Abstract
The transforming growth factor β (TGFβ) superfamily proteins are principle regulators of numerous biological functions. Although recent studies have gained tremendous insights into this growth factor family in female reproduction, the functions of the receptors in vivo remain poorly defined. TGFβ type 1 receptor (TGFBR1), also known as activin receptor-like kinase 5, is the major type 1 receptor for TGFβ ligands. Tgfbr1 null mice die embryonically, precluding functional characterization of TGFBR1 postnatally. To study TGFBR1–mediated signaling in female reproduction, we generated a mouse model with conditional knockout (cKO) of Tgfbr1 in the female reproductive tract using anti-Müllerian hormone receptor type 2 promoter-driven Cre recombinase. We found that Tgfbr1 cKO females are sterile. However, unlike its role in growth differentiation factor 9 (GDF9) signaling in vitro, TGFBR1 seems to be dispensable for GDF9 signaling in vivo. Strikingly, we discovered that the Tgfbr1 cKO females develop oviductal diverticula, which impair embryo development and transit of embryos to the uterus. Molecular analysis further demonstrated the dysregulation of several cell differentiation and migration genes (e.g., Krt12, Ace2, and MyoR) that are potentially associated with female reproductive tract development. Moreover, defective smooth muscle development was also revealed in the uteri of the Tgfbr1 cKO mice. Thus, TGFBR1 is required for female reproductive tract integrity and function, and disruption of TGFBR1–mediated signaling leads to catastrophic structural and functional consequences in the oviduct and uterus.
Author Summary
Approximately 20% of infertile couples in the United States have unexplained causes. Many vital aspects of female fertility are regulated by a family of growth factors called the transforming growth factor β (TGFβ) superfamily. These factors exert their functions via specific receptors and downstream signal mediators. Perturbation of components in this pathway can lead to reproductive dysfunction. We identified a novel role for a TGFβ receptor (called TGFBR1) in female fertility. We demonstrated that female mice with disruption of Tgfbr1 in the reproductive tract are unable to successfully conceive, although they can ovulate and produce fertilizable oocytes. Most importantly, these mice have a striking deformity in the oviduct, marked by the formation of oviductal outpouchings (diverticula) that prevent embryos from reaching the uterus. Concomitant aberrations in the uterine smooth muscle layers are additional features of mice lacking TGFBR1. Therefore, TGFBR1 is critical for the structural integrity and function of the female reproductive tract. Our model can be further exploited to study the development of smooth muscle cells of the female reproductive tract. Genetic mutations in TGFBR1 or other TGFβ signaling machinery may lead to fertility defects in women.
Introduction
The transforming growth factor β (TGFβ) superfamily, the largest family of secreted growth factors in mammals, is a conserved family of proteins that play key roles in diverse physiological and pathological processes [1]–[6]. The pathway consists of ligands, receptors, and SMAD transducers, and is tightly controlled by various regulatory layers such as ligand traps (e.g., noggin, follistatin, and gremlin), inhibitory SMADs (i.e., SMAD6 and SMAD7), as well as multiple interactive pathways that cross talk with TGFβ signaling proteins in a context-specific manner [7]–[9]. TGFβ ligands bind to their type 2 and type 1 receptors and activate intracellular SMAD proteins including receptor-regulated SMADs and common SMAD (SMAD4) to initiate signal transduction. Although more than 40 TGFβ family members have been discovered to date, there are only seven type 1 receptors (ACVRL1, ACVR1, BMPR1A, ACVR1B, TGFBR1, BMPR1B, ACVR1C) and five type 2 receptors (TGFBR2, AMHR2, ACVR2, ACVR2B, and BMPR2) in mammals [10], [11]. The receptor-regulated SMADs can be divided into TGFβ/activin responsive SMADs (i.e., SMAD2/3) and bone morphogenetic protein (BMP) responsive SMADs (i.e., SMAD1/5/8) based on the ligands with which they are associated in the signal transduction cascades [2], [7].
Recent studies have revealed that the TGFβ signaling pathway is critically involved in multiple reproductive events including, but not limited to, ovarian folliculogenesis [3], [12]–[15], cumulus cell expansion and ovulation [16]–[18], uterine decidualization [19], and embryo implantation [20]. Disturbances in TGFβ signaling have been shown to lead to severe pathological conditions such as cancer [1], [2], [4]–[6], [21]–[26], making it an appealing candidate pathway for therapeutic interventions. Early studies in our laboratory demonstrated that inhibin α is a tumor suppressor specific to the gonad and adrenal glands [21], highlighting the functional importance of TGFβ family proteins. Subsequent studies demonstrated that the BMP signaling pathway serves as a brake for ovarian tumor development [23], [26]. During recent years, significant progress has been made toward understanding the roles of this growth factor family in female reproduction [2], [3], [27]–[30]; however, the functions of the receptors in vivo remain poorly defined, partially due to receptor redundancy [26], [31]–[34] or lethal phenotypes of genetically engineered ubiquitous null mouse models.
TGFBR1 is the type 1 receptor for TGFβ ligands [2]. In vitro, TGFBR1 can also mediate the signaling of growth differentiation factor 9 (GDF9) [35], an oocyte-secreted protein required for early ovarian folliculogenesis, cumulus cell functions [12], [36]–[41], and oocyte developmental competence [42], [43]. The above evidence points to a possible role of TGFBR1 in female reproduction in vivo. However, the functional significance of TGFBR1 in the female reproductive tract is unknown because Tgfbr1 null mice die embryonically [44].
Advances in gene targeting technology make it possible to dissect gene functions in specific tissues using a conditional gene inactivation (Cre-loxP) strategy [45]. Our laboratory has successfully utilized this technique to expand the understanding of reproductive functions of TGFβ signaling components [18], [23], [26], [46], [47]. In the current study, we generated a conditional knockout (cKO) of Tgfbr1 in the female reproductive tract using anti-Müllerian hormone receptor type 2 (Amhr2)-Cre. We found that Tgfbr1 cKO mice are sterile. Interestingly, instead of manifesting an overt ovarian phenotype, these mice develop striking oviductal and uterine phenotypes, thereby uncovering a novel role of TGFBR1–mediated signaling in female reproductive tract development and function.
Results
Generation of Tgfbr1 Conditional Knockout Mice
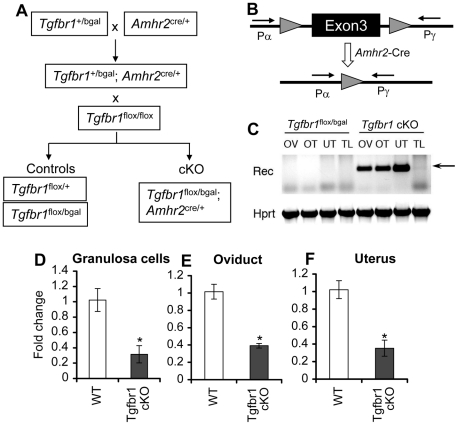
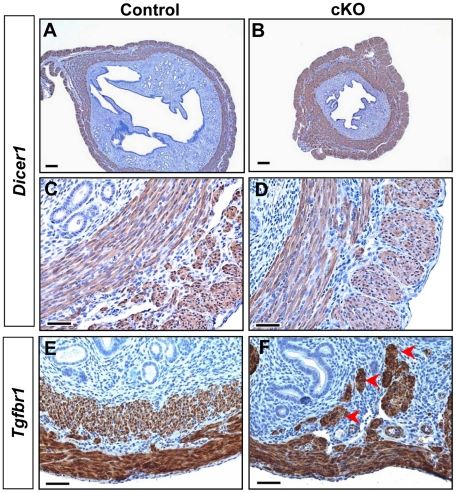
Tgfbr1 null mice die embryonically [44], precluding functional characterization of TGFBR1 postnatally. To study TGFBR1–mediated signaling in female reproduction, we used a Tgfbr1 flox allele and a Tgfbr1 bgal allele, in which a β-galactosidase (β-gal) reporter was inserted into the Tgfbr1 locus to create a null allele and to monitor spatiotemporal expression of Tgfbr1. To ensure maximal deletion of the Tgfbr1 gene, the Tgfbr1 bgal null allele was used in the breeding scheme to produce Tgfbr1 mutant mice. Mice carrying these alleles were crossed with mice harboring the Amhr2-Cre allele [48], which recombines floxed alleles in granulosa cells [49] and Müllerian duct derived tissues (e.g., the smooth muscle layers of the uterus and oviduct but not the epithelial compartment) [50]–[52] to produce Tgfbr1 cKO mice (Tgfbr1 flox/bgal; Amhr2 cre/+) (Figure 1A). Recombination of the Tgfbr1 flox allele and reduction of Tgfbr1 mRNA transcripts were confirmed in the ovary, oviduct, and uterus (Figure 1B–1F).
Figure 1. Generation of Tgfbr1 conditional knockout mice.
(A) Schematic representation of the breeding strategy for generating Tgfbr1 cKO and control mice. To ensure maximal deletion of Tgfbr1 and to visualize the localization of Tgfbr1 in the female reproductive tract, the Tgfbr1 bgal allele was used in the breeding. (B) Illustration of the Tgfbr1 conditional allele with exon 3 flanked by two loxP sites. Primers Pα and Pγ detect the recombined allele of Tgfbr1. (C) Recombination of Tgfbr1 floxed alleles in the genomic DNA of Amhr2-Cre target tissues. The recombined bands were detectable in the ovary (OV), oviduct (OT), and uterus (UT) of the Tgfbr1 cKO mice, but not in those of the controls and tail (TL) DNA samples. (D to F) Relative Tgfbr1 mRNA levels in the granulosa cells (D; n = 3), oviduct (E; n = 5), and uterus (F; n = 5) of control and Tgfbr1 cKO mice. Data are presented as mean ± SEM. *P<0.05 compared with controls.
Tgfbr1 cKO Mice Are Sterile and Develop Prominent Oviductal Diverticula
Whereas control female mice (Tgfbr1 flox/bgal) lacking Amhr2 cre/+ demonstrated normal fertility and fecundity during a 6-month breeding period (8.5±0.2 pups/litter and 1.1±0.0 litter/month), the Tgfbr1 cKO female mice were sterile (Table 1). Copulatory plugs were found in the Tgfbr1 cKO females, indicating the infertility was not due to disrupted mating behavior. These results suggest TGFBR1 is required for female fertility.
Table 1. Fertility test of Tgfbr1 cKO mice.
| Genotype | n | Pups/litter | Litter/month |
| Tgfbr1 flox/bgal | 10 | 8.5±0.2 | 1.1±0.0 |
| Tgfbr1 flox/bgal; Amhr2 cre/+ | 7 | 0 | 0 |
The Tgfbr1 cKO female mice were sterile during a 6-month breeding period compared with control female mice lacking Amhr2 cre/+, which showed normal fertility and fecundity. Data are presented as mean ± SEM.
Data are presented as mean ± SEM.
