Abstract
The planktonic foraminiferal morpho-species Globoconella inflata is widely used as a stratigraphic and paleoceanographic index. While G. inflata was until now regarded as a single species, we show that it rather constitutes a complex of two pseudo-cryptic species. Our study is based on SSU and ITS rDNA sequence analyses and genotyping of 497 individuals collected at 49 oceanic stations covering the worldwide range of the morpho-species. Phylogenetic analyses unveil the presence of two divergent genotypes. Type I inhabits transitional and subtropical waters of both hemispheres, while Type II is restricted to the Antarctic subpolar waters. The two genetic species exhibit a strictly allopatric distribution on each side of the Antarctic Subpolar Front. On the other hand, sediment data show that G. inflata was restricted to transitional and subtropical environments since the early Pliocene, and expanded its geographic range to southern subpolar waters ∼700 kyrs ago, during marine isotopic stage 17. This datum may correspond to a peripatric speciation event that led to the partition of an ancestral genotype into two distinct evolutionary units. Biometric measurements performed on individual G. inflata from plankton tows north and south of the Antarctic Subpolar Front indicate that Types I and II display slight but significant differences in shell morphology. These morphological differences may allow recognition of the G. inflata pseudo-cryptic species back into the fossil record, which in turn may contribute to monitor past movements of the Antarctic Subpolar Front during the middle and late Pleistocene.
Introduction
Planktonic foraminifera are pelagic protists whose calcareous shells have built up one of the most complete and continuous fossil archive of biodiversity changes over the last 180 Myrs. The biogeography of planktonic foraminiferal morpho-species appears to correlate to hydrographic conditions of latitudinal oceanic provinces [1], [2]. Paleoceanographers derive reconstructions of past climates based on empirical relationships between extant environmental parameters of the surface oceans (e.g., temperature, primary production) and the abundance or chemical composition of shells of individual morpho-species from surface sediment samples [3], [4]. These correlations lie on the working assumption that each species has its own, stable habitat preferences that are transferable back into the past to reconstruct changes of water masses physical properties. The accurate recognition of individual species is therefore mandatory for the use of planktonic foraminifera as paleoceanographic proxies.
Extant species of planktonic foraminifera have been defined based on diagnostic characters of their shell (the so-called morpho-species concept), primarily described from fossil specimens extracted from sediments [5], [6]. Yet, molecular analyses applied to living specimens have challenged this concept by demonstrating that this classical taxonomy underestimates planktonic foraminiferal diversity. Each morpho-species analyzed so far actually comprises up to seven distinct genotypes [7], most of which exhibiting a distinct biogeography and/or ecology [8]–[10], and even sometimes subtle but statistically significant differences in shell morphology [11]–[14]. Although strictly identical ribosomal DNA (rDNA) genotypes range across huge geographic distances, different genotypes within a single morpho-species generally display peculiar distributions related to water masses properties [8], [10], [13], [14]. Together with molecular clock analyses calibrated on the fossil record [9]–[11], these results strongly support the hypothesis that distinct genotypes within the classical morpho-species actually correspond to cryptic, or rather pseudo-cryptic biological species, i. e., sibling species that can be differentiated based on subtle morphological features [15]. In this study, we investigate the rDNA genetic diversity and biogeography within the morpho-species Globoconella inflata d'Orbigny 1839.
Globoconella inflata is a macro-perforate, non-spinose morpho-species of planktonic foraminifera (Globorotalioidea), which originated 4.14 Ma ago in transitional waters of the Southwest Pacific [16], and which spread 2.09 Ma ago into the world oceans [17]. Today, this thermocline-dweller species [18], [19] is particularly abundant in transitional and subtropical waters of both hemispheres [1].
Our study focuses on the genetic variations in Small SubUnit (SSU) and Internal Transcribed Spacer (ITS) rDNA sequences of specimens collected worldwide from plankton tows (Figure 1). The SSU rDNA sequences have been used in most studies dealing with foraminiferal genetic diversity [7], [20], [21]. The ITS rDNA sequences evolve significantly faster than SSU rDNA and can be used to discriminate recent events of pseudo-cryptic speciation [10]. In this study, we document the occurrence of two distinct genotypes of Globoconella inflata, supported by both SSU and ITS analyses. These genotypes display specific, strictly allopatric biogeographic distributions on a global scale, as well as statistically significant differences in shell morphology. Ultimately, morpho-genetic delineation of both pseudo-cryptic species of G. inflata may be used to improve paleoceoanographic reconstructions, especially the glacial-interglacial latitudinal swings of the Antarctic Subpolar Front since the mid-Pleistocene transition.
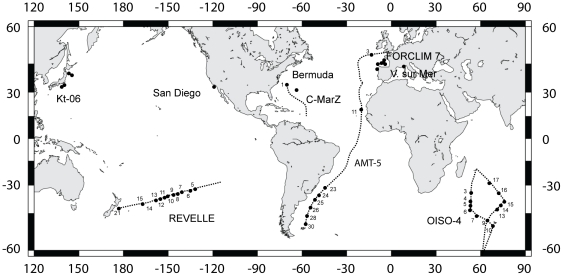
Figure 1. Sample map.
Geographic location and labels of the oceanic stations sampled during the cruises FORCLIM 7, AMT-5, C-MarZ, OISO-4, REVELLE, KT-06-11 and offshore Villefranche-sur-mer, San Diego and Bermuda. Dashed lines represent ship routes and black circles are collecting stations from where Globoconella inflata specimens have been genetically analyzed.
Results
Genetic Variation in Globoconella inflata
SSU rDNA sequences of 21 Globoconella inflata individuals randomly selected from contrasted water masses (Table 1, Figure 1) reveal small but robust molecular differences between specimens collected north and south of the Antarctic Subpolar Front (Table 2, Figure S1). This pattern of genetic diversity is confirmed based on the analysis of the complete ITS array (ITS-1, 5.8S and ITS-2) of 80 specimens from 41 stations that cover the entire environmental range of the morpho-species. In order to assess the intra-individual level of genetic variations, clones from single cells were also characterized in both SSU (33) and ITS (55) rDNA (Table 3). All together, our dataset includes 50 SSU rDNA sequences and 135 ITS rDNA sequences from 83 individuals. The remaining 414 individuals were genetically characterized through Restriction Fragment Length Polymorphism analysis (RFLP).
Table 1. Location of the sampling stations, with indications of the number of sequenced and genotyped specimens of Globoconella inflata (in parenthesis, number of copies from the same individual).
