophan-rich antigen (Pv-fam-a) | Immune evasion | - | [3], [30] |
| PVX_093680 | Phist protein (Pf-fam-b) | Immune evasion | [3] | |
| PVX_118695 PVX_101520 | Pv-fam-d protein | Immune evasion | - | [3] |
| PVX_089835 | RAD protein (Pv-fam-e) | Immune evasion | - | [3] |
| PVX_110810 | Duffy receptor precursor | Invasion | - | [28] |
| PVX_086040 | Aspartic Protease Plasmepsin IV | Hemoglobin degradation | C 2-symmetric compounds encompassing the 1,2-dihydroxyethylene scaffold and a variety of elongated P1/P1′ side chains | [13] |
| PVX_122425 | M1-family aminopeptidase, putative | hemoglobin digestion | hPheP[CH2]Phe, Co4, and Bestatin | [14], [15] |
| PVX_092065 | Spermidine synthase, putative | Spermine synthesis | cyclohexylamine, dicyclohexylamine | [16] |
| PVX_118255 | Fructose 1,6-bisphosphate aldolase, putative | Glycolysis and Invasion | - | [17]–[19] |
| PVX_113935 | Pyridoxal kinase | Biotin Synthesis | 6-diazo-5-oxo-L-norleucine | [20] |
| PVX_118495 | Triosephosphate isomerase, putative | Glycolysis | Synthetic interface peptides | [21], [22] |
| PVX_116630 | Lactate dehydrogenase | Glycolysis | Gossypol derivatives, oxamate derivatives | [23] |
| PVX_087950 | Hsp90 | Signal transduction, Cell proliferation | Geldanamycin, 17AAG | [24], [25] |
| PVX_089425 | Hsp70 | Chaperone activity | pyrimidinone-amides | [26], [27] |
Comparison of P. vivax proteome with transcriptome
All studies on P. vivax till date have been carried out with either parasites directly isolated from malaria patients or with isolates adapted to short term culture. The amount of information available about the gene expression and protein expression profiles of P. vivax is limited to two transcriptome studies [5], [6] and this proteomics study. Upon closer examination/inspection of proteins reported in this study with two transcriptome datasets we found no transcript evidence for three proteins. These are PVX_114832 (Elongation factor 1 alpha, putative), PVX_250300 (ADP/ATP transporter on adenylate translocase, putative) and PVX_220290 (putative cyclophilin). This observation is intriguing as the previous two studies have used parasite materials from two different geographically distinct regions (one from Thailand and the other from Peru) [5], [6]. Furthermore, these studies included the stage specific transcriptome of intraerythrocytic developmental cycle. Individual differences in gene expression profiles among field isolates have been proposed to reflect parasites adaptability in response to host environment [32], [33]. Based on proteomic evidence from the field isolates of P. vivax, we propose that it is possible that these three genes, for which no transcript data exists, either reflect an altered physiological state or are expressed specifically in Indian isolates. This interpretation is based on the assumption that probes for all P. vivax genes have been included in the transcriptome analysis.
In addition, we found 14 proteins which show differential expression at transcript levels in different individuals from the two transcriptome studies mentioned above, which may relate to different physiological states [5], [6]. These are PVX_122850 (Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, putative), PVX_099275 (hypothetical protein), PVX_080555 (hypothetical protein), PVX_086990 (vacuolar ATP synthase subunit E), PVX_095195 (ATP-dependent RNA helicase, putative), PVX_099095 (elongation factor 1B, putative), PVX_121930 (hypothetical protein), PVX_092630 (hypothetical protein), PVX_098620 (hypothetical protein), PVX_124190 (hypothetical protein), PVX_113635 (hypothetical protein), PVX_101480 (hypothetical protein), PVX_087690 (hypothetical protein) and PVX_110810 (Duffy receptor precursor). As is evident, nine of these are annotated as hypothetical in PlasmoDB database. Domain analysis of these hypothetical proteins revealed that two of them have DNA repair (PVX_099275) and chromatin assembly (PVX_092630) as a putative function. These could be proteins involved in responding to environmental cues and modulating pathways that may affect parasite survival.