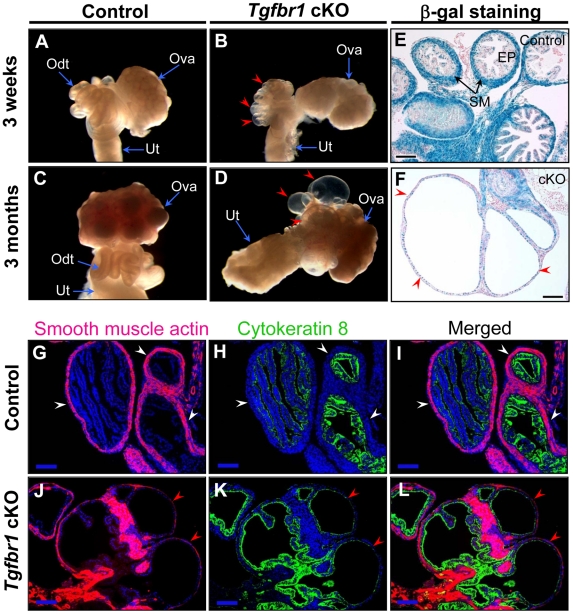
To examine the structural integrity of the reproductive tract and determine possible causes of sterility in the Tgfbr1 cKO females, we performed morphological and histological analyses of Tgfbr1 cKO and control mice. Strikingly, we found the development of bilateral oviductal diverticula (i.e., clear fluid-filled outpouchings that are present throughout the length of each oviduct) in 100% of the Tgfbr1 cKO females examined (Figure 2A–2D). This phenotype highlights the importance of TGFBR1 in the oviduct where its expression was detected in both smooth muscle and epithelial compartments (Figure 2E). Deletion of Tgfbr1 was expected only in the smooth muscle compartment due to the presence of Amhr2-Cre activity in the mesenchymal cells that give rise to the smooth muscle cells but not the epithelial cells. The oviductal diverticula enlarged with age and were characterized by a single layer of flattened epithelium and disrupted smooth muscle layers, as demonstrated by β-gal staining (Figure 2F) and immunofluorescence using antibodies against smooth muscle α-actin (ACTA2) and cytokeratin 8 (KRT8)(Figure 2G–2L) as well as calponin 1 (CNN1; Figure S1), a smooth muscle-specific protein implicated in contraction.
Figure 2. Oviductal diverticula development in Tgfbr1 cKO mice.
(A to F) Gross morphology (A to D) and β-gal staining (E and F) of oviducts or oviductal diverticula in control and Tgfbr1 cKO mice. (G to L) Immunofluorescence of ACTA2 and KRT8 in the oviducts of 3-week-old control and Tgfbr1 cKO mice. White arrowheads indicate the normal oviductal structure, while red arrowheads indicate oviductal diverticula. Odt, oviduct; Ova, ovary; Ut, uterus; SM, smooth muscle; EP, epithelium. Scale bars = 100 µm.
Tgfbr1 cKO Mice Demonstrate Minimal Ovarian Defects
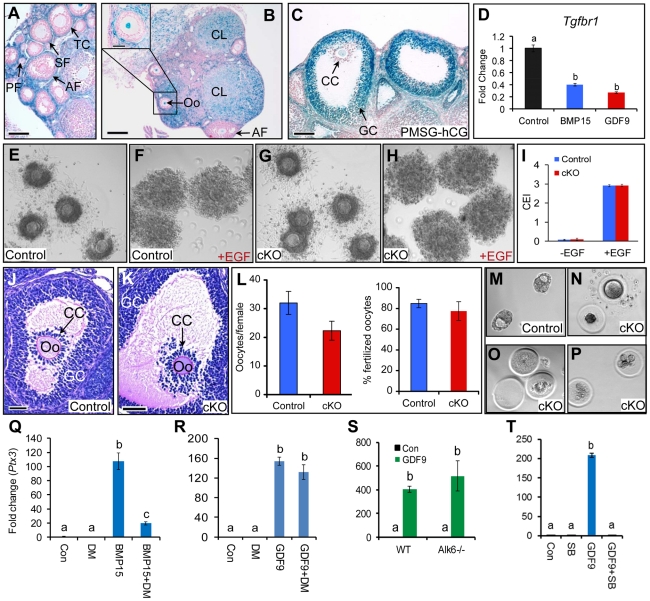
To define the causes of female sterility, we examined the ovaries of the Tgfbr1 cKO mice. In contrast to the marked oviductal phenotype, the ovaries of Tgfbr1 cKO mice were grossly normal and contained follicles at various follicular stages (Figure S2A and S2B). To address the cellular distribution of TGFBR1 in the mouse ovary, we performed β-gal staining and found that TGFBR1 was predominantly localized to the thecal layers of developing follicles (Figure 3A), corpora lutea (Figure 3B), oocytes (Figure 3B), and mural granulosa cells of preovulatory follicles induced by gonadotropins (Figure 3C). TGFBR1 expression signals in the granulosa cells of developing follicles and cumulus cells of preovulatory follicles were close to the background level (Figure 3A–3C). Furthermore, we found that GDF9 and its oocyte paralog BMP15 reduced the expression of Tgfbr1 mRNA in mouse granulosa cells cultured in vitro (Figure 3D).
Figure 3. Cellular distribution and functional characterization of TGFBR1 in mouse ovary.
(A to C) β-gal staining of ovaries from immature [(A; untreated) and (C; PMSG-hCG treated)] and adult (B) Tgfbr1 +/bgal mice. (D) Suppression of Tgfbr1 mRNA in mouse granulosa cells by recombinant BMP15 or GDF9 after 5 h treatment (n = 3). (E to H) Expansion of COCs from control and Tgfbr1 cKO mice in vitro in the absence (E and G) or presence (F and H) of EGF (10 ng/ml). (I) Cumulus expansion index (CEI) of the in vitro cultured COCs from control (n = 4) and Tgfbr1 cKO (n = 5) mice. (J and K) Preovulatory follicles from Tgfbr1 cKO mice demonstrating cumulus expansion. PF, primary follicle; SF, secondary follicle; AF, antral follicle; Oo, oocyte; TC, thecal cell; GC, granulosa cell; CC, cumulus cell; CL, corpus luteum; COC, cumulus-oocyte complex. Scale bars = 50 µm (J and K); 100 µm (A and C); and 200 µm (B). (L) Ovulation and oocyte fertilization potential of Tgfbr1 cKO mice (n = 8–10). (M to P) Blastocysts or degenerated oocytes/embryos recovered from control (M) and Tgfbr1 cKO mice (N to P). (Q to T) Identification of receptor preference for GDF9 signaling in mouse granulosa cells using small molecule inhibitors and Alk6 −/− granulosa cells. Dorsomorphin (DM; 4 µM) was preincubated with granulosa cells for 1 h before BMP15 (100 ng/ml) or GDF9 (15 ng/ml) was added. DM markedly reduced Ptx3 induction by BMP15 (Q), while a similar effect was not observed on GDF9-induced Ptx3 mRNA expression (R). Induction of Ptx3 mRNA by GDF9 (100 ng/ml) was not attenuated in Alk6−/− granulosa cells (S). However, SB-505124 (SB; 1 µM) suppressed GDF9-induced Ptx3 mRNA expression (T). Con, control. n = 3 for each group. Relative mRNA levels of Tgfbr1 and Ptx3 were normalized to Gapdh. Data are presented as mean ± SEM. Bars without a common letter are significantly different at P<0.05.
Because development of preovulatory follicles occurred in Tgfbr1 cKO mice exposed to exogenous gonadotropins (Figure S2C and S2D), we examined cumulus expansion, a critical event in ovulation, in these follicles. We found that cumulus cells from the Tgfbr1 cKO mice underwent normal expansion both in vitro and in vivo (Figure 3E–3K). We next conducted superovulation analysis to evaluate ovulatory potential and found that Tgfbr1 cKO mice could ovulate although a trend of reduced ovulation rate was observed in these mice (Figure 3L). Similar to controls (Figure S3A), the ovaries of the superovulated Tgfbr1 cKO mice contained corpora lutea (Figure S3B), which were capable of synthesizing 3-β-hydroxysteroid dehydrogenase (3β-HSD) (Figure S3D). As further evidence of the presence of functional corpora lutea in the Tgfbr1 cKO mice, serum progesterone levels were comparable between the control and Tgfbr1 cKO mice at 20 h after hCG injection (gonadotropin primed immature mice) or at 3.5 days post coitum (dpc; adult females) (Figure S3F). Moreover, oocytes could be located and recovered from the oviductal diverticula of the Tgfbr1 cKO mice and were fertilizable (Figure 3L).
TGFBR1, also known as activin receptor-like kinase 5 (ALK5), had been proposed to mediate GDF9 signaling in vitro [35]. Based on the lack of a prominent ovarian phenotype in the Tgfbr1 cKO mice and the minimal, if any, expression of TGFBR1 in the granulosa cells of preantral follicles, our results suggest that TGFBR1 is at least not the sole physiological type 1 receptor for GDF9 in mouse ovary. As an initial step toward exploring the potential type 1 receptor(s) for GDF9, we performed in vitro studies using Alk6 null granulosa cells as well as small molecule inhibitors for ALK2/3/6 (Dorsomorphin; DM) and ALK4/5/7 (SB-505124; SB). While dorsomorphin potently suppressed BMP15-induced Ptx3 expression as expected (Figure 3Q), a dramatic effect of this inhibitor on GDF9-induced Ptx3 expression was not observed when GDF9 was applied at a concentration (15 ng/ml) that induced Ptx3 mRNA expression with closer amplitude to that stimulated by 100 ng/ml of recombinant BMP15 (Figure 3R). Furthermore, GDF9 signaling remains intact in Alk6 null granulosa cells as measured by the ability of GDF9 to induce the expression of cumulus expansion-related transcripts such as Ptx3 (Figure 3S). In contrast, the ALK4/5/7 inhibitor, SB-505124, completely blocked the induction of Ptx3 in mouse granulosa cells by GDF9 (Figure 3T). However, SB-505124 also reduced recombinant BMP15-induced Ptx3 expression (data not shown), potentially due to its effect on basal gene expression in mouse granulosa cells. The above in vitro studies combined with our in vivo data support the hypothesis that GDF9 does not signal through type 1 BMP receptors (ALK2/3/6), but more likely signals through ALK4 and/or ALK7 in the mouse ovary.
Deleterious Effects of Oviductal Diverticula on Female Fertility
Since ovulation and fertilization occurred in the Tgfbr1 cKO and control females, we next assessed whether the formation of oviductal diverticula was detrimental to embryo development and/or transit of embryos to the uterus. After timed matings of adult Tgfbr1 cKO females with proven fertile wild type (WT) males, we could recover blastocysts (7.75±0.63) at 3.5 dpc from the uteri of controls (Figure 3M), but not Tgfbr1 cKO females (Table 2). Instead, degenerating oocytes/embryos and their zona pellucida remnants were recovered from the oviductal diverticula (Figure 3N–3P), indicating that embryo development and embryo transit to the uterus were severely compromised in the Tgfbr1 cKO female mice. Since the oviduct is the site where sperm complete their maturation and undergo capacitation [53], sperm transport and/or capacitation could also be impeded in the adult Tgfbr1 cKO mice due to the severe oviductal phenotype.
Table 2. Embryo recovery from 3.5 dpc uteri of control and Tgfbr1 cKO mice.
| Genotype | Dpc | n | No. of embryo |
| Tgfbr1 flox/bgal | 3.5 | 4 | 7.75±0.63 |
| Tgfbr1 flox/bgal; Amhr2 cre/+ | 3.5 | 5 | 0 |
After timed matings, blastocysts could be recovered at 3.5 dpc from the uteri of controls, but not Tgfbr1 cKO females. Data are presented as mean ± SEM.
Data are presented as mean ± SEM.