| Cruise | Station | Latitude | Longitude | Environmental data | Number of sequenced specimens (SSU) | Number of sequenced specimens (ITS) | Number of RFLP identifications | Genotype |
| C-MarZ | 1 | 33°54′N | 69°93′W | CTD (T) | 2 | 5 (3; 16) | 79 | I |
| OISO-4 | 3 | 35°00′S | 53°30′E | CTD (T, F) | 2 | 1 (17) | 3 | I |
| OISO-4 | 4 | 40°01′S | 52°53′E | CTD (T, F) | 2 (13) | 3 | 8 | I |
| OISO-4 | 5 | 42°31′S | 52°29′E | CTD (T, F) | 1 | 7 | I | |
| OISO-4 | 6 | 45°00′S | 52°05′E | CTD (T, F) | 4 | 5 (19) | 0 | II |
| OISO-4 | 7 | 47°40′S | 58°00E | CTD (T, F) | 8 | 8 | II | |
| OISO-4 | 9 | 48°31′S | 64°59′E | CTD (T, F) | 3 (2) | 2 | 0 | II |
| OISO-4 | 10 | 50°40′S | 68°24′E | CTD (T, F) | 0 | 2 | II | |
| OISO-4 | 13 | 44°58′S | 73°21′E | CTD (T, F) | 2 | 8 | I | |
| OISO-4 | 14 | 42°30′S | 74°53′E | CTD (T, F) | 2 | 18 | I | |
| OISO-4 | 15 | 40°00′S | 76°24′E | CTD (T, F) | 3 | 13 | I | |
| OISO-4 | 16 | 34°59′E | 73°28′E | CTD (T, F) | 0 | 3 | I | |
| OISO-4 | 17 | 29°59′S | 66°24′E | CTD (T, F) | 1 | 1 | I | |
| FORCLIM 7 | I | 42°37′N | 10°02′W | CTD (T, F) | 0 | 15 | I | |
| FORCLIM 7 | II | 44°20′N | 8°45′W | CTD (T, F) | 1 | 9 | I | |
| FORCLIM 7 | III | 45°35′N | 7°33′W | CTD (T, F) | 1 (2) | 1 | 9 | I |
| FORCLIM 7 | V | 45°37′N | 7°33′W | CTD (T, F) | 0 | 3 | I | |
| FORCLIM 7 | VIII | 45°38′N | 7°36′W | CTD (T, F) | 1 | 4 | I | |
| FORCLIM 7 | XV | 45°06′N | 5°38′W | CTD (T, F) | 3 | 17 | I | |
| FORCLIM 7 | XXII | 45°56′N | 6°15′W | CTD (T, F) | 0 | 50 | I | |
| FORCLIM 7 | XXVII | 46°36′N | 5°49′W | CTD (T, F) | 0 | 17 | I | |
| KT-06 | C | 39°00′N | 145°00′E | CTD (T,F) | 1 | 2 | 32 | I |
| KT-06 | E | 34°00′N | 140°00′E | CTD (T,F) | 1 | 1 | 1 | I |
| KT-06 | F | 33°00N | 139°00E | CTD (T,F) | 3 | 9 | I | |
| KT-06 | G | 33°00N | 139°00E | CTD (T,F) | 0 | 1 | I | |
| REVELLE | 5 | 31°35′S | 127°83′W | SST | 1 | 3 | I | |
| REVELLE | 6 | 32°04′S | 130°98′W | SST | 1 | 2 | I | |
| REVELLE | 7 | 33°39′S | 137°12′W | CTD (T,F) | 0 | 14 | I | |
| REVELLE | 8 | 34°03′S | 140°05′W | SST | 1 | 6 | I | |
| REVELLE | 9 | 34°73′S | 143°27′W | CTD (T,F) | 1 | 1 | 2 | I |
| REVELLE | 10 | 35°39′S | 146°29′W | SST | 1 | 1 (3) | 1 | I |
| REVELLE | 11 | 36°07′S | 149°48′W | CTD (T,F) | 1 | 8 | I | |
| REVELLE | 12 | 36°73′S | 152°58′W | SST | 1 | 5 | I | |
| REVELLE | 13 | 37°45′S | 156°00′W | CTD (T,F) | 1 | 6 | I | |
| REVELLE | 15 | 39°62′S | 166°69′W | CTD (T,F) | 1 | 5 | I | |
| REVELLE | 21 | 42°95′S | 176°26′E | SST | 1 | 45 | I | |
| Villefranche-sur-Mer | N/A | 43°40′N | 07°15′E | N/A | 1 (2) | 0 | I | |
| San Diego | N/A | 33°16′N | 118°08′E | N/A | 1 (2) | 0 | I | |
| Bermuda | 1 | 32°08′N | 64°33′W | N/A | 1 | 0 | I | |
| Bermuda | 2 | 32°20′N | 64°33′W | N/A | 1 | 0 | I | |
| Bermuda | 3 | 32°20′N | 64°33′W | N/A | 1 | 0 | I | |
| AMT-5 | 3 | 47°98′N | 13°20′W | CTD (T,F) | 2 | 0 | I | |
| AMT-5 | 11 | 19°71′N | 20°50′W | CTD (T,F) | 1 | 0 | I | |
| AMT-5 | 23 | 31°37′S | 44°52′W | CTD (T,F) | 3 | 0 | I | |
| AMT-5 | 24 | 35°29′S | 48°52′W | CTD (T,F) | 2 | 0 | I | |
| AMT-5 | 25 | 38°50′S | 51°55′W | CTD (T,F) | 1 | 3 | 0 | I |
| AMT-5 | 26 | 42°14′S | 54°27′W | CTD (T,F) | 3(2) | 0 | I | |
| AMT-5 | 28 | 46°03′S | 56°42′W | CTD (T,F) | 2 (8; 9) | 4 | 0 | II |
| AMT-5 | 30 | 49°79′S | 57°62′W | CTD (T,F) | 1 | 0 | II | |
| Total | 21 (50) | 80 (135) | 414 |
Table 2. Inter-individual patristic distances (substitutions per site) measured on phylogenetic trees obtained from the five datasets analyzed (see Methods for details about the co nstruction of the datasets).
| SSU | CleITS | CloITS | LarITS | ComITS | |||||||
| Min | Max | Min | Max | Min | Max | Min | Max | Min | Max | ||
| Type I | Median | 4.77E-03 | 3.84E-02 | 1.01E-02 | 1.56E-02 | 1.37E-02 | |||||
| 95% CI | 9.25E-08 | 1.23E-02 | 1.41E-02 | 8.21E-02 | 3.30E-03 | 3.76E-02 | 2.60E-03 | 3.52E-02 | 2.00E-03 | 3.18E-02 | |
| Min-Max | 2.00E-10 | 1.44E-02 | 1.46E-07 | 1.02E-01 | 2.00E-10 | 5.55E-02 | 2.00E-10 | 5.68E-02 | 2.00E-10 | 4.32E-02 | |
| Type II | Median | 7.67E-03 | 5.24E-02 | 1.17E-02 | 1.83E-02 | 1.37E-02 | |||||
| 95% CI | 2.51E-07 | 1.42E-02 | 1.90E-02 | 9.06E-02 | 8.40E-07 | 2.82E-02 | 2.60E-03 | 4.33E-02 | 8.10E-07 | 3.21E-02 | |
| Min-Max | 7.25E-08 | 1.65E-02 | 9.60E-03 | 9.59E-02 | 2.00E-10 | 3.54E-02 | 2.00E-10 | 5.91E-02 | 2.00E-10 | 4.44E-02 | |
| Type I vs. Type II | Median | 1.38E-01 | 4.37E-02 | 6.70E-02 | 4.83E-02 | ||||||
| 95% CI | 1.34E-02 | 3.13E-02 | 1.03E-01 | 1.80E-01 | 3.10E-02 | 5.72E-02 | 4.88E-02 | 8.86E-02 | 3.54E-02 | 6.75E-02 | |
| Min-Max | 1.03E-02 | 3.47E-02 | 8.26E-02 | 2.11E-01 | 5.00E-03 | 6.90E-02 | 3.82E-02 | 1.13E-01 | 2.87E-02 | 8.54E-02 | |
The median, 95% non-parametric confidence interval, and minimum and maximum patristic distance values are given within and among genotypes.