The P. vivax interactome
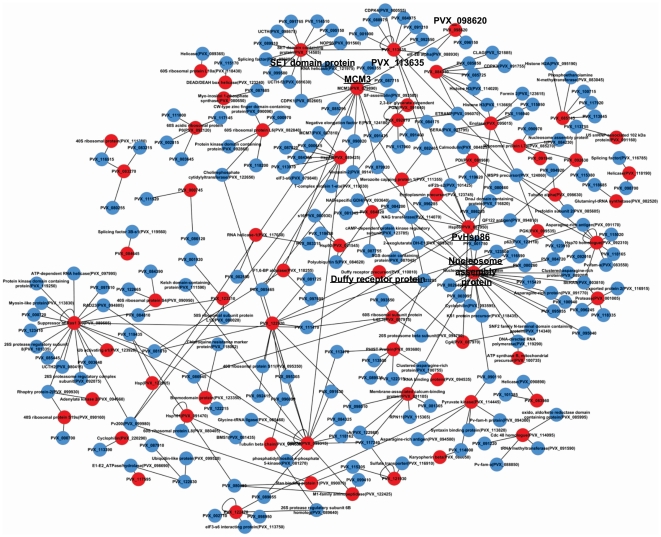
In order to gain further insight into the functions of the P. vivax proteins detected in our study, we have constructed an interactome of the P. vivax proteome. For this, we first looked for P. falciparum homologs for the proteins detected in our study. We then selected those homologs for which interaction data was available in literature and found homologs for their interactors in the P. vivax genome [34], [35]. We then constructed an interactome consisting of P. vivax proteins detected in our study with other P. vivax proteins (Figure 3). Of the 153 proteins detected in our study, we found P. falciparum homologs for 137, 16 proteins were specific for P. vivax. Interactors for only 68 homologs could be found in literature and these were extrapolated to form the P. vivax interactome. In total, 309 interactions were included. Interestingly, many of the proteins found in our study form highly interconnected and busy networks in the parasite with unannotated hypothetical proteins indicating that these could be Plasmodium-specific pathways. For instance, MCM3, involved in chromosome maintenance, interacts directly with PVX_098620 and forms a network indirectly with SET domain containing protein and PVX_113635. Another highly interconnected hub is formed by Hsp86, nucleosome assembly protein 1 and Cg4, all of which have been detected in our study. Interestingly, Hsp86 is indirectly associated with Duffy receptor protein through Cg4 (member of Hsp70 family) indicating its involvement in virulence of the parasite. The P. vivax interactome for the proteins detected in our study indicate that highly interconnected and possibly active regulatory networks exist in the parasite, and several vivax-specific networks are awaiting discovery.
Figure 3. Interaction network of the proteins identified in P. vivax from patient.
Interaction network of P. vivax proteins detected in our study has been constructed based on the presence of interactions of their P. falciparum homologs. The P. vivax proteins which have been detected in our study have been colored as red nodes. Many proteins detected in P. vivax from malaria patients form highly interconnected hubs showing the regulatory role of these proteins in several processes in vivax malaria. The major nodes have been indicated with an enlarged font and are hyperlinked to PlasmoDB.
Discussion
Research on human malaria caused by P. vivax has not received the attention that it deserves due to several factors. Firstly, unlike P. falciparum, P. vivax is not amenable to laboratory culture. Secondly (and more importantly), P. vivax, untill recently, was considered to be a parasite that causes mild and non-fatal disease. However, recent studies have shown that, in malaria endemic countries, like India, Indonesia, Papuea New Guinea, approximately 27% of the patients with severe malaria symptoms are due to P. vivax infection [36]. As with P. falciparum, drug resistance poses a severe problem in the treatment of vivax-malaria. Drug resistant strains of P. vivax have evolved against commonly used anti-malarials such as chloroquine, primaquine and sulphadoxine-pyrimethamine [37]–[39]. Notwithstanding the recent advances in global studies of P. vivax by transcriptomics and next generation sequencing [5], [6], [40], the amount of knowledge that exists about P. vivax is insufficient to understand its biology that will enable discovery of newer drug targets against the parasite.