Loss of TGFBR1–Mediated Signaling Results in Defective Smooth Muscle Development in Mouse Uterus
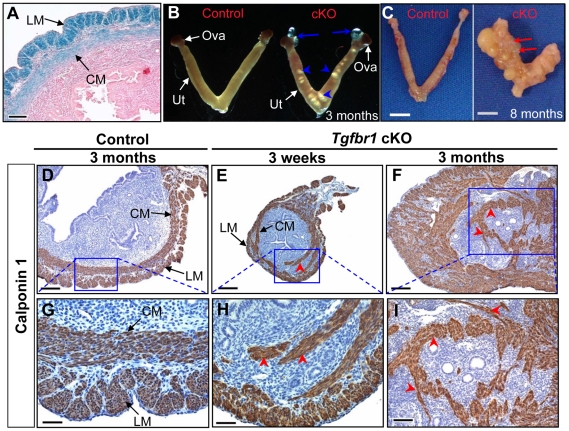
Because TGFBR1 expression was also detected in smooth muscle cells of the uterus (Figure 4A) where Amhr2-Cre activity is present, we also examined the consequences of deletion of Tgfbr1 in the uterus. Grossly, the uteri of the Tgfbr1 cKO mice were comparable in size to those of controls up through 3 months (Figure S4A and S4B). However, the Tgfbr1 cKO uteri contained multiple smooth muscle-defective areas, as evidenced by transillumination (Figure 4B). By 8 months of age, the uterine pathology in the Tgfbr1 cKO mice culminated in uterine cyst formation and an almost unrecognizable mass of tissue (Figure 4C). The severely disrupted smooth muscle structure was evident by immunostaining of ACTA2 (Figure S4C–S4H) and CNN1 (Figure 4D–4I and Figure S5). In contrast to controls (Figure 4D and 4G), the myometrium of the Tgfbr1 cKO mice was disorganized with poorly formed smooth muscle layers and intermingled with the endometrial components (Figure 4E, 4F, 4H, and 4I). Our data demonstrated that loss of TGFBR1–mediated signaling causes defective smooth muscle development in the oviduct and uterus.
Figure 4. Tgfbr1 cKO mice demonstrate disrupted uterine smooth muscle development.
(A) Localization of TGFBR1 to myometrium by β-gal staining. (B and C) Gross uterine morphology of control and Tgfbr1 cKO mice. Blue arrows and arrowheads indicate the respective oviductal diverticula and the smooth muscle defective areas, whereas red arrows point to uterine cysts. (D to I) Immunostaining of CNN1 in the uteri of control (D and G) and Tgfbr1 cKO (E, F, H, and I) mice. LM, longitudinal muscle layer; CM, circular muscle layer; Ova, ovary; Ut, uterus. Red arrowheads point to disorganized uterine smooth muscle structures. Scale bars = 50 µm (G and H); 100 µm (A and I); 200 µm (D to F); and 5 mm (C).
To determine if the disorganization of uterine smooth muscle layers can affect stromal cell function, we performed an artificial decidualization study. Both the control and Tgfbr1 cKO mice demonstrated responses to uterine scratches (Figure S6A and S6B). To quantitatively compare the decidual response between the control and Tgfbr1 cKO groups, we calculated the weight ratio of stimulated (scratched) horn versus unstimulated horn. There was no significant difference between the two groups (Figure S6C; P>0.05), although disrupted uterine smooth muscle layers were visualized by immunostaining of CNN1 (Figure S6D–S6G). These results suggest that the compromised smooth muscle development in Tgfbr1 cKO uterus does not prevent the uterine decidual response, although the decidualization occurs in an overall abnormal uterine environment where stromal cells were segregated by dispersed smooth muscle structure.
Smooth Muscle Gene Expression in Tgfbr1 cKO Oviducts
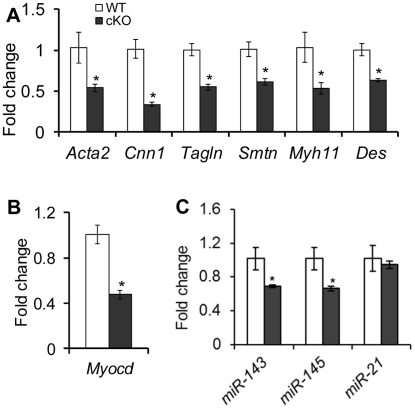
To examine if loss of TGFBR1 affects the expression of smooth muscle genes, we compared the mRNA levels of select genes between control and Tgfbr1 cKO oviducts from 3–4 week old mice. Accompanying the defective smooth muscle phenotype, transcript levels of all smooth muscle genes examined, including Acta2, Cnn1, transgelin (Tagln), smoothelin (Smtn), smooth-muscle myosin heavy chain (Myh11), and desmin (Des), were reduced in the oviducts of Tgfbr1 cKO mice compared with controls (Figure 5A). Concomitantly, the mRNA level of myocardin (Myocd), a smooth muscle and cardiac muscle-specific transcriptional co-activator and a master modulator of smooth muscle gene expression [54], was also decreased in the Tgfbr1 cKO oviducts (Figure 5B).
Figure 5. Dysregulation of smooth muscle genes and miR-143 and miR-145 in the oviducts of Tgfbr1 cKO mice at 3–4 weeks of age.
(A) Global reduction of smooth muscle gene transcripts in the oviducts of Tgfbr1 cKO mice. (B) Down-regulation of Myocd mRNA in the Tgfbr1 cKO oviducts. (C) Alterations of miR-143 and miR-145, but not miR-21, in the oviducts of Tgfbr1 cKO mice. Relative mRNA levels of smooth muscle genes (A) and Myocd (B) were normalized to Gapdh, while levels of miR-143, miR-145, and miR-21 (C) were normalized against snoRNA202. n = 3–5 independent pools of oviducts. Data are presented as mean ± SEM. *P<0.05 versus controls.
Interestingly, a similar oviductal phenotype was also observed in the Amhr2-Cre mediated conditional deletion of DICER1 [51], [52], [55], the RNase III involved in microRNA (miRNA) processing in cytoplasm. A recent study [56] demonstrated that TGFβ signaling can induce the maturation of a subset of miRNAs. An intriguing question was whether TGFβ signaling is linked to the miRNA pathway in the female reproductive tract. We found that two newly identified vascular smooth muscle associated miRNAs, miR-143 and miR-145 [57]–[60], were down-regulated in the oviducts of Tgfbr1 cKO mice (Figure 5C). However, miR-21, a known target of TGFβ ligands [56], was not significantly affected in the Tgfbr1 cKO oviducts (Figure 5C). Moreover, the downstream targets of miR-143/145 (i.e., transcription factors Elk1, Klf4, and Camk2d) were not altered in the oviducts of Tgfbr1 cKO mice (Figure S7).
Since the smooth muscle genes and miR-143/145 were predominantly expressed in the smooth muscle compartments, loss of smooth muscle tissues in the oviducts during the formation of oviductal diverticula could potentially lead to reduced expression of smooth muscle genes or associated miRNAs in the Tgfbr1 cKO oviducts. To address this possibility, we collected and analyzed postnatal day 7 oviducts prior to significant smooth muscle loss. However, dramatic reductions in the expression of smooth muscle genes and miR-143 and miR-145 were not detected at this stage (data not shown).
Tgfbr1 cKO Mice Develop a Uterine Phenotype Distinct from Dicer1 cKO Mice
Despite the occurrence of oviductal diverticula in both Tgfbr1 cKO and Dicer1 cKO mice, we found that the uterine phenotype of Tgfbr1 cKO mice is distinct from that of the Dicer1 cKO mice. Consistent with our previous report [55], Dicer1 cKO mice have smaller uteri than controls (Figure 6A and 6B), as is in contrast to Tgfbr1 cKO mice. Unlike Tgfbr1 cKO mice (Figure 6F), immunostaining showed that mice with a conditional deletion of Dicer1 developed normal smooth muscle layers (Figure 6B and 6D). The divergence in the uterine phenotypes between Tgfbr1 cKO and Dicer1 cKO mice also suggests that the development of oviductal diverticula in these two mouse models may not be in a linear pathway. As further support of this concept, we found that mRNA levels for genes up-regulated in Dicer1 cKO oviducts (Wnt5a, Wnt7a, Hoxa9, Hoxa10, etc.) [55] were not increased in Tgfbr1 cKO mice (Figure S8).
Figure 6. Distinct uterine phenotypes between Dicer1 cKO and Tgfbr1 cKO mice.
(A–D) Adult Dicer1 cKO mice (B and D) have smaller uteri but normal smooth muscle structure compared with controls (A and C) by immunostaining of uterine cross sections with CNN1. (E and F) Disruption of uterine smooth muscle layers in Tgfbr1 cKO mice (F; arrowheads) versus controls (E) by immunostaining of uterine longitudinal sections with CNN1.
Molecular Alterations in the Oviduct of Tgfbr1 cKO Mice
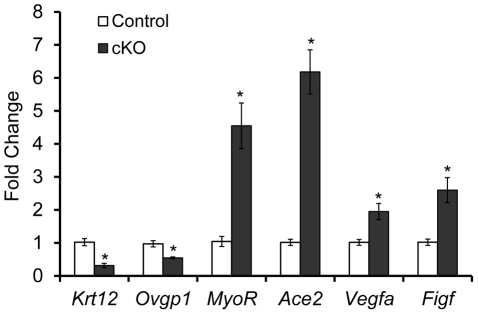
To further study the molecular basis of the striking oviductal diverticulum phenotype in Tgfbr1 cKO mice, we performed real-time PCR analyses of oviducts from postnatal day 7 mice. We herein uncovered the dysregulation of candidate genes in the Tgfbr1 cKO oviducts that are associated with cell differentiation (Krt12 and MyoR) and migration (Ace2, Vegfa, and Figf ).
The keratins are intermediate filament proteins that are important structural components of epithelial cells [61], [62]. In the Tgfbr1 cKO oviducts, expression of Krt12, a member of the keratins, was markedly reduced (P<0.05; Figure 7). Another epithelial gene, the oviductal glycoprotein 1 (Ovgp1), was also down-regulated (P<0.05; Figure 7). The altered expression of epithelial genes suggests the importance of TGFBR1–mediated signaling in the maintenance of the mesenchymal-epithelial interactions critical for oviductal development.
Figure 7. Dysregulation of genes in the oviducts from postnatal day 7 Tgfbr1 cKO mice.
Real-time PCR analyses using oviducts from postnatal day 7 mice demonstrated down-regulation of Krt12 and Ovgp1 and up-regulation of MyoR, Ace2, Vegfa, and Figf in Tgfbr1 cKO mice (n = 5 independent pools of oviducts) versus controls (n = 5 independent pools of oviducts). Relative mRNA levels were normalized to Gapdh. Data are presented as mean ± SEM. *P<0.05 versus respective controls.
TGFβ signaling can regulate the differentiation of vascular smooth muscle cells [63]–[65]. We found that mRNA encoding the skeletal muscle differentiation associated basic helix-loop-helix (bHLH) transcription factor, MyoR/musculin, was increased more than 4-fold in Tgfbr1 cKO oviducts versus controls (P<0.05; Figure 7).