Table 3. Intra-individual patristic distances (substitutions per site) measured on phylogenetic trees obtained from four of the five analyzed datasets (no available distances for the CleITS dataset; see Methods for details about the construction of the datasets).
| SSU | CloITS | LarITS | ComITS | ||||||
| Min | Max | Min | Max | Min | Max | Min | Max | ||
| Oi 375 | Median | 6.39E-03 | |||||||
| 95% CI | 2.00E-10 | 1.27E-02 | |||||||
| Min-Max | 2.00E-10 | 1.43E-02 | |||||||
| AM605 | Median | 6.51E-03 | |||||||
| 95% CI | 7.25E-08 | 1.12E-02 | |||||||
| Min-Max | 7.80E-08 | 1.12E-02 | |||||||
| AM609 | Median | 9.07E-03 | |||||||
| 95% CI | 8.25E-08 | 1.33E-02 | |||||||
| Min-Max | 8.24E-08 | 1.34E-02 | |||||||
| CM115 | Median | 6.67E-03 | 1.03E-02 | 7.79E-03 | |||||
| 95% CI | 4.15E-07 | 1.50E-02 | 3.98E-10 | 2.34E-02 | 2.16E-07 | 1.56E-02 | |||
| Min-Max | 2.00E-10 | 1.50E-02 | 2.00E-10 | 2.60E-02 | 2.00E-10 | 1.77E-02 | |||
| Oi265 | Median | 1.16E-02 | 1.30E-02 | 1.17E-02 | |||||
| 95% CI | 1.04E-07 | 1.95E-02 | 1.41E-07 | 2.11E-02 | 3.33E-07 | 1.78E-02 | |||
| Min-Max | 2.00E-10 | 2.18E-02 | 2.00E-10 | 2.12E-02 | 7.06E-08 | 1.79E-02 | |||
| Oi689 | Median | 5.01E-03 | 7.74E-03 | 5.83E-03 | |||||
| 95% CI | 1.99E-07 | 1.03E-02 | 1.92E-07 | 1.82E-02 | 2.05E-07 | 1.37E-02 | |||
| Min-Max | 2.00E-10 | 1.17E-02 | 2.00E-10 | 2.08E-02 | 2.00E-10 | 1.56E-02 | |||
| Re1010 | Median | 4.94E-03 | |||||||
| 95% CI | 1.09E-03 | 5.79E-03 | |||||||
| Min-Max | 8.90E-04 | 5.83E-03 | |||||||
The median, 95% non-parametric confidence interval, and minimum and maximum patristic distance values are given within 7 cloned individuals. Specimen Re-1010 displays a Type I-like sequence with a Type II RFLP pattern.
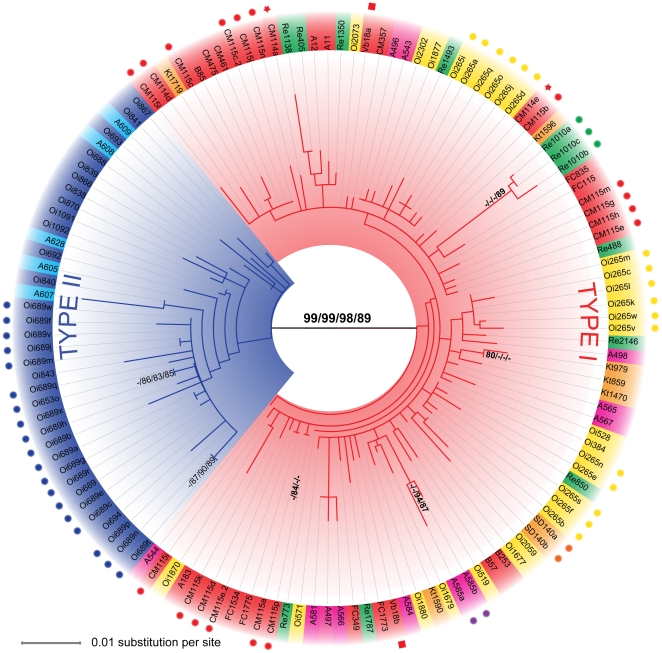
SSU and ITS rDNA sequences of Globoconella inflata cluster into two clearly distinct groups called here Type I and Type II (Figure 2 and Figure S1). In datasets analyzed (1 SSU and 4 ITS; see Material and Methods), patristic distances between the two genotypes are markedly higher than those measured within each genotype (Table 2). Moreover, genetic variations within individuals are equal or greater than differentiations amongst populations from the three oceanic basins, making these sequences uninformative at the population level (Table 3). The node separating Types I and II of G. inflata is supported by bootstrap values ≥98% (except for the comITS dataset, 89%), whereas no branch support higher than 80% is observed within phylotypes, with only few exceptions for small clusters of terminal branches (Figure 2).
Figure 2. Phylogenetic analysis of Globoconella inflata.
ITS-based evolutionary relationships between 135 clones of G. inflata from 41 localities in the world oceans (see Table 1 and Figure 1 for station names and locations). This Maximum Likelihood inference shows the relationships between the two phylotypes (Type I in red and Type II in blue). The bootstrap scores (500 replicates) greater than 80% are given next to branches for each dataset following a CleITS/CloITS/LarITS/ComITS-dataset order. The scale and branch lengths are given in % of nucleotide substitution per site. The colors associated to leaf labels indicate geographic area of collection: blue = subpolar Indian Ocean; light blue = subpolar South Atlantic; Pink = South Atlantic north of the Subpolar Front; Yellow = Indian Ocean north of the Subpolar Front; red = North Atlantic; green = South Pacific; Orange = North Pacific. Circles, stars and squares associated to specific colors indicate clones sequenced from the same individuals.
A unique specimen (Re-1010) collected in the subtropical south Pacific during the cruise REVELLE (Figure 1, 3), displays an atypical Type I ITS rDNA sequence but a Type II RFLP pattern. Based on the CleITS alignment (652 unambiguously aligned sites), sequences obtained from this specimen (3 clones were characterized from two distinct PCR, showing observed pairwise distances between 0.006676 and 0.002387 substitution per site) share 13 synapomorphous sites with Type I, 4 with Type II (all found in a 28 bp-long region), and possess 5 autapomorphous sites when compared to the 60% consensus sequences of Types I and II as inferred by SEAVIEW [22]. Even if rather ambiguous in the studied dataset, Indel locations do seem to roughly follow the same pattern. Despite its RFLP pattern, this individual clearly clusters within Type I in ITS phylogeny, although being slightly divergent and decreasing the bootstrap support between the two genotypes (Figure 2). Moreover, its SSU rDNA sequence clearly belongs to Type I. Since we found no clear evidence of hybridization but could not reject it based on the available data, we currently favor the hypothesis that this isolated specimen coming from a well-sampled oceanic area (Table 1) could be the representative of a distinct Type I-population, which remains underrepresented in our dataset.