Although transcriptome analysis documents a complete picture of the mRNA complement of a cell, they provide an inadequate view of the operational cellular protein networks. The absence of long term cultures of P. vivax both necessitates and provides us with an opportunity to study this parasite in the wild, i.e. directly from malaria-infected patients. In this context, it is essential to study the proteome of P. vivax in order to identify the molecular factors involved in P. vivax patho-biology. Proteomic analysis of the clinical isolates of Plasmodium is a difficult task owing to the low parasitemia and masking of parasite proteins by abundant host proteins.
Despite these technical shortcomings, we were able to isolate P. vivax parasites from patient's samples to enable a proteomic analysis. Although we could not completely eliminate host protein contamination, we were able to detect 153 P. vivax proteins in our study. Besides parasite proteins, we detected about 315 proteins of host-origin (Table S3). Of these, about 212 (Table S5) have been detected at transcript levels in the human reticulocyte transcriptome [41]. The other proteins may be from reticulocytes or remnants of other contaminating human blood cells such as leukocytes that have not been successfully removed from the sample.
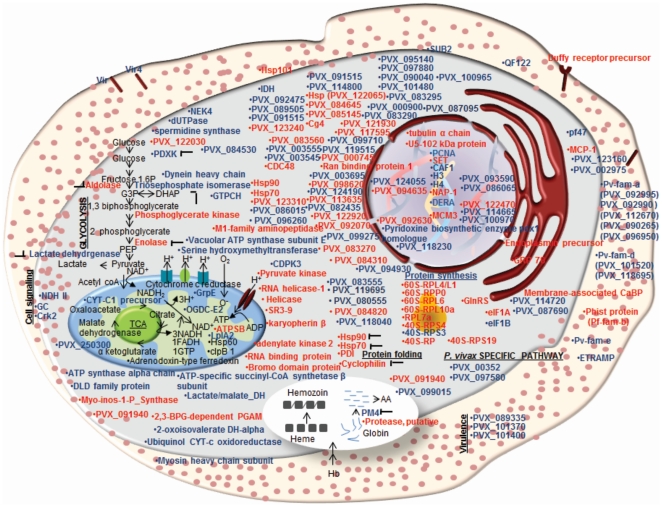
Additionally, 29 P. vivax proteins were detected for which no MS based evidence is available in any other Plasmodium species (Table S3). Majority (about 36%) of the detected P. vivax proteins consisted of hypothetical proteins. Further domain analysis by Pfam and SMART databases revealed putative function for 16 out of 55 hypothetical proteins detected in this study (Table S2). Amongst the 99 proteins with known function, enzymes involved in general metabolic functions were heavily represented (22 proteins), followed by proteins involved in chaperoning function (14) and translation (13) (Figure 2A). Several of these proteins are targets for chemotherapeutic interventions (Table 2). Notably, some of these proteins like PfA-M1, Spermidine synthase, Lactate dehydrogenase and hsp90 have been validated as drug targets. Besides some of the known drug targets, several proteins with immunogenic properties were detected which can be further exploited for designing vaccines. Most importantly, we found that the members of Pv-fam-a family (Tryptophan rich antigens) of proteins were abundantly expressed in the field isolates of P.vivax which warrants further evaluation of their candidature as vaccines. Figure 4 schematically describes the localization of the identified proteins in P. vivax and also gives a snap-shot of the biochemical pathways that would be operational in the clinical state of P. vivax.
Figure 4. Schematic representations of pathways operational in the P. vivax from patient.
The figure depicts the cellular localization of the identified proteins. Proteins indicated in blue represent the ones detected in the clinical isolate of P. vivax and black shows the pathways they are involved in. ‘ ’ indicates potential targets for anti-malarials.
’ indicates potential targets for anti-malarials.