Overexpression of angiotensin-converting enzyme 2 (ACE2), a member of the renin-angiotensin system, is associated with cell migration [66]. In the Tgfbr1 cKO oviducts, the expression of Ace2 mRNA was markedly increased in the Tgfbr1 cKO oviducts (P<0.05; Figure 7). Even at 21 days of age, levels of Ace2 mRNA were consistently higher in Tgfbr1 cKO oviducts than controls (data not shown). Other migration related genes such as vascular endothelial growth factor A (Vegfa) and c-fos induced growth factor (Figf/Vegfd) were also up-regulated in the Tgfbr1 cKO oviducts (P<0.05; Figure 7).
Discussion
Despite the progress made on functional characterization of TGFβ family ligands in female reproduction, the in vivo roles of individual receptors in this pathway have remained elusive. Because conventional inactivation of Tgfbr1 results in embryonic lethality [44], the functional understanding of this receptor in female reproductive tissues was hampered. In the current study, conditional deletion of Tgfbr1 in the female reproductive tract using Amhr2-Cre expressed in granulosa cells and mesenchymal compartments of the oviduct and uterus [51], [55] led to female sterility. Histological analysis revealed that Tgfbr1 cKO mice had minimal defects in their ovaries, which contain morphologically normal follicles at various developmental stages. To determine if the Tgfbr1 cKO mice have normal ovarian function, we conducted superovulation and fertilization experiments. Our results showed that Tgfbr1 cKO mice could ovulate, and the ovulated oocytes were fertilizable. These data suggest that TGFBR1 in mouse granulosa cells may not be essential for ovulation and oocyte fertilization.
TGFBR1 can mediate GDF9 signaling in granulosa cells in vitro [35]. Since GDF9 regulates folliculogenesis and cumulus cell expansion [12], [36], [42], [67], we were interested to know if cumulus cell function was impaired in the Tgfbr1 cKO mice. We found that Tgfbr1 cKO cumulus cells could expand in vivo and in vitro. Moreover, TGFBR1 was predominantly localized to thecal cells and corpora lutea, but not granulosa cells of developing follicles at preantral stage, the known sites for GDF9 action. After PMSG-hCG injection, the mural granulosa cells but not cumulus cells of preovulatory follicles highly expressed β-galactosidase from the Tgfbr1 bgal knockin allele. Recombinant GDF9 or BMP15, another oocyte-derived factor implicated in follicular development [68]–[70], could markedly reduce Tgfbr1 expression in mouse granulosa cells, which might explain the low intensity of TGFBR1 signals in cumulus cells adjacent to the oocyte. The data indicate that TGFBR1 might not be a physiological receptor for GDF9, or at least not the sole GDF9 type 1 receptor in mouse ovarian somatic cells. To further explore the potential GDF9 type 1 receptor(s) in mouse ovary, we utilized mouse granulosa cell culture and took advantage of Alk6 null granulosa cells and small molecule inhibitors for ALK2/3/6 [71] and ALK4/5/7 [72]. Consistent with ALK6 as the BMP15 type 1 receptor [73], [74], the ability of recombinant BMP15 to induce cumulus expansion-related transcript expression is completely lost in mouse granulosa cells lacking ALK6 (data not shown). However, GDF9 signaling remains intact in Alk6 null cells, excluding ALK6 as a GDF9 receptor. The small molecule inhibitor studies further helped to identify potential candidate receptors for GDF9 in mouse ovary (i.e., ALK4 and/or ALK7), although SB-505124 cannot precisely distinguish the type 1 receptor through which GDF9 signals. Despite these findings, future functional studies using conditional deletion of one or more type 1 receptors are needed to pinpoint the physiological receptor(s) for GDF9 in mouse ovary.
Because Amhr2 is also expressed in mesenchyme-derived tissues in the oviduct and uterus [75], we examined whether the observed sterility is a phenotypic consequence of conditional knockout of Tgfbr1 in the oviduct and/or uterus. We found that the Tgfbr1 cKO mice develop a striking oviductal phenotype marked by the formation of bilateral diverticula. Histologically, a well-formed diverticulum comprises a single layer of smooth muscle cells and epithelium. The presence of degenerating oocytes/embryos in the oviductal diverticula but absence of blastocysts in the uteri (3.5 dpc) of the Tgfbr1 cKO mice strongly indicate that development of oviductal diverticula is sufficient to cause female infertility in the Tgfbr1 cKO mice, though it is plausible that disruption of the uterine smooth muscle development might sequentially confound the pregnancy outcome if pregnancy could occur in these mice. It is known that the myometrium plays an important role in key pregnancy-associated reproductive events, although current knowledge of myometrial causes of reproductive disorders is limited [76]. A successful labor is dependent on the synchronous myometrial contractions, which are regulated by a series of coordinated events at both hormonal and molecular levels during pregnancy [77], [78]. The disruption of uterine smooth muscle structure in the Tgfbr1 cKO mice could potentially impede the contractility of the uterus or cause uterine rupture, with an adverse impact on pregnancy outcome. Emerging evidence suggests the involvement of the Wnt pathway in the maintenance of myometrium organization and integrity [79], [80]. Further investigations on the potential link between TGFBR1–mediated signaling and the Wnt pathway as well as the direct impact of the myometrial abnormalities resulting from loss of TGFβ/Wnt signaling components on reproductive potential may shed mechanistic light on reproductive disorders associated with smooth muscle pathology.
It is noteworthy that the oviductal phenotype of the TGFBR1-deficient mice resembles that of the conditional deletion of Dicer1, a key gene involved in miRNA and small interfering RNA (siRNA) biogenesis pathways [81]. MicroRNAs are non-coding small RNAs that regulate gene expression by inducing translational repression or mRNA degradation of target genes [81]. Recent studies in vascular smooth muscle cells suggest that TGFβ signaling can induce the maturation of a subset of miRNAs through the interactions between SMADs and the consensus RNA sequence of miRNAs within the DROSHA microprocessor complex [56], [82], [83]. Based on these findings and the similarity of the oviductal phenotype between Tgfbr1 and our previously described Dicer1 cKO mice [55], we proposed a potential link between the TGFβ signaling and miRNA pathways in the female reproductive tract. To test this hypothesis, we examined the expression of select genes/miRNAs in the oviducts of 3–4 week old Tgfbr1 cKO and control mice. We found a global reduction of expression of smooth muscle genes, as well as two miRNAs, miR-143 and miR-145, in the Tgfbr1 cKO oviducts compared with controls. These two miRNAs are expressed in smooth muscle cells and have debatable roles in specifying smooth muscle phenotype [57]–[60], [84]. However, miR-21, which is regulated by TGFβ signaling in vascular smooth muscle cells [56], was not altered in the Tgfbr1 cKO oviducts. The defective oviductal smooth muscle phenotype of the Tgfbr1 cKO mice raised the possibility that the reductions of smooth muscle genes and smooth muscle associated miRNAs could be a consequence of reduced muscle components in the oviductal samples. To further address this question, we collected and analyzed oviductal samples from both control and Tgfbr1 cKO mice at the age of 7 days prior to significant smooth muscle loss. We confirmed by quantitative PCR that miR-143 was not significantly altered in the Tgfbr1 cKO oviducts. Consistently, alteration of smooth muscle gene expression was not found in 7-day-old oviducts of Tgfbr1 cKO mice. Therefore, the decreased expression of miR-143/145 and smooth muscle genes in the 3–4 week old Tgfbr1 cKO mice is likely caused by reduced smooth muscle components. Although Dicer1 cKO mice develop oviductal diverticula [55], they have distinct uterine phenotypes (i.e., small uteri but histologically normal smooth muscle layers) and oviductal gene expression patterns compared to the Tgfbr1 cKO mice. Moreover, the phenotype of Tgfbr1 cKO mice is distinct from that of conditional deletion of Smad2 and Smad3 [18], suggesting the involvement of SMAD-independent pathway(s) downstream of TGFBR1. Collectively, the oviductal phenotype observed in Tgfbr1 cKO mice is likely not a direct consequence of miRNA dysregulation.
Molecular analysis of the postnatal day 7 oviducts from Tgfbr1 cKO mice demonstrated dysregulation of genes associated with cell differentiation and migration. Keratins have recently been highlighted as vital regulators of diverse cellular properties and functions (e.g., apico-basal polarization, motility, etc.), rather than simple epithelial markers [62]. KRT12 is a member of epithelial intermediate filament proteins which generally consist of two types of keratins (type 1 and type 2) as heterodimeric polymers [61], [62]. Dysregulation of epithelial genes in the oviducts of Tgfbr1 cKO mice suggests that mesenchymal-epithelial interactions, which are potentially vital for smooth muscle development [85], could be affected when TGFBR1–mediated signaling is disrupted in the smooth muscle compartment although potentially functional TGFBR1 might still be present in the epithelial compartment due to the lack of Amhr2-Cre activity. As evidence of potentially altered smooth muscle cell differentiation, we found that MyoR/musculin was substantially up-regulated in the Tgfbr1 cKO oviducts. Despite the fact that MyoR is also expressed in other cell types and can regulate their differentiation [86], the significance of MyoR up-regulation in Tgfbr1 cKO oviducts awaits further investigation as current understanding of MyoR-regulated cell differentiation has been confined to the skeletal muscle lineage [87]. Beyond the aspects of cell differentiation, our data also point to the potential aberration of cell migration in the Tgfbr1 cKO oviducts. It is well established that the renin-angiotensin system serves as a physiological system regulating blood pressure. Renin catalyzes the conversion of angiotensinogen to angiotensin I, which can be converted by ACE into angiotensin II. ACE2 is a newly described member of the renin-angiotensin system that can cleave angiotensin II into angiotensin 1–7, or angiotensin I into angiotensin 1–9 [66]. The renin-angiotensin system has been implicated in vascular smooth muscle cell proliferation and migration [88], [89], and ACE2 overexpression-induced alterations in cell migration have been documented [88]. In further support of aberrant cell migrations in the Tgfbr1 cKO oviducts, we found increased expression of Vegfa and Figf/Vegfd. VEGFA and VEGFD are known regulators of smooth muscle cell migration [90], [91], and VEGF receptors are expressed in vascular smooth muscle cells [92]. Interestingly, Vegf is induced by TGFβ in mouse macrophages [93]. However, given the highly context-dependent nature of gene regulation by TGFβ signaling [94] as well as the diversity of ligands which signal via TGFBR1, it is not surprising that Vegf is up-regulated in mouse oviducts lacking TGFBR1. Moreover, it is not clear if the dysregulation of the aforementioned genes in the oviducts are direct or indirect effects of the loss of TGFBR1. Thus, our results indicate that profound molecular changes may occur in the smooth muscle and/or epithelial compartments of the oviduct in the absence of TGFBR1–mediated signaling. These alterations may developmentally affect the structural, migratory, and differentiating properties of the smooth muscle/epithelial cells, ultimately leading to the formation of the deleterious oviductal diverticula.
In summary, this study provides genetic evidence that TGFBR1–mediated signaling controls the integrity and function of the female reproductive tract. Disruption of TGFBR1–mediated signaling leads to catastrophic structural and functional consequences. Further in-depth understanding of the functional and regulatory significance of TGFBR1–mediated signaling in female reproductive physiology and pathology may help to discover novel therapeutic approaches for infertility treatment.
Materials and Methods
Ethics Statement
All mouse lines were manipulated according to the NIH Guide for the Care and Use of Laboratory Animals. All procedures have been approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine. We took all necessary steps to minimize suffering of mice during the experiments.