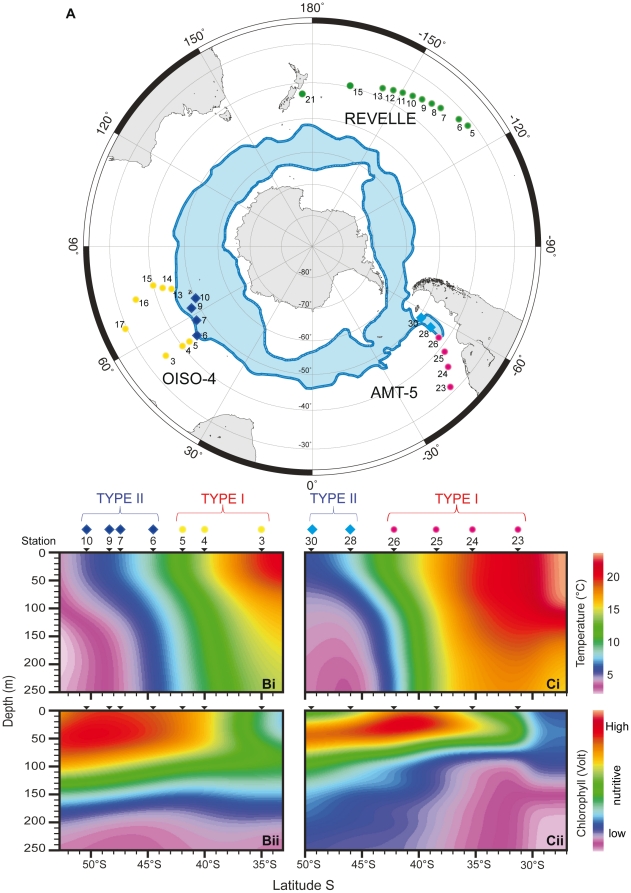
Figure 3. Geographical and ecological distribution of genotypes.
Latitudinal distribution of Types I and II of Globoconella inflata in the South Hemisphere (the North Hemisphere contains representatives of Type I only). (A) Polar projection of the cruises AMT-5 (September-October 1997), OISO-4 (January–February 2000) and REVELLE (January–february 2004); the position of the Antarctic Circumpolar Current is shown in blue [61]. Temperature and fluorescence profiles (0–250 m) are given for the cruise OISO-4 (Bi and Bii) and AMT-5 (Ci and Cii); occurrences of genotypes (circles for Type I; diamonds for Type II) are given for each station, positioned with latitudes. Colors as in Figure 1.
Geographic Distribution of Globoconella inflata Genotypes
Our molecular dataset, including both sequence and RFLP analyses, originates from 49 sampled stations that cover the entire biogeographic/environmental spectrum known for Globoconella inflata, except for northern high-latitudes (see next section). Type I and Type II exhibit a strictly allopatric distribution. Type I is found within the subtropical and transitional water masses of the world oceans, whereas Type II is restricted to the cold, vertically mixed and nutrient-rich water masses of the Subpolar Southern Ocean (Figure 3A). This distribution pattern is apparently not primarily controlled by water productivity. Indeed, surface waters with the highest fluorescence values matching maximum chlorophyll concentrations yield different genotypes in the South Atlantic (Type I in stations 25 and 26 of AMT-5; Figure 3C) and South Indian Ocean (Type II in stations 7, 9 and 10 of OISO-4; Figure 3B). However, in both Atlantic and Indian Ocean basins, the biogeographic boundary between the two genotypes corresponds to the location of the North Subpolar Front, where Sea Surface Temperatures (SSTs) range between 8°C and 12°C (Figure 3B, C).
Bipolar Distribution of Globoconella inflata
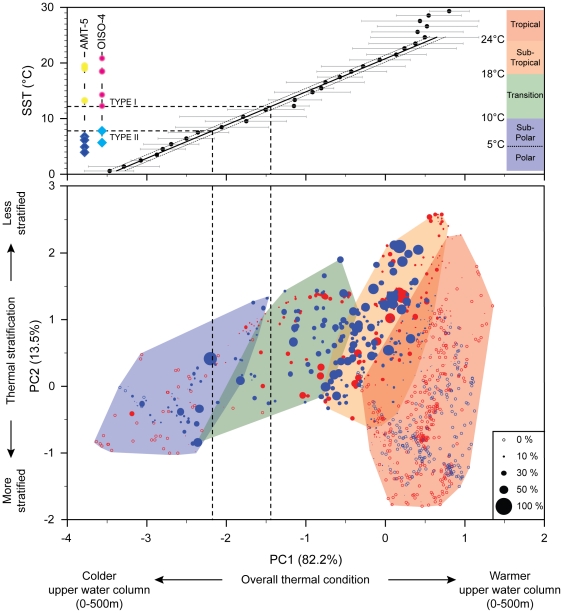
Given the lack of plankton tow samples from northern high latitudes in our dataset, we cannot check whether the subpolar waters of the Northern Hemisphere host the Type II of Globoconella inflata, as it would be expected in a bipolar biogeographic pattern [23], [24], or, alternatively, another genotype. In order to circumvent this sampling limitation, we performed a biogeo-hydrographic analysis of the overall distribution of G. inflata in surface sediments samples from the Brown University Foraminiferal Database [25]. The BFD records the absolute abundances of 37 extant morpho-species of planktonic foraminifera at 1265 core-top sites of the global oceans. For each BFD site, thirteen mean annual temperatures measured between sea surface and 500 m depth, i.e., the potential depth-habitat range of G. inflata, were extracted from the World Ocean Atlas [26]. A principal component analysis was performed on these 13 temperatures values×1265 localities allowing the direct comparison between the synthetic descriptors of the thermal structure of the water column (the resulting principal components) and the abundance of G. inflata (Figure 4). The resulting PC1 is a mean thermal state of the upper 500 m of the water column; PC2 contrasts the mean annual temperatures of the first 125 m (negative weight) with the temperatures recorded between 150 and 500 m (positive weight; Figure S2). Excepted a very few (11) core-top samples located in the vicinity of the Gulf Stream [27] where G. inflata is sparsely recorded, the morpho-species is absent in northern subpolar waters at thermal conditions where Type II occurs in southern high latitudes. The latitudes at which G. inflata disappears in the Northern Hemisphere exhibit mean SST values identical to those measured for the biogeographic boundary between the two genotypes in the Southern Hemisphere (Figure 4). Although we acknowledge that further samples from the Northern Hemisphere (Figure 1) would be helpful to definitely exclude the presence of another genotype in this area, our observations clearly support an absence of a specific or shared arctic subpolar genotype.
Figure 4. Morpho-species vs. genotype distributions.
The bubble biplot allows the direct comparison of the relative abundance of Globoconella inflata (proportional to filled circle size; empty circle: absent) in surface sediments of 1265 localities from the North (in red) and South (in blue) Hemisphere with the thermal structure of the 500 upper meters of the water column. A least square regression between the mean values of PC1 for each 1 SST-°C interval and SST (Sea Surface Temperature = 10 meters temperature following [3]; standard deviation indicated by a gray line) shows the good relationship between these two descriptors in the 0–25°C-SST interval and illustrates the clear thermal boundary (between 8 and 12°C based on the studied samples) between the two genotypes for the cruises AMT-5 and OISO-4 (colors and symbols as in Figure 2).