We provide evidence for expression of four proteins which were hitherto not detected at the transcript level in earlier genome-wide analysis. This discrepancy can be explained in part due to an altered physiological state of the parasite in response to the host environment, though differential expression of genes by parasites from geographically distinct regions cannot be ruled out. The chief difference between the previous transcriptome analysis and this proteomics study lies in the regions of the world from where P. vivax samples have been collected. The two transcriptome analyses have collected samples from Thailand and Peru whereas our samples are from Mangalore, India. It is possible that different strains of P. vivax express marginally different protein complements.
An important finding of our study was the presence of 16 proteins that are entirely unshared by P. falciparum indicating that these proteins are likely involved in P. vivax specific infection. This is interesting considering the significant differences between P. falciparum and P. vivax infection such as preference for different host cells and extent of sequestration within host capillaries. It is possible that the unique proteins detected in our study are involved in important vivax-specific pathways.
This is the first proteomics analysis of P. vivax-an important first step towards understanding of P. vivax cell biology. Through this study, we have identified the most abundant proteins expressed in clinical P. vivax during its infection of the host such as possible regulatory proteins and strain specific proteins (Figure 4). These proteins provide a glimpse of the cellular pathways operational in the parasite. One limitation of this study is that, it was carried out with P. vivax samples pooled from different donors and hence we could not associate in-vivo parasite biological states with host environment, such as immune status of the patient. To better appreciate the host-parasite dynamics, additional cohort studies involving patients with asymptomatic parasitemia, mild and severe disease are required. This will not only facilitate identification of molecular factors important for clinical manifestation of malaria but will also lead to designing of better chemotherapeutic or vaccine intervention strategies.
Supporting Information
List of P. vivax proteins identified in clinical malaria sample and peptides identified.
(XLS)
Functional classification of hypothetical proteins based on predicted PFAM domains.
(DOC)
P. vivax proteins which do not have Mass Spectrometric evidence in any species of Plasmodium.
(XLS)
Host proteins identified along with the proteome of P. vivax.
(XLS)
Plasmodium vivax host proteins which are present at transcriptome level.
(XLS)
P. vivax proteins detected in this study which have a score less than 0.47.
(XLS)
Acknowledgments
The authors would like to thank Ms. Shweta Chaubey for critical reading of the manuscript. They would also like to express their gratitude to the patients who participated in this study.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Grant No: DBTO201, “A Systems Biology Approach towards Understanding Severe Malaria with Special Reference to the Transcriptome and Proteome of the Parasite and the Host” funded by Department of Biotechnology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 2.Galinski MR, Barnwell JW. Plasmodium vivax: who cares? Malar J. 2008;7(Suppl 1):S9. doi: 10.1186/1475-2875-7-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 5.Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci U S A. 2008;105:16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westenberger SJ, McClean CM, Chattopadhyay R, Dharia NV, Carlton JM, et al. A systems-based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PLoS Negl Trop Dis. 2010;4:e653. doi: 10.1371/journal.pntd.0000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roobsong W, Roytrakul S, Sattabongkot J, Li J, Udomsangpetch, et al. Determination of the Plasmodium vivax schizont stage proteome. J Proteomics. Apr. 2011;13 doi: 10.1016/j.jprot.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huh Ae-J, Kwak YE, Kim ES, Lee KS, Yeom Joon-S, et al. Parasitemia Characteristics of Plasmodium vivax Malaria Patients in the Republic of Korea. J Korean Med Sci. 2011;26:42–46. doi: 10.3346/jkms.2011.26.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acharya P, Pallavi R, Chandran S, Chakravarti H, Middha S, et al. A glimpse into the clinical proteome of human malaria parasites Plasmodium falciparum and Plasmodium vivax. Proteomics Clin. 2009;appl 3:1314–1325. doi: 10.1002/prca.200900090. [DOI] [PubMed] [Google Scholar]