Animals and Cell Lines
All mouse lines were maintained on a mixed genetic background (C57BL/6/129S6/SvEv). The Tgfbr1 flox allele was constructed by flanking the Tgfbr1 exon 3 which encodes the transmembrane domain and the glycine/serine-rich (GS) domain with two loxP sites [44]. Mice harboring a Tgfbr1 bgal allele (null allele) were generated and characterized by Deltagen and obtained from the Jackson laboratory. The function of the Tgfbr1 gene was disrupted by insertion of a bacterial LacZ into the Tgfbr1 gene (http://jaxmice.jax.org/strain/005847.html). The Amhr2 cre/+ mice were created by inserting a Cre-Neo cassette into the fifth exon of the Amhr2 locus [48]. HEK-293 and HEK-293T cells were obtained from the Tissue Culture Core at Baylor College of Medicine. Dicer1 cKO mice were generated as previously described [55].
Production of Purified Recombinant BMP15 and GDF9
Recombinant human BMP15 was produced from HEK-293 stable cell lines as described previously [95]. Recombinant mouse GDF9 was constructed, produced, and purified using a similar strategy for engineering recombinant human BMP15 [95]. Briefly, we cloned the mouse GDF9 cDNA from 3-week old mouse ovaries. PCR-based mutations and introduction of restriction sites were performed using Phusion Hot Start High-Fidelity DNA polymerase (Finnzymes). Optimization of the cleavage and surrounding sequence (SHRSKRSLSGG) and introduction of a FLAG-tag (DYKDDDDK) were conducted by using overlap extension PCR. The genetically modified GDF9 sequence was cloned into pEFIRES-P, a bicistronic expression vector driven by human polypeptide chain elongation factor 1α promoter [96]. The GDF9 expression construct was then transfected into HEK-293T cells, and cell clones stably expressing recombinant GDF9 were selected in the presence of puromycin (Invitrogen) and used for the production of recombinant proteins. As a rigorous control for the purified recombinant GDF9, a “control buffer” was produced from the culture medium of non-transfected cells under the same purification approach as previously described [95].
Generation of Tgfbr1 cKO Mice
Using a Cre-loxP system [55], we first generated Tgfbr1 +/bgal; Amhr2 cre/+ mice. We subsequently crossed these mice with Tgfbr1 flox/flox (homozygotes) to produce mice with the following genotypes: Tgfbr1 flox/bgal; Amhr2 cre/+ (cKO; experimental mice) and controls (Tgfbr1 flox/bgal and Tgfbr1 flox/+). For the fertility tests, each female cKO or control mouse was caged with a WT male with known fertility at the age of 6 weeks for a 6-month period. The genotypes of the mice were analyzed by PCR using gene specific primers (Table 3).
Table 3. Primers for conventional PCR and SYBR green-based real-time PCR.
| Name | Sequence (5′-3′) | Reference | |
| Tgfbr1 flox | Forward | ACTCACATGTTGGCTCTCACTGTC | [102] |
| Reverse | AGTCATAGAGCATGTGTTAGAGTC | ||
| Tgfbr1 +/− | moIMR0003 | GGGCCAGCTCATTCCTCCCACTCAT | Tgfbr1 tm1Dgen |
| moIMR0843 | CCTGTGGAGCTGGCAGCTGTCATTG | Jackson lab | |
| moIMR0844 | ACTATCCGGGTCAGACAGCAAGCTC | ||
| Tgfbr1 Rec | Forward | ATTTCTTCTGCTATAATCCTGCAG | [102] |
| Reverse | AGTCATAGAGCATGTGTTAGAGTC | ||
| Amhr2 Cre | Forward | CGCATTGTCTGAGTAGGTGT | [18] |
| Reverse | GAAACGCAGCTCGGCCAGC | ||
| Hprt | Forward | GGACCTCTCGAAGTGTTGGATAC | N/A |
| Reverse | CTTGCGCTCATCTTAGGCTT | ||
| Tagln | Forward | CGATGGAAACTACCGTGGAGA | [55] |
| Reverse | TGAAGGCCAATGACGTGCT | ||
| Acta2 | Forward | CCACCGCAAATGCTTCTAAGT | [55] |
| Reverse | GGCAGGAATGATTTGGAAAGG | ||
| Cnn1 | Forward | GGTGAAACCCCACGACATCTT | [55] |
| Reverse | TTTGTCTTGGCCATGCTGG | ||
| Camk2d | Forward | CAGCAGGCATGGTTTGGTT | N/A |
| Reverse | CATAAGGATCTTTACGCAGGACTTC | ||
| Myh11 | Forward | CATCCTGACCCCACGTATCAA | [55] |
| Reverse | ATCGGAAAAGGCGCTCATAGG | ||
| Des | Forward | TACACCTGCGAGATTGATGCC | [55] |
| Reverse | GCGCAATGTTGTCCTGATAGC | ||
| Smtn | Forward | TCACTACCTTCAGCCATGCCT | [55] |
| Reverse | GCCATTAGCTGCTTCCACTGT | ||
| Gapdh | Forward | CAATGTGTCCGTCGTGGATCT | [55] |
| Reverse | GCCTGCTTCACCACCTTCTT | ||
| Tgfbr1 | Forward | TGCCATAACCGCACTGTCA | N/A |
| Reverse | AATGAAAGGGCGATCTAGTGATG | ||
| Myocd | Forward | CCAACACCTTGCCCAGTTATC | N/A |
| Reverse | GGAGCTTGTGCTGCCAAAG | ||
| Hoxa9 | Forward | CCTTCTCCGAAAACAATGCC | [55] |
| Reverse | TCCTTCTCCAGTTCCAGCGT | ||
| Hoxa10 | Forward | TGGAGAAGGAGTTTCTATTCAACATG | N/A |
| Reverse | TGGACGCTACGGCTGATCT | ||
| Elk1 | Forward | CATGACAGCCACGAATATGAAGA | N/A |
| Reverse | TCTGACATATGCCATCATTTTGC | ||
| Klf4 | Forward | CTCACACAGGCGAGAAACCTT | N/A |
| Reverse | GAGCGGGCGAATTTCCA | ||
Primer sequences are listed along with available references. All real-time PCR primers were designed using Primer Express software (Applied Biosystems).
Histology and β-Gal Staining
For histological studies, mouse samples (ovaries, oviducts, and uteri) were fixed in 10% neutral buffered formalin (NBF) overnight. The samples were washed with 70% ethanol and embedded in paraffin by the Pathology Core Services Facility at Baylor College of Medicine. The samples were further processed for hematoxylin and eosin (H&E) or periodic acid Schiff (PAS)-hematoxylin staining using standard procedures.
For β-gal staining, mouse samples were fixed in fixation solution (2% paraformaldehyde, 0.2% glutaraldehyde, and 0.1 M phosphate pH 7.4) for 10–15 min. The samples were then rinsed 3 times for 30 min each in rinse buffer (0.01% sodium deoxycholate, 0.02% NP-40, 2 mM MgCl2, and 0.1 M phosphate pH 7.4). The β-gal staining was performed overnight at room temperature in staining buffer (0.01% sodium deoxycholate, 0.02% NP-40, 2 mM MgCl2, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/ml X-gal, and 0.1 M phosphate pH 7.4). After staining, the samples were fixed in 10% NBF for at least 4 h, washed with 70% ethanol, and processed for sectioning. The sections were counterstained with fast red (Vector lab).
Immunohistochemistry and Immunofluorescence
Paraffin-embedded sections (5 µm) were deparaffinized in xylene and rehydrated in graded alcohol. Antigen retrieval was performed by boiling the sections in 10 mM citrate buffer (pH 6.0) for 20 min. To quench endogenous peroxidase, the sections were treated with 0.3% (v/v) hydrogen peroxide, and then blocked with 3% goat serum for 30 min, followed by incubation with rabbit anti smooth muscle α-actin (Abcam; 1∶500) or rabbit anti-calponin 1 (Millipore; 1∶200) at 4°C overnight. After primary antibody incubation, the sections were washed and sequentially incubated with biotinylated anti-rabbit IgG and ABC reagent (Vector Labs). Immunoreactive signals were developed using a DAB substrate kit (Vector Labs). The sections were counterstained with hematoxylin.
Immunofluorescence was conducted using a similar protocol except that the hydrogen peroxide treatment was omitted and the secondary antibodies were Alexa fluor 488 (Invitrogen) and Alexa fluor 555 (Molecular Probes). The rat anti-cytokeratin-8 antibody was obtained from the Developmental Studies Hybridoma Bank (DSHB). The sections were mounted with Vectashield mounting medium containing DAPI.
Superovulation, Embryo Culture, and Timed Mating
Superovulation experiments were performed to assess the ovulatory potential of the Tgfbr1 cKO mice as described [18], [46]. In brief, the gonadotropin-primed female Tgfbr1 cKO and control mice were mated with WT males. The cumulus-oocyte complexes (COCs) were recovered, exposed to M2 medium containing 1 mg/ml hyaluronidase, and counted. They were then cultured in M16 medium (Sigma). The number of embryos at the 2-cell stage was determined and recorded at the end of culture [55]. For timed matings, the control and Tgfbr1 cKO females (6–8 week old) were mated with WT males, and copulatory plugs were checked the next morning. The oocytes or embryos were collected at 3.5 dpc from the oviducts or uteri.
Hormone Analyses
Blood samples were collected by cardiac puncture from mice anesthetized with isoflurane. The serum was separated from the blood and stored at −20°C until assayed. Serum progesterone (P4) levels were measured by the Ligand Assay and Analysis Core at the Center for Research in Reproduction, University of Virginia. The detection limit was 0.1 ng/ml. Assay details can be found at http://www.healthsystem.virginia.edu/internet/crr/ligand.cfm.
Cumulus Expansion Assays
Cumulus expansion analysis was conducted as previously described [18]. For in vitro cumulus expansion assay, the COCs were collected and cultured in the presence or absence of epidermal growth factor (EGF; 10 ng/ml). Cumulus expansion was examined after 16 h of culture and scored. The cumulus expansion index (CEI) was calculated based on the degree of COC expansion using a scale from 0 (no expansion) to 4 (complete expansion) [97]. For in vivo cumulus expansion analysis, immature mice (21–23 d) were treated with PMSG (46 h) and hCG (7 h). The ovaries were collected and processed for PAS staining, and the preovulatory follicles were examined microscopically for cumulus cell expansion.
Primary Granulosa Cell Culture
Immature WT or Alk6 null female mice [98], [99] were treated with 5 IU PMSG (i.p.). Large antral follicles were punctured to collect granulosa cells after 44–46 h [18], [95]. The cells were filtered through a 40 µm nylon mesh (Becton, Dickinson and Company), washed, and resuspended for the following experiments. (1) Tgfbr1 mRNA regulation by oocyte-produced factors: The WT mouse granulosa cells were treated with control buffer [95], recombinant BMP15 (100 ng/ml), or recombinant GDF9 (100 ng/ml). The cells were collected in lysis buffer after 5 h treatment. (2) Gene induction assay (Ptx3): The WT and Alk6 null granulosa cells were collected and treated with recombinant BMP15 (100 ng/ml) or GDF9 (100 ng/ml), and the cells were collected 5 h later. (3) Small molecule inhibitor assay: Mouse granulosa cells were preincubated for 1 h with dorsomorphin (Calbiochem; 4 µM) or SB-505124 (Sigma; 1 µM), and BMP15 or GDF9 was then added to the culture and further incubated for 5 h before the cells were collected. Total RNA was isolated from the cells harvested in the above experiments, and real-time PCR analyses were performed to determine the Tgfbr1 or Ptx3 mRNA expression.