Biometry
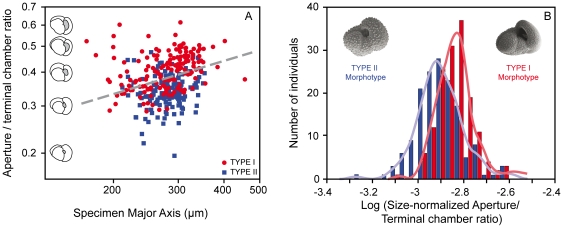
A biometric analysis was performed on 306 non-genetically characterized Globoconella inflata specimens from plankton tows across the Antarctic subpolar frontal system (AMT-5 cruise). Rather than direct morpho-genetic comparisons, we favored this approach because of the highly unbalanced molecular sampling between Types I and II. Based on the procedure by [28] and the available genetic dataset, the probability to get a mix of Type I and Type II specimens in a given locality north or south of the Subpolar Front is <0.035% at the 95% confidence level. This clearly indicates that non-genotyped individuals currently living, and thus collected north (n = 154) and south (n = 152) of the front are very likely to be representatives of Type I and Type II, respectively. Measurements of overall shell size and apertural relative size descriptors reveal a weak, but highly significant difference among the two sets of individuals (genotypes) sampled on each side of the front (Figure 5). A discriminant analysis involving both populations indicates a highly significant differentiation among genotypes (Wilk's λ = 0.858; F = 25.06; df = 2, 303; p = 8.48×10−11), which is also non-parametrically evidenced based on the log-ratio between the aperture/terminal chamber ratio (a size-normalized apertural length) and the specimen major axis (Figure 5B; equal-median Mann-Whitney test: U = 6607, p = 4.5×10−11; same-distribution Kolmogorov-Smirnov test: D = 0.358, p = 3.4×10−9). At an identical overall shell size, specimens collected north of the front display a significantly higher size-normalized apertural length than those collected south of the front. This morphological distinction of two varieties of G. inflata agrees with surface sediment data by [29], which differentiated, in the SW Pacific, a low to mid latitudes morphotype and a subpolar morphotype, this later being characterized by a smaller umbilico-ventral aperture. Even if our simple morphometric descriptor does not yet allow a powerful discrimination between the two genotypes (the resulting discriminant function allows the correct genotype classification of only 66% of the analyzed specimens), our results suggest that further, more sophisticated morphometric analyses will probably make possible to morphologically characterize the two genotypes in fossil assemblages.
Figure 5. Morphological differences between Globoconella inflata genotypes.
A. Log-Log Biplot of specimen major axis vs. aperture/terminal chamber length ratio for 306 specimens collected during the cruise AMT-5 in the South Atlantic. All specimens collected north of the Subpolar Front are considered to be representatives of Type I; all others are considered as Type II. The discriminant boundary that maximizes the separation between the two genotypes is represented by a gray dashed line. B. Histograms and Gaussian kernel densities of the log-ratio between the aperture/terminal chamber length ratio and the specimen major axis.
Discussion
Ribosomal DNA for Identifying Genetic Variability in Planktonic Foraminifera
Ribosomal DNA provides useful markers for phylogenetic systematics in protist taxa, especially at the species level [30]–[35]. However, ITS rDNA-based phylogenetic reconstructions have to be interpreted cautiously because intragenomic variations can lead to overestimates of the diversity or to the incorrect finding of cryptic species, as shown for example for the dinoflagellate cluster Symbiodinium spp. [36]. In the case of the planktonic foraminifera Globoconella inflata, the branch separating the two phylogroups we evidence is highly supported by both SSU and ITS analyses. In addition, no clone from individual assigned to Type II clustered within the Type I, and inversely. These two lines of evidence strongly suggest that the two phylogroups evolved as separated evolutionary units that split off during the past.
The biology of planktonic foraminifera remains poorly understood, especially because none was able yet to obtain complete life cycles of these organisms in laboratory cultures. It is therefore not possible to conduct mating experiments to test those genotypes of G. inflata for the Mayr's biological species concept (groups of interbreeding natural populations that are reproductively isolated from other such groups). Consequently, the appropriate way to prove the reliability of the rDNA-defined phylogroups as different biological species is to combine evidences from different life history traits that directly or indirectly relate to the interbreeding criterion [37]. The highly supported genetic characterization, the complete geographical and ecological disruption, and the morphological differences evidenced among genotypes of G. inflata (Figure 3, 4, 5) argue together for a biological species-level differentiation among Types I and II.
Abundant sediment data make G. inflata a well known planktonic foraminiferal morpho-species in the fossil record, both in terms of origination and paleogeographic distribution [38], [16]. It initially appeared during the early Pliocene (4.14 Ma; [16]) in the transitional Southwest Pacific before definitely invading the transitional and subtropical waters of both hemispheres 2.09 myrs ago [17]. Restricted to these environments during the first million of years of its evolution, G. inflata expanded its geographic range to the southern subpolar waters at ∼700 ka [39], [40]. Previous studies interpreted this event as an intra-specific migration associated with an adaptation to cold waters of the high latitudes. Such interpretation implied that the morpho-species exhibits a wider temperature tolerance in the southern than in the northern Hemisphere, where its distribution does not range as far north as the transitional waters. Alternatively, our data strongly suggest that this invading datum may correspond to a peripatric [41] speciation in which an ancestral genotype evolved into two evolutionary significant units.
Cryptic Diversity as a Tool for Monitoring Past Migrations of the Antarctic Subpolar Front
In planktonic foraminifera, co-occurrences of distinct genotypes of the same morpho-species may induce significant bias in paleoceanographic reconstructions [3], [8], [10], [13], [28], [42]–[46]. Consequently to its evolutionary history, Globoconella inflata falls in a unique case because no co-occurrence of cryptic species has been observed to date, in an almost global dataset. As a consequence, the cryptic diversity in G. inflata does not affect paleoceanographic reconstructions as soon as they rely on calibrations that have been completed based on modern specimens collected within the biogeographic range of a single genotype. For example, most stable isotope studies focusing on the ecology of current G. inflata are based on the analysis of specimens that have been collected within the geographic range of Type I [19], [47]–[51]. These studies are consequently not affected by the existence of two genotypes with different ecologies. Stable isotope analyses (and their paleoceanographic inferences) based on current specimens collected through the Antarctic Subpolar Front [52]–[53], should however be interpreted cautiously, because the observed isotopic signals may be biased by the way each of the two genotypes fractionate oxygen and/or carbon isotopes. Furthermore, the transfer functions used by paleoceanographers to predict past sea surface temperatures are calibrated based on abundances of current planktonic foraminiferal species in surface sediments. Most of these calibrations being basin-wide based, they mix data originating from the ranges of Type I and Type II of G. inflata in the South Hemisphere, then potentially affecting the resolving power of the transfer functions [13].
On the other hand, cryptic diversity in Globoconella inflata may be a powerful tool to monitor past migrations of the Antarctic Subpolar Front during the glacial and interglacial stages of the middle and late Pleistocene. Though biometric studies of G. inflata specimens from surface sediments are needed to test our hypothesis, several arguments argue for a direct recognition of Type I and Type II back into the middle and late Pleistocene. First, only two extant cryptic species have been described to date, making feasible to transfer the genetic information to the interpretation of the fossil record. Second, since the Type II of G. inflata has permanently inhabited the subantarctic waters since ∼700 ka, Types I and II have probably maintained their structural sensitivity to temperature conditions and fluctuations. Third, our rough preliminary biometrical analysis clearly suggests that the genotype of shell samples could be statistically inferred, e.g., based on a geometric morphometry analysis of the apertural/final chamber relationship. Based on the two first points, application of this morphometric approach should work equally in the fossil record, making the identification of cryptic species in G. inflata a useful paleontological proxy. Such identification should therefore be considered as a promising tool to track the past glacial/interglacial oscillations of the Antarctic Subpolar Front over the last ∼700 ka, a critical parameter for global paleoceanographic and paleoclimatic reconstructions [54], [55].
Materials and Methods
Sample Collection
Living Globoconella inflata were collected with plankton tows (64 µm or 100 µm mesh sizes) from the world oceans (Figure 1): (i) in the Atlantic Ocean during the cruises AMT-5 (Sept–Oct 1997), C-MarZ (April 2006), FORCLIM 7 (April 2009) and offshore Bermuda; (ii) in the Pacific Ocean during the cruises REVELLE (January–February 2004), KT-06-11 (June 2007), and offshore San Diego; (iii) in the Indian Ocean during the cruise OISO-4 (January–February 2000). Additional material was collected offshore Villefranche-sur-mer (France) in the Mediterranean Sea. No specific permits were required for the described field studies. The specimens were individually cleaned with a fine brush, isolated on the day of collection into a DNA extraction buffer (see below), and stored at −20°C. In total, we genetically analyzed 497 specimens from 49 open oceanic stations. At most sampling sites, temperature, salinity, and chlorophyll-a fluorescence vertical profiles down to 250 m were obtained by CTD casts.