- 10.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 11.Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fried M, Hixson KK, Anderson L, Ogata Y, Mutabingwa TK, et al. The distinct proteome of placental malaria parasites. Mol Biochem Parasitol. 2007;155:57–65. doi: 10.1016/j.molbiopara.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, et al. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 14.Ersmark K, Nervall M, Gutierrez-de-Teran H, Hamelink E, Janka LK, et al. Macrocyclic inhibitors of the malarial aspartic proteases plasmepsin I, II, and IV. Bioorg Med Chem. 2006;14:2197–2208. doi: 10.1016/j.bmc.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 15.McGowan S, Porter CJ, Lowther J, Stack CM, Golding SJ, et al. Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase. Proc Natl Acad Sci U S A. 2009;106:2537–2542. doi: 10.1073/pnas.0807398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trenholme KR, Brown CL, Skinner-Adams TS, Stack C, Lowther J, et al. Aminopeptidases of malaria parasites: new targets for chemotherapy. Infect Disord Drug Targets. 2010;10:217–225. doi: 10.2174/187152610791163363. [DOI] [PubMed] [Google Scholar]
- 17.Becker JV, Mtwisha L, Crampton BG, Stoychev S, van Brummelen AC, et al. Plasmodium falciparum spermidine synthase inhibition results in unique perturbation-specific effects observed on transcript, protein and metabolite levels. BMC Genomics. 2010;11:235. doi: 10.1186/1471-2164-11-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itin C, Burki Y, Certa U, Dobeli H. Selective inhibition of Plasmodium falciparum aldolase by a tubulin derived peptide and identification of the binding site. Mol Biochem Parasitol. 1993;58:135–143. doi: 10.1016/0166-6851(93)90097-h. [DOI] [PubMed] [Google Scholar]
- 19.Kim H, Certa U, Dobeli H, Jakob P, Hol WG. Crystal structure of fructose-1,6-bisphosphate aldolase from the human malaria parasite Plasmodium falciparum. Biochemistry. 1998;37:4388–4396. doi: 10.1021/bi972233h. [DOI] [PubMed] [Google Scholar]
- 20.Wanidworanun C, Nagel RL, Shear HL. Antisense oligonucleotides targeting malarial aldolase inhibit the asexual erythrocytic stages of Plasmodium falciparum. Mol Biochem Parasitol. 1999;102:91–101. doi: 10.1016/s0166-6851(99)00087-0. [DOI] [PubMed] [Google Scholar]
- 21.Wrenger C, Eschbach ML, Muller IB, Warnecke D, Walter RD. Analysis of the vitamin B6 biosynthesis pathway in the human malaria parasite Plasmodium falciparum. J Biol Chem. 2005;280:5242–5248. doi: 10.1074/jbc.M412475200. [DOI] [PubMed] [Google Scholar]
- 22.Velanker SS, Ray SS, Gokhale RS, Suma S, Balaram H, et al. Triosephosphate isomerase from Plasmodium falciparum: the crystal structure provides insights into antimalarial drug design. Structure. 1997;5:751–761. doi: 10.1016/s0969-2126(97)00230-x. [DOI] [PubMed] [Google Scholar]
- 23.Joubert F, Neitz AW, Louw AI. Structure-based inhibitor screening: a family of sulfonated dye inhibitors for malaria parasite triosephosphate isomerase. Proteins. 2001;45:136–143. doi: 10.1002/prot.1133. [DOI] [PubMed] [Google Scholar]
- 24.Granchi C, Bertini S, Macchia M, Minutolo F. Inhibitors of lactate dehydrogenase isoforms and their therapeutic potentials. Curr Med Chem. 2010;17:672–697. doi: 10.2174/092986710790416263. [DOI] [PubMed] [Google Scholar]
- 25.Banumathy G, Singh V, Pavithra SR, Tatu U. Heat shock protein 90 function is essential for Plasmodium falciparum growth in human erythrocytes. J Biol Chem. 2003;278:18336–18345. doi: 10.1074/jbc.M211309200. [DOI] [PubMed] [Google Scholar]