Reverse Transcription, Real-Time PCR, and microRNA Analysis
Total RNA from mouse granulosa cells, oviducts, or uteri was isolated using Qiagen RNeasy Micro or Mini Kit. Two hundred nanograms of total RNA were reverse transcribed using Superscript III reverse transcriptase (Invitrogen) [95]. Real-time PCR was performed using Taqman gene expression assay (Applied Biosystems) and Taqman PCR Master Mix or customized primers and SYBR green master mix [55]. Primer information is listed in Table 3 and Table 4.
Table 4. Taqman gene expression and miRNA assays.
| miRNA | Taqman assay ID |
| Tgfbr1 | Mm00436971_m1 |
| Ptx3 | Mm00477267_g1 |
| Wnt5a | Mm00437347_m1 |
| Wnt7a | Mm00437355_m1 |
| hsa-mir-143 | 002249 |
| hsa-mir-145 | 002278 |
| hsa-miR-21 | 000397 |
| snoRNA202 | 001232 |
Taqman assays were obtained from Applied Biosystems, and assay names and their corresponding IDs are listed.
For microRNA analysis, total RNA was isolated from mouse oviducts using mirVana miRNA isolation kit (Ambion). Levels of mature miRNA were measured using a two-step TaqMan MicroRNA Assay (Applied Biosystems). First, reverse transcription was performed using 10 ng of total RNA and stem-loop primers specific for miR-143, miR-145, and miR-21. Then, quantitative real-time PCR was conducted using specific Taqman probes for these miRNAs and Taqman universal PCR master mix (No AmpErase UNG).
Real-time PCR was carried out based on a protocol consisting of 40 cycles: 95°C for 10 min (hold), 95°C for 15 s (denature), and 60°C for 1 min (anneal/extend). All real-time PCR assays were performed in duplicate or triplicate for each sample. Gapdh was used as an internal control for the quantification of gene expression, while snoRNA202 was used for normalization of miRNA levels. Relative mRNA abundance was calculated using ΔΔCT method [100].
Uterine Decidualization
The uterine decidualization experiment was performed as described elsewhere [101]. Briefly, adult Tgfbr1 cKO and control mice were subjected to ovariectomy followed by 3 days of estradiol treatment (100 ng/mouse) and 2 days of rest. The mice were subsequently treated with both estradiol (6.7 ng/mouse) and progesterone (1.0 mg/mouse) for 3 days. To artificially induce uterine decidualization, one uterine horn was traumatized/scratched using a needle on the antimesometrial side. The other uterine horn was not traumatized and used as a control. After that, the mice were continuously primed with estradiol and progesterone for 5 more days before sacrifice. Both uterine horns were weighed, and the tissues were collected and fixed in paraformaldehyde for histological and immunohistochemical assays.
Statistical Analyses
A one-way analysis of variance (ANOVA) was applied to determine the difference of means among groups, and the difference between two means was further assessed by Tukey's HSD test. Comparison of means between two groups was conducted using t-test. Data are presented as mean ± standard error of the mean (SEM). Statistical significance was defined at P<0.05.
Supporting Information
Tgfbr1 cKO mice have defective smooth muscle formation in the oviduct. (A to C) Calponin 1 is expressed in the smooth muscle layers of the control oviduct (A and B). Negative control in which the primary antibody was substituted with rabbit IgG is shown in (C). (D to F) Defective smooth muscle formation in the oviducts of Tgfbr1 cKO mice. Higher magnification view of the selected regions within the blue and red rectangles in (D) were depicted in (E) and (F), respectively. Arrowheads demonstrate smooth muscle defects. SM, smooth muscle; EP, epithelium. Scale bars = 50 µm (E and F); 100 µm (B and C); and 200 µm (A and D).
(TIF)
Follicular development in Tgfbr1 cKO mice. (A and B) Ovaries of 3-week-old control and Tgfbr1 cKO mice containing follicles at various developmental stages. (C and D) PMSG-induced follicular development in control (C) and Tgfbr1 cKO (D) mice. Arrows indicate preovulatory follicles. Scale bars = 200 µm.
(TIF)
Corpora lutea formation in Tgfbr1 cKO mice. (A and B) PAS staining of ovaries from PMSG-hCG treated control (A) and Tgfbr1 cKO (B) mice. Corpora lutea formed in both control and Tgfbr1 cKO mice after 46 h of PMSG and 20 h of hCG treatment. (C to E) Immunohistochemical staining of 3β-HSD in the corpora lutea of control (C) and Tgfbr1 cKO (D) mice. A representative negative control of the Tgfbr1 cKO mouse ovary was shown in (E). Scale bars = 200 µm. CL, corpus luteum. (F) Serum progesterone levels in gonadotropin-primed and natural pregnant mice (n = 3–4). Data are presented as mean ± SEM.
(TIF)
Tgfbr1 cKO mice have defects in uterine smooth muscle formation. (A and B) Gross uterine morphology of Tgfbr1 cKO mice at 3 weeks and 3 months of age. (C to H) Immunostaining of ACTA2 in the uteri of control (C and F) and Tgfbr1 cKO (D, E, G, and H) mice. Note the disrupted smooth muscle layers in the Tgfbr1 cKO mice, and the unusual appearance of the smooth muscle structure in the endometrium. Ut, uterus; LM, longitudinal muscle layer; CM, circular muscle layer. Red arrowheads point to the disorganized smooth muscle layers. Scale bars = 50 µm (F to H); 200 µm (C to E); and 5 mm (A and B).
(TIF)
Smooth muscle defects lead to severe disruption of uterine structures in Tgfbr1 cKO mice. (A) Uterine abnormalities in an 8-month-old Tgfbr1 cKO female compared to an age-matched control mouse. Blue arrows indicate the uteri. (B to F) Immunostaining of CNN1 in the uteri of 8-month-old control (B) and Tgfbr1 cKO mice (C to F). CNN1 staining demonstrated multiple structural abnormalities in the uterine smooth muscle layers of the Tgfbr1 cKO mice. Defective smooth muscle development leads to uterine cyst formation (arrow heads; C, D, and E), and complete loss of normal uterine structure in the severe cases (D and F). Scale bars = 200 µm (B to F).
(TIF)
The uteri of Tgfbr1 cKO mice can undergo artificial decidualization. (A and B) Gross morphology of the uteri in the control (A) and Tgfbr1 cKO (B) mice 5 days after the decidual stimulus. The left horns (control horns) of the uteri were unstimulated while the right ones (decidual horns) were traumatized. Note that the uteri of both control and Tgfbr1 cKO mice can undergo artificially induced decidualization. (C) Weight ratio of decidual horn to control horn in control (n = 3) and Tgfbr1 cKO mice (n = 4). Data are presented as mean ± SEM. (D–G) Immunostaining of control and decidual horns of Tgfbr1 cKO mice using CCN1. Higher magnification views of (D) and (F) are depicted in (E) and (G), respectively. Note the disruption of smooth muscle structure in both unstimulated and decidual horns of the Tgfbr1 cKO mice. Scale bars = 200 µm (E and G); 400 µm (D and F), and 10 mm (A).
(TIF)
Expression of target genes of miR-143/145 in Tgfbr1 cKO mice. Real-time PCR analyses using 3–4 week old oviducts demonstrated that Elk1, Klf4, and Camk2d are expressed at comparable levels between Tgfbr1 cKO mice and controls. n = 3–4 independent pools of oviducts. Relative mRNA levels were normalized to Gapdh. Data are presented as mean ± SEM.
(TIF)
Messenger RNA levels of genes up-regulated in Dicer1 cKO oviducts are not elevated in the oviducts of Tgfbr1 cKO mice. Wnt5a, Wnt7a, Hoxa9, and Hoxa10 are significantly up-regulated genes in the 3–4 week old oviducts of Dicer1 cKO mice [55]. Real-time PCR analyses using age-matched Tgfbr1 cKO and control oviducts did not demonstrate up-regulation of these genes in the Tgfbr1 cKO mice. n = 3–4 independent pools of oviducts. Relative mRNA levels were normalized to Gapdh. Data are presented as mean ± SEM.
(TIF)
Acknowledgments
We are grateful to Dr. Stefan Karlsson for generously contributing the Tgfbr1 floxed mouse line, Dr. Richard Behringer for the gift of the Amhr2-Cre mice, Dr. Karen Lyons for generously providing the Alk6 null mice, Dr. Paul Oh for help with importing the Tgfbr1 mutant mice to Baylor College of Medicine, and Ms. Luz Martinez for technical assistance.
Footnotes
The authors have declared that no competing interests exist.