DNA Extraction, Amplification and Sequencing
DNA extractions of 497 specimens were performed using the GITC [20] and GITC* [13] extraction buffers. Molecular analyses were carried out using the conserved 18S SSU and the more variable ITS (ITS-1, 5.8S, ITS-2) rDNA sequences. For 21 specimens originating from water masses of contrasted properties (Table 1), we amplified a ∼620 pb fragment localized at the 3′ end of the SSU rDNA based on PCR using the foraminiferal specific primers S15rf (5′ GTG CAT GGC CGT TCT TAG TTC 3′) - S19f (5′ CCC GTA CRA GGC ATT CCT AG 3′). For 80 specimens, we further amplified the whole ITS enclosing ITS-1, 5.8S, and ITS-2 (∼1000 pb) using the primers S30f (5′ AAGAGAAGTCGTAACAAGGC 3′) - L5f (5′ TCGCCGTTACTAAGGRAATC 3′). All PCR products were cloned using the TOPO TA cloning kit (Invitrogen). The 50 clones for the SSU and 135 clones for the ITS were sequenced using an ABI prism sequencer (Applied Biosystem) at the Station Biologique de Roscoff. The new sequences obtained in this study were deposited in Genbank with accession numbers JN164368 to JN164502 and JN164503 to JN164552, for the ITS and SSU regions, respectively.
Datasets, Alignment and DNA Sequence Analysis
The 50 SSU rDNA sequences obtained were manually aligned using SEAVIEW 4.0 [22]. Best fitted model of evolution was selected by jModeltest v 0.1.1 [56]. Using the selected model of substitution (HKY+G), four discrete categories for the gamma distributions and NNI+SPR tree improvements, a Maximum Likelihood approach implemented in PhyML software [57] with non-parametric bootstrapping (500 pseudo-replicates) was used to assess the most-likely tree topologies.
For the ITS array, four steps of alignments were successively used in our phylogenetic analysis. A first alignment (cleITS) included only the strictly unambiguous sequences from 54 specimens. In a second alignment (cloITS), clones from single specimens were added to test the strength of non-concerted evolution in Globoconella inflata ITS rDNA. Sequences that included a few ambiguous sites were added in a third alignment (larITS) in order to increase the geographic coverage of the dataset. Finally, three sequences cloned from an atypical individual displaying a Type II RFLP pattern within a Type I population (Re-1010, from REVELLE station 10) were included into a fourth dataset (comITS). Each dataset was aligned both manually and automatically using the MUSCLE software implemented in SEAVIEW 4.0 [22]. For both alignments of each dataset, poorly (highly variable) aligned regions were removed from the final alignments using GBLOCKS v. 0.91b [58] with the options allowing “smaller final blocks”, “gap positions within the final blocks” and “less strict flancking positions”. Finally, a consensus of both the manual and automatic alignments after GBLOCKS treatment was built, and the ambiguous positions between both methods were removed from the final alignment. We then used jModeltest v 0.1.1 [56] to select the most appropriate nucleotide substitution models. The (HKY+I+G) model was selected under the Akaike information criterion for the CleITS and LarITS datasets, while the (TrN+G) and the (HKY+G) were favored for the CloITS and the ComITS datasets, respectively. Using these models of substitution, four discrete categories for the gamma distributions and NNI+SPR tree improvements, a Maximum Likelihood approach [57] with non-parametric bootstrapping (500 pseudo-replicates) was used to assess the most-likely trees topologies.
RFLP Analysis
We developed a RFLP protocol based on the ITS region to rapidly recognize the genotypes of the 417 Globoconella inflata specimens that were not sequenced. After single-cell PCR, the products were digested with the endonuclease BstNI (New England Biolabs), which cuts at the sequence CC/WGG, according to the following protocol: 5 µl of the ITS rDNA PCR product were directly digested for 2 h at 60°C in a total volume of 10 µl containing 0.1 µl of the enzyme (1 U), 1 µl of the 10× buffer (New England Biolabs), and 3.9 µl of distilled water. Distinct patterns for each genotype were UV-detected after migration of the digested PCR products on 1.5% agarose gel, and ethidium bromide staining. The Type I typically produces a two-band pattern at 400 bp and 600 pb, and sometimes a third band occurs when specimens display length polymorphism between ITS copies. The Type II is not cut.
Biogeography of Globoconella inflata from Surface Sediments
Based on the BFD [25], we analyzed the global, environmental distribution of Globoconella inflata. The BFD records the absolute abundances of ∼551,600 intact individuals distributed within 37 extant morpho-species over 1265 sample localities from the world surface sediments (median sample size: 379 individuals; 95% Confidence Interval: 235–1033). Globoconella inflata is recorded in 566 (44.7%) localities, where it represents an average of 11.4% of the total assemblage (95% C.I.: 0.2%–54.7%).
For each of the 1265 core-tops, we extracted temperature data (annual mean) from the World Ocean Atlas 2005 [26] at the following water depths: 0, 10, 20, 30, 50, 75, 100, 125, 150, 250, 300, 400 and 500 m. In 3.27% (561 over 17,710) of the sample locality×water depth couples analyzed, temperature values were not directly available in the WOA for the 1°2 target-cell where the BFD sample station is located. In those cases, we interpolated the missing values from the ≤8 1°2 cells immediately surrounding the target-cell (forming a 3°×3° surface area). Interpolated values are weighted averages (weighting factor: inverse angular distance to the target-cell).
Based on this directly available and interpolated mean annual temperature dataset, the synthetic descriptors of the thermal structure of the water column for each of the 1265 sample localities were estimated through a correlation matrix-based Principal Component Analysis of the 13 temperature variables. A bubble biplot of the two first components, representing 95.7% of the overall annual 3D thermal variability, illustrates the distributions of the morpho-species abundances and genotypes occurrences throughout the main climatic provinces of the world oceans (Figure 4).
Biometrics
Looking at potential morphological differences between the two genotypes of Globoconella inflata, we conducted biometric measurements on 306 specimens collected during the cruise AMT-5 across the subpolar frontal zone. Specimens collected north (n = 154) and south (n = 152) of the Antarctic Subpolar Front were assumed to be representatives of Type I and Type II, respectively. The robustness of such an assumption was estimated based on the procedure by [28], which computes the probability that the observed distribution is biased by overlooking rare specimens of the presumably absent genotype. We used the equations:
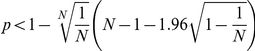 |
Where p is the relative abundance of such a rare genotype, q the probability of not having collected a rare genotype among the sampled material, and N the total number of collected individuals.
All specimens were mounted on glass cover slips with double side tape and similarly oriented on the umbilical and edge views of their shells, and then digitized under the microscope using an optical image analyzer (OPTIMAS v. 6.51). Length of the major axis of each specimen in edge view (a simple and robust estimator of individual size), together with lengths of the major axes of the terminal chamber and aperture were extracted from digitized outlines. The ratio between the lengths of major axes of the aperture and terminal chamber (a size-normalized apertural length) was plotted against individual size in a Log-Log diagram (Figure 5A), and differences between the specimens located north and south of the Antarctic Subpolar Front were quantified through a two-group discriminant analysis. Alternatively, histograms and Gaussian kernel densities of the log-ratio between the size-normalized apertural length and the specimen major axis were plotted for both groups of specimens (Figure 5B). Resulting non-gaussian distributions were compared non-parametrically using Mann-Whitney U-test (H0: the two genotypes are taken from populations with equal median) and Kolmogorov-Smirnov D-test (H0: the two genotypes are taken from populations with equal distribution) [59]. All the computations were done using PAST v 2.00 [60].