- 26.Pallavi R, Roy N, Nageshan RK, Talukdar P, Pavithra SR, et al. Heat shock protein 90 as a drug target against protozoan infections: biochemical characterization of HSP90 from Plasmodium falciparum and Trypanosoma evansi and evaluation of its inhibitor as a candidate drug. J Biol Chem. 2010;285:37964–37975. doi: 10.1074/jbc.M110.155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pesce ER, Cockburn IL, Chiang AN, Valderramos JC, Balachandran R, et al. Select pyrimidinones inhibit the propagation of the malarial parasite, Plasmodium falciparum. Bioorg Med Chem. 2009;17:1527–1533. doi: 10.1016/j.bmc.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pesce ER, Cockburn IL, Goble JL, Stephens LL, Blatch GL. Malaria heat shock proteins: drug targets that chaperone other drug targets. Infect Disord Drug Targets. 2010;10:147–157. doi: 10.2174/187152610791163417. [DOI] [PubMed] [Google Scholar]
- 29.Fraser T, Michon P, Barnwell JW, Noe AR, Al-Yaman F, et al. Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect Immun. 1997;65:2772–2777. doi: 10.1128/iai.65.7.2772-2777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Becerra C, Yamamoto MM, Vencio RZ, Lacerda M, Rosanas-Urgell A, et al. Plasmodium vivax and the importance of the subtelomeric multigene vir superfamily. Trends Parasitol. 2009;25:44–51. doi: 10.1016/j.pt.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Jalah R, Sarin R, Sud N, Alam MT, Parikh N, et al. Identification, expression, localization and serological characterization of a tryptophan-rich antigen from the human malaria parasite Plasmodium vivax. Mol Biochem Parasitol. 2005;142:158–169. doi: 10.1016/j.molbiopara.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 32.Daily JP, Scanfeld D, Pochet N, Le Roch K, Plouffe D, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450:1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 33.LeRoux M, Lakshmanan V, Daily JP. Plasmodium falciparum biology: analysis of in vitro versus in vivo growth conditions. Trends Parasitol. 2009;25:474–481. doi: 10.1016/j.pt.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 34.LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 35.Date SV, Stoeckert CJ., Jr Computational modeling of the Plasmodium falciparum interactome reveals protein function on a genome-wide scale. Genome Res. 2006;16:542–549. doi: 10.1101/gr.4573206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price RN, Douglas NM, Anstey NM. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr Opin Infect Dis. 2009;22:430–435. doi: 10.1097/QCO.0b013e32832f14c1. [DOI] [PubMed] [Google Scholar]
- 37.Baird JK, Schwartz E, Hoffman SL. Prevention and treatment of vivax malaria. Curr Infect Dis Rep. 2007;9:39–46. doi: 10.1007/s11908-007-0021-4. [DOI] [PubMed] [Google Scholar]
- 38.Collins WE, Jeffery GM. Primaquine resistance in Plasmodium vivax. Am J Trop Med Hyg. 1996;55:243–249. doi: 10.4269/ajtmh.1996.55.243. [DOI] [PubMed] [Google Scholar]
- 39.Korsinczky M, Fischer K, Chen N, Baker J, Rieckmann K, et al. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob Agents Chemother. 2004;48:2214–2222. doi: 10.1128/AAC.48.6.2214-2222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dharia NV, Bright AT, Westenberger SJ, Barnes SW, Batalov S, et al. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc Natl Acad Sci U S A. 2010;107:20045–20050. doi: 10.1073/pnas.1003776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goh SH, Josleyn M, Lee YT, Danner RL, Gherman RB, et al. The human reticulocyte transcriptome. Physiol Genomics. 2007;30:172–178. doi: 10.1152/physiolgenomics.00247.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of P. vivax proteins identified in clinical malaria sample and peptides identified.
(XLS)
Functional classification of hypothetical proteins based on predicted PFAM domains.
(DOC)
P. vivax proteins which do not have Mass Spectrometric evidence in any species of Plasmodium.
(XLS)
Host proteins identified along with the proteome of P. vivax.
(XLS)
Plasmodium vivax host proteins which are present at transcriptome level.
(XLS)
P. vivax proteins detected in this study which have a score less than 0.47.
(XLS)