Serum progesterone analysis was performed at the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core, supported by NICHD (SCCPRR) grant U54-HD28934. This research was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NIH grant HD033438 to MMM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Massague J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 2.Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian transforming growth factor-β superfamily. Endocr Rev. 2002;23:787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- 3.Juengel JL, McNatty KP. The role of proteins of the transforming growth factor-beta superfamily in the intraovarian regulation of follicular development. Hum Reprod Update. 2005;11:144–161. doi: 10.1093/humupd/dmh061. [DOI] [PubMed] [Google Scholar]
- 4.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 5.Bellam N, Pasche B. TGF-beta signaling alterations and colon cancer. Cancer Treat Res. 2010;155:85–103. doi: 10.1007/978-1-4419-6033-7_5. [DOI] [PubMed] [Google Scholar]
- 6.Ikushima H, Miyazono K. TGF-beta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 7.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 8.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 9.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 10.Massague J. Receptors for the TGF-beta family. Cell. 1992;69:1067–1070. doi: 10.1016/0092-8674(92)90627-o. [DOI] [PubMed] [Google Scholar]
- 11.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 12.Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, et al. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383:531–535. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi M, McGee EA, Min G, Klein C, Rose UM, et al. Recombinant growth differentiation factor-9 (GDF-9) enhances growth and differentiation of cultured early ovarian follicles. Endocrinology. 1999;140:1236–1244. doi: 10.1210/endo.140.3.6548. [DOI] [PubMed] [Google Scholar]
- 14.Myers M, Pangas SA. Regulatory roles of transforming growth factor beta family members in folliculogenesis. Wiley Interdiscip Rev Syst Biol Med. 2010;2:117–125. doi: 10.1002/wsbm.21. [DOI] [PubMed] [Google Scholar]
- 15.Nagaraja AK, Middlebrook BS, Rajanahally S, Myers M, Li Q, et al. Defective gonadotropin-dependent ovarian folliculogenesis and granulosa cell gene expression in inhibin-deficient mice. Endocrinology. 2010;151:4994–5006. doi: 10.1210/en.2010-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragovic RA, Ritter LJ, Schulz SJ, Amato F, Thompson JG, et al. Oocyte-secreted factor activation of SMAD 2/3 signaling enables initiation of mouse cumulus cell expansion. Biol Reprod. 2007;76:848–857. doi: 10.1095/biolreprod.106.057471. [DOI] [PubMed] [Google Scholar]
- 17.Diaz FJ, Wigglesworth K, Eppig JJ. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J Cell Sci. 2007;120:1330–1340. doi: 10.1242/jcs.000968. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Pangas SA, Jorgez CJ, Graff JM, Weinstein M, et al. Redundant roles of SMAD2 and SMAD3 in ovarian granulosa cells in vivo. Mol Cell Biol. 2008;28:7001–7011. doi: 10.1128/MCB.00732-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee KY, Jeong JW, Wang J, Ma L, Martin JF, et al. Bmp2 is critical for the murine uterine decidual response. Mol Cell Biol. 2007;27:5468–5478. doi: 10.1128/MCB.00342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roelen BA, Lin HY, Knezevic V, Freund E, Mummery CL. Expression of TGF-βs and their receptors during implantation and organogenesis of the mouse embryo. Dev Biol. 1994;166:716–728. doi: 10.1006/dbio.1994.1350. [DOI] [PubMed] [Google Scholar]
- 21.Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- 22.Li Q, Graff JM, O'Connor AE, Loveland KL, Matzuk MM. SMAD3 regulates gonadal tumorigenesis. Mol Endocrinol. 2007;21:2472–2486. doi: 10.1210/me.2007-0147. [DOI] [PubMed] [Google Scholar]
- 23.Pangas SA, Li X, Umans L, Zwijsen A, Huylebroeck D, et al. Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol Cell Biol. 2008;28:248–257. doi: 10.1128/MCB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Middlebrook BS, Eldin K, Li X, Shivasankaran S, Pangas SA. Smad1-Smad5 ovarian conditional knockout mice develop a disease profile similar to the juvenile form of human granulosa cell tumors. Endocrinology. 2009;150:5208–5217. doi: 10.1210/en.2009-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edson MA, Nalam RL, Clementi C, Franco HL, Demayo FJ, et al. Granulosa cell-expressed BMPR1A and BMPR1B have unique functions in regulating fertility but act redundantly to suppress ovarian tumor development. Mol Endocrinol. 2010;24:1251–1266. doi: 10.1210/me.2009-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang H, Lau AL, Matzuk MM. Studying TGF-beta superfamily signaling by knockouts and knockins. Mol Cell Endocrinol. 2001;180:39–46. doi: 10.1016/s0303-7207(01)00513-5. [DOI] [PubMed] [Google Scholar]
- 28.Pangas SA, Matzuk MM. The TGFβ family in the reproductive tract. In: Derynck R, Miyazono K, editors. TGFβ family. New York: Cold Spring Harbor Laboratory Press; 2008. pp. 861–888. [Google Scholar]
- 29.Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr Rev. 2009;30:624–712. doi: 10.1210/er.2009-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards JS, Pangas SA. The ovary: basic biology and clinical implications. J Clin Invest. 2010;120:963–972. doi: 10.1172/JCI41350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matzuk MM, Kumar TR, Bradley A. Different phenotypes for mice deficient in either activins or activin receptor type II. Nature. 1995;374:356–360. doi: 10.1038/374356a0. [DOI] [PubMed] [Google Scholar]
- 32.Song J, Oh SP, Schrewe H, Nomura M, Lei H, et al. The type II activin receptors are essential for egg cylinder growth, gastrulation, and rostral head development in mice. Dev Biol. 1999;213:157–169. doi: 10.1006/dbio.1999.9370. [DOI] [PubMed] [Google Scholar]
- 33.Lee SJ, Reed LA, Davies MV, Girgenrath S, Goad ME, et al. Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc Natl Acad Sci USA. 2005;102:18117–18122. doi: 10.1073/pnas.0505996102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orvis GD, Jamin SP, Kwan KM, Mishina Y, Kaartinen VM, et al. Functional redundancy of TGF-beta family type I receptors and receptor-Smads in mediating anti-Mullerian hormone-induced Mullerian duct regression in the mouse. Biol Reprod. 2008;78:994–1001. doi: 10.1095/biolreprod.107.066605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazerbourg S, Klein C, Roh J, Kaivo-Oja N, Mottershead DG, et al. Growth differentiation factor-9 signaling is mediated by the type I receptor, activin receptor-like kinase 5. Mol Endocrinol. 2004;18:653–665. doi: 10.1210/me.2003-0393. [DOI] [PubMed] [Google Scholar]
- 36.Elvin JA, Clark AT, Wang P, Wolfman NM, Matzuk MM. Paracrine actions of growth differentiation factor-9 in the mammalian ovary. Mol Endocrinol. 1999;13:1035–1048. doi: 10.1210/mend.13.6.0310. [DOI] [PubMed] [Google Scholar]
- 37.Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science. 2002;296:2178–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- 38.Su YQ, Wu X, O'Brien MJ, Pendola FL, Denegre JN, et al. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol. 2004;276:64–73. doi: 10.1016/j.ydbio.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 39.Gui L-M, Joyce IM. RNA interference evidence that growth differentiation factor-9 mediates oocyte regulation of cumulus expansion in mice. Biol Reprod. 2005;72:195–199. doi: 10.1095/biolreprod.104.033357. [DOI] [PubMed] [Google Scholar]
- 40.Su YQ, Sugiura k, Wigglesworth k, O'Brien MJ, Affourtit JP, et al. Oocyte regulation of metabolic cooperativity between mouse cumulus cells and oocytes: BMP15 and GDF9 control cholesterol biosynthesis in cumulus cells. Development. 2008;135:111–121. doi: 10.1242/dev.009068. [DOI] [PubMed] [Google Scholar]
- 41.Myllymaa S, Pasternack A, Mottershead DG, Poutanen M, Pulkki MM, et al. Inhibition of oocyte growth factors in vivo modulates ovarian folliculogenesis in neonatal and immature mice. Reproduction. 2010;139:587–598. doi: 10.1530/REP-09-0391. [DOI] [PubMed] [Google Scholar]
- 42.Li Q, McKenzie LJ, Matzuk MM. Revisiting oocyte-somatic cell interactions: in search of novel intrafollicular predictors and regulators of oocyte developmental competence. Mol Hum Reprod. 2008;14:673–678. doi: 10.1093/molehr/gan064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilchrist RB. Recent insights into oocyte-follicle cell interactions provide opportunities for the development of new approaches to in vitro maturation. Reprod Fertil Dev. 2011;23:23–31. doi: 10.1071/RD10225. [DOI] [PubMed] [Google Scholar]
- 44.Larsson J, Goumans M-J, Sjostrand LJ, van Rooijen MA, Ward D, et al. Abnormal angiogenesis but intact hematopoietic potential in TGF-β type I receptor-deficient mice. EMBO J. 2001;20:1663–1673. doi: 10.1093/emboj/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kos CH. Cre/loxP system for generating tissue-specific knockout mouse models. Nutr Rev. 2004;62:243–246. doi: 10.1301/nr2004.jun243-246. [DOI] [PubMed] [Google Scholar]
- 46.Pangas SA, Li X, Robertson EJ, Matzuk MM. Premature luteinization and cumulus cell defects in ovarian-specific Smad4 knockout mice. Mol Endocrinol. 2006;20:1406–1422. doi: 10.1210/me.2005-0462. [DOI] [PubMed] [Google Scholar]
- 47.Pangas SA, Jorgez CJ, Tran M, Agno J, Li X, et al. Intraovarian activins are required for female fertility. Mol Endocrinol. 2007;21:2458–2471. doi: 10.1210/me.2007-0146. [DOI] [PubMed] [Google Scholar]
- 48.Jamin SP, Arango NA, Mishina Y, Hanks MC, Behringer RR. Requirement of Bmpr1a for Mullerian duct regression during male sexual development. Nat Genet. 2002;32:408–410. doi: 10.1038/ng1003. [DOI] [PubMed] [Google Scholar]
- 49.Jorgez CJ, Klysik M, Jamin SP, Behringer RR, Matzuk MM. Granulosa cell-specific inactivation of follistatin causes female fertility defects. Mol Endocrinol. 2004;18:953–967. doi: 10.1210/me.2003-0301. [DOI] [PubMed] [Google Scholar]
- 50.Petit FG, Jamin SP, Kurihara I, Behringer RR, DeMayo FJ, et al. Deletion of the orphan nuclear receptor COUP-TFII in uterus leads to placental deficiency. Proc Natl Acad Sci USA. 2007;104:6293–6298. doi: 10.1073/pnas.0702039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong X, Luense LJ, McGinnis LK, Nothnick WB, Christenson LK. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149:6207–6212. doi: 10.1210/en.2008-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabriel G, Richard RB. Dicer is required for female reproductive tract development and fertility in the mouse. Mol Reprod Dev. 2009;76:678–688. doi: 10.1002/mrd.21010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller DJ. Physiology and endocrinology symposium: sperm-oviduct interactions in livestock and poultry. J Anim Sci. 2011;89:1312–1314. doi: 10.2527/jas.2010-3796. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, Wang D-Z, Pipes GCT, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagaraja AK, Andreu-Vieyra C, Franco HL, Ma L, Chen R, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22:2336–2352. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng Y, Liu X, Yang J, Lin Y, Xu D-Z, et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishida K, Honma Y, Dota A, Kawasaki S, Adachi W, et al. Isolation and chromosomal localization of a cornea-specific human keratin 12 gene and detection of four mutations in Meesmann corneal epithelial dystrophy. Am J Hum Genet. 1997;61:1268–1275. doi: 10.1086/301650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karantza V. Keratins in health and cancer: more than mere epithelial cell markers. Oncogene. 2011;30:127–138. doi: 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 64.King KE, Iyemere VP, Weissberg PL, Shanahan CM. Kruppel-like factor 4 (KLF4/GKLF) is a target of bone morphogenetic proteins and transforming growth factor β1 in the regulation of vascular smooth muscle cell phenotype. J Biol Chem. 2003;278:11661–11669. doi: 10.1074/jbc.M211337200. [DOI] [PubMed] [Google Scholar]