Supporting Information
SSU rDNA based phylogenetic tree of Globoconella inflata . Evolutionary relationships between 50 SSU rDNA clones of G. inflata from 13 localities in the Atlantic, Pacific and Indian Oceans (see Table 1 and Figure 1 for station names and locations). This Maximum Likelihood inference shows the relationships between the two phylotypes (Type I in red and Type II in blue). The bootstrap scores (500 replicates) greater than 80% are given next to branches. The scale and branch lengths are given in % of nucleotide substitution per site. The colors associated to leaf labels indicate geographic area of collection: blue = subpolar Indian Ocean; light blue = subpolar South Atlantic; Pink = South Atlantic north of the Subpolar Front; Yellow: Indian Ocean north of the Subpolar Front; red = North Atlantic; green = South Pacific; Orange = North Pacific. Circles associated to specific colors indicate clones sequenced from the same individuals.
(TIF)
Principal components correlation coefficients. PCA loading histograms, showing the correlations between the 13 depth-temperatures and the two first resulting Principal Components, representing 82.2% and 13.5% of the total variance, respectively. PC1 is a mean thermal state of the upper 500 m of the water column, whereas PC2 contrasts the temperatures of the first 125 m (negative weight) with the temperatures recorded between 150 and 500 m (positive weight) (weak contrast correspond to high PC2-value; high contrast correspond to low PC2-value).
(TIF)
Acknowledgments
We are grateful to D.B. Robbins, J. Aiken, N. Metzl, E. Goetze, P. Wiebe, H. Howa and Y. Okazaki for their invitation to participate in the different cruises. Officers, crew and scientists on board are acknowledged for their assistance. Special thanks to the Institut Polaire Français Paul Emile Victor (IPEV) for the organization of the OISO cruises. We also thank Y. Ujiié, L. Bittner and F. Mahé for technical support and discussions, and two anonymous referees for their helpful reviews.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors declare funding from the following programs (see details for the role of each funders): Grants from the IFR41 of Université Lyon 1 (Molecular analysis and morphometric analysis); INSU INTERRVIE program (Molecular analysis and morphometric analysis); Fondation pour la Recherche sur la Biodiversité: (morphometric analysis); ANR-06-JCJC-0142 PALEO-CTD (sample collection); ANR Blanc FORCLIM (sample collection); ANR-09-BLAN-0348 POSEIDON (molecular analysis); BIOMARKS – Biodiversity of marine eukaryotes, funded by the FP7 Eranet Biodiversa (Molecular analysis). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bé AWH. An ecological, zoogeographic and taxonomic review of recent planktonic foraminifera; A.T.S., editor. London: Academic Press; 1977. [Google Scholar]
- 2.Brayard A, Escarguel G, Bucher H. Latitudinal gradient of taxonomic richness: combined outcome of temperature and geographic mid-domains effects? Journal of Zoological Systematics and Evolutionary Research. 2005;43:178–188. [Google Scholar]
- 3.Kucera M, Rosell-Melé A, Schneider R, Waelbroeck C, Weinelt M. Multiproxy approach for the reconstruction of the glacial ocean surface (MARGO). Quaternary Science Reviews. 2005;24:813–819. [Google Scholar]
- 4.Katz ME, Cramer BS, Franzese A, Hönisch B, Miller KG, et al. Traditional and emerging geochemical proxies in foraminifera. Journal of Foraminiferal Research. 2010;40:165–192. [Google Scholar]
- 5.Bolli HM, Saunders JB. Oligocene to Holocene low latitude planktic foraminifera. In: Bolli HM, Saunders JB, Perch-Nielsen K, editors. Plankton Stratigraphy. Cambridge: Cambridge University Press; 1985. pp. 228–301. [Google Scholar]
- 6.Kennett JP, Srinivasan MS. Neogene Planktonic Foraminifera: A Phylogenetic Atlas. PA (Hutchinson Ross): Stroudsburg; 1983. [Google Scholar]
- 7.Darling KF, Wade CM. The genetic diversity of planktic foraminifera and the global distribution of ribosomal RNA genotypes. Marine Micropaleontology. 2008;67:216–238. [Google Scholar]
- 8.Vargas Cd, Bonzon M, Rees NW, Pawlowski J, Zaninetti L. A molecular approach to biodiversity and biogeography in the planktonic foraminifer Globigerinella siphonifera (d'Orbigny). Marine Micropaleontology. 2002;45:101–116. [Google Scholar]
- 9.Vargas Cd, Norris RD, Zaninetti L, Gibb SW, Pawlowski J. Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proceedings of the National Academy of Sciences U S A. 1999;96:2864–2868. doi: 10.1073/pnas.96.6.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vargas Cd, Renaud S, Hilbrecht H, Pawlowski J. Pleistocene adaptive radiation in Globorotalia truncatulinoides: genetic, morphologic, and environmental evidence. Paleobiology. 2001;27:104–125. [Google Scholar]
- 11.Aurahs R, Treis Y, Darling K, Kucera M. A revised taxonomic and phylogenetic concept for the planktonic foraminifer species Globigerinoides ruber based on molecular and morphometric evidence. Marine Micropaleontology. 2011;79:1–14. [Google Scholar]
- 12.Huber BT, Bijma J, Darling KF. Cryptic speciation in the living planktonic foraminifer Globigerinella siphonifera (d'Orbigny). Paleobiology. 1997;23:33–62. [Google Scholar]
- 13.Morard R, Quillévéré F, Escarguel G, Ujiié Y, De Garidel Thoron T, et al. Morphological recognition of cryptic species in the planktonic foraminifer Orbulina universa. Marine Micropaleontology. 2009;71:148–165. [Google Scholar]
- 14.Quillévéré F, Morard R, Escarguel G, Douady CJ, Ujiié Y, et al. Global scale same-specimen morpho-genetic analysis of Truncorotalia truncatulinoides: A perspective on the morphological species concept in planktonic foraminifera. Palaeogeography, Palaeoclimatology, Palaeoecology. 2011 In Press. [Google Scholar]
- 15.Knowlton N. Sibling Species in the Sea. Annual Review of Ecology and Systematics. 1993;24:189–216. [Google Scholar]
- 16.Scott GH, Kennett JP, Wilson KJ, Hayward BW. Globorotalia puncticulata: Population divergence, dispersal and extinction related to Pliocene-Quaternary water masses. Marine Micropaleontology. 2007;62:235–253. [Google Scholar]
- 17.Berggren WA, Kent DV, Swisher CC, III, Aubry M-P. Berggren WA, Kent DV, Aubry MP, Hardenbol J, editors. A revised cenozoic geochronology and chronostratigraphy. Geochronology, Time Scales, and Global Stratigraphic Correlation: Society of Sedimentary Geology. 1995. pp. 129–212.