- 65.Kennard S, Liu H, Lilly B. Transforming growth factor-β (TGF-β1) down-regulates Notch3 in fibroblasts to promote smooth muscle gene expression. J Biol Chem. 2008;283:1324–1333. doi: 10.1074/jbc.M706651200. [DOI] [PubMed] [Google Scholar]
- 66.Lovren F, Pan Y, Quan A, Teoh H, Wang G, et al. Angiotensin converting enzyme-2 confers endothelial protection and attenuates atherosclerosis. Am J Physiol Heart Circ Physiol. 2008;295:H1377–1384. doi: 10.1152/ajpheart.00331.2008. [DOI] [PubMed] [Google Scholar]
- 67.Pangas SA, Matzuk MM. The art and artifact of GDF9 activity: cumulus expansion and the cumulus expansion-enabling factor. Biol Reprod. 2005;73:582–585. doi: 10.1095/biolreprod.105.042127. [DOI] [PubMed] [Google Scholar]
- 68.Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–866. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- 69.McNatty KP, Juengel JL, Wilson T, Galloway SM, Davis GH. Genetic mutations influencing ovulation rate in sheep. Reprod Fertil Dev. 2001;13:549–555. doi: 10.1071/rd01078. [DOI] [PubMed] [Google Scholar]
- 70.Shimasaki S, Moore RK, Otsuka F, Erickson GF. The bone morphogenetic protein system in mammalian reproduction. Endocr Rev. 2004;25:72–101. doi: 10.1210/er.2003-0007. [DOI] [PubMed] [Google Scholar]
- 71.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DaCosta Byfield S, Major C, Laping NJ, Roberts AB. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2004;65:744–752. doi: 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- 73.Moore RK, Otsuka F, Shimasaki S. Molecular basis of bone morphogenetic protein-15 signaling in granulosa cells. J Biol Chem. 2003;278:304–310. doi: 10.1074/jbc.M207362200. [DOI] [PubMed] [Google Scholar]
- 74.Gilchrist RB, Lane M, Thompson JG. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update. 2008;14:159–177. doi: 10.1093/humupd/dmm040. [DOI] [PubMed] [Google Scholar]
- 75.Arango NA, Kobayashi A, Wang Y, Jamin SP, Lee HH, et al. A mesenchymal perspective of Mullerian duct differentiation and regression in Amhr2-lacZ mice. Mol Reprod Dev. 2008;75:1154–1162. doi: 10.1002/mrd.20858. [DOI] [PubMed] [Google Scholar]
- 76.Aguilar HN, Mitchell BF. Physiological pathways and molecular mechanisms regulating uterine contractility. Hum Reprod Update. 2010;16:725–744. doi: 10.1093/humupd/dmq016. [DOI] [PubMed] [Google Scholar]
- 77.Tong D, Lu X, Wang HX, Plante I, Lui E, et al. A dominant loss-of-function GJA1 (Cx43) mutant impairs parturition in the mouse. Biol Reprod. 2009;80:1099–1106. doi: 10.1095/biolreprod.108.071969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, et al. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc Natl Acad Sci USA. 2010;107:20828–20833. doi: 10.1073/pnas.1008301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller C, Sassoon DA. Wnt-7a maintains appropriate uterine patterning during the development of the mouse female reproductive tract. Development. 1998;125:3201–3211. doi: 10.1242/dev.125.16.3201. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y, Jia Y, Franken P, Smits R, Ewing PC, et al. Loss of APC function in mesenchymal cells surrounding the Mullerian duct leads to myometrial defects in adult mice. Mol Cell Endocrinol. 2011;341:48–54. doi: 10.1016/j.mce.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 81.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 82.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davis-Dusenbery BN, Hata A. Smad-mediated miRNA processing: A critical role for a conserved RNA sequence. RNA Biol. 2011;8:71–76. doi: 10.4161/rna.8.1.14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DiSandro MJ, Li Y, Baskin LS, Hayward S, Cunha G. Mesenchymal-epithelial interactions in bladder smooth muscle development: epithelial specificity. J Urol. 1998;160:1040–1046. doi: 10.1097/00005392-199809020-00022. [DOI] [PubMed] [Google Scholar]
- 86.Yu L, Mikloucich J, Sangster N, Perez A, McCormick PJ. MyoR is expressed in nonmyogenic cells and can inhibit their differentiation. Exp Cell Res. 2003;289:162–173. doi: 10.1016/s0014-4827(03)00252-0. [DOI] [PubMed] [Google Scholar]
- 87.Lu J, Webb R, Richardson JA, Olson EN. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc Natl Acad Sci USA. 1999;96:552–557. doi: 10.1073/pnas.96.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang F, Hu Y, Xu Q, Ye S. Different effects of angiotensin II and angiotensin-(1–7) on vascular smooth muscle cell proliferation and migration. PLoS ONE. 2010;5:e12323. doi: 10.1371/journal.pone.0012323. doi: 10.1371/journal.pone.0012323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yaghini FA, Song CY, Lavrentyev EN, Ghafoor HU, Fang XR, et al. Angiotensin II-induced vascular smooth muscle cell migration and growth are mediated by cytochrome P450 1B1-dependent superoxide generation. Hypertension. 2010;55:1461–1467. doi: 10.1161/HYPERTENSIONAHA.110.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roy H, Bhardwaj S, Yla-Herttuala S. Biology of vascular endothelial growth factors. FEBS Lett. 2006;580:2879–2887. doi: 10.1016/j.febslet.2006.03.087. [DOI] [PubMed] [Google Scholar]
- 91.Banerjee S, Mehta S, Haque I, Sengupta K, Dhar K, et al. VEGF-A165 induces human aortic smooth muscle cell migration by activating neuropilin-1-VEGFR1-PI3K axis. Biochemistry. 2008;47:3345–3351. doi: 10.1021/bi8000352. [DOI] [PubMed] [Google Scholar]
- 92.Ishida A, Murray J, Saito Y, Kanthou C, Benzakour O, et al. Expression of vascular endothelial growth factor receptors in smooth muscle cells. J Cell Physiol. 2001;188:359–368. doi: 10.1002/jcp.1121. [DOI] [PubMed] [Google Scholar]
- 93.Jeon SH, Chae BC, Kim HA, Seo GY, Seo DW, et al. Mechanisms underlying TGF-beta1-induced expression of VEGF and Flk-1 in mouse macrophages and their implications for angiogenesis. J Leukoc Biol. 2007;81:557–566. doi: 10.1189/jlb.0806517. [DOI] [PubMed] [Google Scholar]
- 94.Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007;8:970–982. doi: 10.1038/nrm2297. [DOI] [PubMed] [Google Scholar]
- 95.Li Q, Rajanahally S, Edson MA, Matzuk MM. Stable expression and characterization of N-terminal tagged recombinant human bone morphogenetic protein 15. Mol Hum Reprod. 2009;15:779–788. doi: 10.1093/molehr/gap062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hobbs S, Jitrapakdee S, Wallace JC. Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1alpha promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem Biophys Res Commun. 1998;252:368–372. doi: 10.1006/bbrc.1998.9646. [DOI] [PubMed] [Google Scholar]
- 97.Vanderhyden BC, Caron PJ, Buccione R, Eppig JJ. Developmental pattern of the secretion of cumulus expansion-enabling factor by mouse oocytes and the role of oocytes in promoting granulosa cell differentiation. Dev Biol. 1990;140:307–317. doi: 10.1016/0012-1606(90)90081-s. [DOI] [PubMed] [Google Scholar]
- 98.Yi SE, Daluiski A, Pederson R, Rosen V, Lyons KM. The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development. 2000;127:621–630. doi: 10.1242/dev.127.3.621. [DOI] [PubMed] [Google Scholar]
- 99.Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, et al. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci USA. 2005;102:5062–5067. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 101.Lee K, Jeong J, Kwak I, Yu CT, Lanske B, et al. Indian hedgehog is a major mediator of progesterone signaling in the mouse uterus. Nat Genet. 2006;38:1204–1209. doi: 10.1038/ng1874. [DOI] [PubMed] [Google Scholar]
- 102.Park SO, Lee YJ, Seki T, Hong K-H, Fliess N, et al. ALK5- and TGFBR2-independent role of ALK1 in the pathogenesis of hereditary hemorrhagic telangiectasia type 2. Blood. 2008;111:633–642. doi: 10.1182/blood-2007-08-107359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tgfbr1 cKO mice have defective smooth muscle formation in the oviduct. (A to C) Calponin 1 is expressed in the smooth muscle layers of the control oviduct (A and B). Negative control in which the primary antibody was substituted with rabbit IgG is shown in (C). (D to F) Defective smooth muscle formation in the oviducts of Tgfbr1 cKO mice. Higher magnification view of the selected regions within the blue and red rectangles in (D) were depicted in (E) and (F), respectively. Arrowheads demonstrate smooth muscle defects. SM, smooth muscle; EP, epithelium. Scale bars = 50 µm (E and F); 100 µm (B and C); and 200 µm (A and D).
(TIF)
Follicular development in Tgfbr1 cKO mice. (A and B) Ovaries of 3-week-old control and Tgfbr1 cKO mice containing follicles at various developmental stages. (C and D) PMSG-induced follicular development in control (C) and Tgfbr1 cKO (D) mice. Arrows indicate preovulatory follicles. Scale bars = 200 µm.
(TIF)
Corpora lutea formation in Tgfbr1 cKO mice. (A and B) PAS staining of ovaries from PMSG-hCG treated control (A) and Tgfbr1 cKO (B) mice. Corpora lutea formed in both control and Tgfbr1 cKO mice after 46 h of PMSG and 20 h of hCG treatment. (C to E) Immunohistochemical staining of 3β-HSD in the corpora lutea of control (C) and Tgfbr1 cKO (D) mice. A representative negative control of the Tgfbr1 cKO mouse ovary was shown in (E). Scale bars = 200 µm. CL, corpus luteum. (F) Serum progesterone levels in gonadotropin-primed and natural pregnant mice (n = 3–4). Data are presented as mean ± SEM.
(TIF)
Tgfbr1 cKO mice have defects in uterine smooth muscle formation. (A and B) Gross uterine morphology of Tgfbr1 cKO mice at 3 weeks and 3 months of age. (C to H) Immunostaining of ACTA2 in the uteri of control (C and F) and Tgfbr1 cKO (D, E, G, and H) mice. Note the disrupted smooth muscle layers in the Tgfbr1 cKO mice, and the unusual appearance of the smooth muscle structure in the endometrium. Ut, uterus; LM, longitudinal muscle layer; CM, circular muscle layer. Red arrowheads point to the disorganized smooth muscle layers. Scale bars = 50 µm (F to H); 200 µm (C to E); and 5 mm (A and B).
(TIF)
Smooth muscle defects lead to severe disruption of uterine structures in Tgfbr1 cKO mice. (A) Uterine abnormalities in an 8-month-old Tgfbr1 cKO female compared to an age-matched control mouse. Blue arrows indicate the uteri. (B to F) Immunostaining of CNN1 in the uteri of 8-month-old control (B) and Tgfbr1 cKO mice (C to F). CNN1 staining demonstrated multiple structural abnormalities in the uterine smooth muscle layers of the Tgfbr1 cKO mice. Defective smooth muscle development leads to uterine cyst formation (arrow heads; C, D, and E), and complete loss of normal uterine structure in the severe cases (D and F). Scale bars = 200 µm (B to F).
(TIF)
The uteri of Tgfbr1 cKO mice can undergo artificial decidualization. (A and B) Gross morphology of the uteri in the control (A) and Tgfbr1 cKO (B) mice 5 days after the decidual stimulus. The left horns (control horns) of the uteri were unstimulated while the right ones (decidual horns) were traumatized. Note that the uteri of both control and Tgfbr1 cKO mice can undergo artificially induced decidualization. (C) Weight ratio of decidual horn to control horn in control (n = 3) and Tgfbr1 cKO mice (n = 4). Data are presented as mean ± SEM. (D–G) Immunostaining of control and decidual horns of Tgfbr1 cKO mice using CCN1. Higher magnification views of (D) and (F) are depicted in (E) and (G), respectively. Note the disruption of smooth muscle structure in both unstimulated and decidual horns of the Tgfbr1 cKO mice. Scale bars = 200 µm (E and G); 400 µm (D and F), and 10 mm (A).
(TIF)
Expression of target genes of miR-143/145 in Tgfbr1 cKO mice. Real-time PCR analyses using 3–4 week old oviducts demonstrated that Elk1, Klf4, and Camk2d are expressed at comparable levels between Tgfbr1 cKO mice and controls. n = 3–4 independent pools of oviducts. Relative mRNA levels were normalized to Gapdh. Data are presented as mean ± SEM.
(TIF)
Messenger RNA levels of genes up-regulated in Dicer1 cKO oviducts are not elevated in the oviducts of Tgfbr1 cKO mice. Wnt5a, Wnt7a, Hoxa9, and Hoxa10 are significantly up-regulated genes in the 3–4 week old oviducts of Dicer1 cKO mice [55]. Real-time PCR analyses using age-matched Tgfbr1 cKO and control oviducts did not demonstrate up-regulation of these genes in the Tgfbr1 cKO mice. n = 3–4 independent pools of oviducts. Relative mRNA levels were normalized to Gapdh. Data are presented as mean ± SEM.
(TIF)