- 18.Fairbanks RG, Sverdlove M, Free R, Wiebe PH, Be AWH. Vertical distribution and isotopic fractionation of living planktonic foraminifera from the Panama Basin. Nature. 1982;298:841–844. [Google Scholar]
- 19.Wilke I, Bickert T, Peeters FJC. The influence of seawater carbonate ion concentration [CO32−] on the stable carbon isotope composition of the planktic foraminifera species Globorotalia inflata. Marine Micropaleontology. 2006;58:243–258. [Google Scholar]
- 20.Pawlowski J. Introduction to the molecular systematics of foraminifera. Micropaleontology. 2000;46:1–12. [Google Scholar]
- 21.Bowser SS, Habura A, Pawlowski J. Molecular evolution of foraminifera. In: Katz LA, Bhattacharya D, editors. Genomics and evolution of microbial eucaryotes. Oxford University Press, Oxford; 2006. pp. 78–93. [Google Scholar]
- 22.Gouy M, Guindon S, Gascuel O. SeaView Version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 23.Pawlowski J, Fahrni J, Lecroq B, Longet D, Cornelius N, et al. Bipolar gene flow in deep sea benthic foraminifera. Molecular Ecology. 2007;16:4089–4096. doi: 10.1111/j.1365-294X.2007.03465.x. [DOI] [PubMed] [Google Scholar]
- 24.Darling KF, Wade CM, Stewart IA, Kroon D, Dingle R, et al. Molecular evidence for genetic mixing of Arctic and Antarctic subpolar populations of planktonic foraminifers. Nature. 2000;405:43–47. doi: 10.1038/35011002. [DOI] [PubMed] [Google Scholar]
- 25.Prell WL, Martin A, Cullen JL, Trend M. The Brown University Foraminiferal Data Base. 1999. IGBP PAGES/World Data Center-A for Paleoclimatology, Data Contribution Series # 1999-027, NOAA/NGDC Paleoclimatology Program, Boulder CO, USA.
- 26.Locarnini RA, Mishonov AV, Antonov JI, Boyer TP, Garcia HE. World Ocean Atlas 2005. 2006. 182 Volume 1: Temperature S Levitus: NOAA Atlas NESDIS 61, U.S. Government Printing Office, Washington, D.C.
- 27.Dietrich G, Kalle K, Krauss W, Sielder G, editors. General Oceanography. 2nd ed. New York: John Wiley and Sons (Wiley-Interscience); 1980. [Google Scholar]
- 28.Aurahs R, Grimm GW, Hemleben V, Hemleben C, Kucera M. Geographical distribution of cryptic genetic types in the planktonic foraminifer Globigerinoides ruber. Molecular Ecology. 2009;18:1692–1706. doi: 10.1111/j.1365-294X.2009.04136.x. [DOI] [PubMed] [Google Scholar]
- 29.Kustanowich S. Distribution of planktonic foraminifera in surface sediments of the south-west Pacific Ocean. New Zealand Journal of Geology and Geophysics. 1963;6:534–565. [Google Scholar]
- 30.Amato A, Kooistra WHCF, Hee Levialdi Ghiron J, Mann DG, Pröschold T, et al. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist. 2007;18:193–207. doi: 10.1016/j.protis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Behnke A, Friedl T, Chepurnov VA, Mann DG. Reproductive compatibility and rDNA sequence analyses in the Sellaphora pupula species complex (Bacillariophyta). Journal of Phycology. 2004;40:193–208. doi: 10.1111/j.1529-8817.2007.00454.x. [DOI] [PubMed] [Google Scholar]
- 32.Beszteri B, Ács É, Medlin LK. Ribosomal DNA sequence variation among sympatric strains of the Cyclotella meneghiniana complex (Bacillariophyceae) reveals cryptic diversity. Protist. 2005;156:317–333. doi: 10.1016/j.protis.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Casteleyn G, Chepurnov VA, Leliaert F, Mann DG, Bates SS, et al. Pseudo-nitzschia pungens (Bacillariophyceae): a cosmopolitan diatom species? Harmful Algae. 2008;7:241–257. [Google Scholar]
- 34.Van Oppen MJH, Miedog JC, Sanchez CA, Fabricius KE. Diversity of algal endosymbionts (zooxanthellae) in octocorals: the roles of geography and host relationships. Molecular Ecology. 2005;14:2403–2417. doi: 10.1111/j.1365-294X.2005.02545.x. [DOI] [PubMed] [Google Scholar]
- 35.Vanormelingen P, Chepurnov VA, Mann DG, Sabbe K, Vyverman W. Genetic divergence and reproductive barriers among morphologically heterogeneous sympatric clones of Eunotia bilunaris sensu lato (Bacillariophyta). Protist. 2008;159:73–90. doi: 10.1016/j.protis.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Thornhill DJ, Lajeunesse TC, Santos SR. Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Molecular Ecology. 2007;16:5326–5340. doi: 10.1111/j.1365-294X.2007.03576.x. [DOI] [PubMed] [Google Scholar]
- 37.De Queiroz K. Species concepts and species delimitation. Systematic Biology. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 38.Norris RD, Corfield RM, Cartlidge JE. Evolutionary ecology of Globorotalia (Globoconella) (planktic foraminifera). Marine Micropaleontology. 1994;23:121–145. [Google Scholar]
- 39.Keany J, Kennett JP. Pliocene-early Pleistocene paleoclimatic history recorded in Antarctic-Subantarctic deep-sea cores. Deep Sea Research. 1972;19:539–548. [Google Scholar]
- 40.Kennett JP. Pleistocene paleoclimates and foraminiferal biostratigraphy in subantarctic deep-sea cores. Deep Sea Research. 1970;17:127–140. [Google Scholar]
- 41.Norris RD. Pelagic species diversity, biogeography and evolution. Paleobiology. 2000;26:236–258. [Google Scholar]
- 42.Darling KF, Kucera M, Kroon D, Wade CM. A resolution for the coiling direction paradox in Neogloboquadrina pachyderma. Paleoceanography. 2006;21:1–14. [Google Scholar]
- 43.Darling KF, Kucera M, Pudsey CJ, Wade CM. Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proceedings of the National Academy of Sciences U S A. 2004;101:7657–7662. doi: 10.1073/pnas.0402401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darling KF, Kucera M, Wade CM. Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist. Proceedings of the National Academy of Sciences U S A. 2007;104:5002–5007. doi: 10.1073/pnas.0700520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darling KF, Kucera M, Wade CM, Langen Pv, Pak D. Seasonal distribution of genetic types of planktonic foraminifer morphospecies in the Santa Barbara Channel and its paleoceanographic implications. Paleoceanography. 2003;18:1032–1042. [Google Scholar]
- 46.Stewart IA, Darling KF, Kroon D, Wade CM, Troelstra SR. Genotypic variability in subarctic Atlantic planktic foraminifera. Marine Micropaleontology. 2001;43:143–153. [Google Scholar]
- 47.Cléroux C, Cortijo E, Duplessy J-C. Deep-dwelling foraminifera as thermocline temperature recorders. Geochemistery, Geophysics, Geosystems. 2007;8:1–19. [Google Scholar]
- 48.Farmer EC, Kaplan A, de Menocal PB, Lynch-Stieglitz J. Corroborating ecological depth preferences of planktonic foraminifera in the tropical Atlantic with the stable oxygen isotope ratios of core top specimens. Paleoceanography. 2007;22:1–14. [Google Scholar]
- 49.Ganssen GM, Kroon D. The isotopic signature of planktonic foraminifera from NE Atlantic surface sediments: implications for the reconstruction of past oceanic conditions. Journal of Geological Society, London. 2000;157:693–699. [Google Scholar]
- 50.Keigwin L, Bice M, Copley N. Seasonality and stable isotopes in planktonic foraminifera off Cape Cod, Massachusetts. Paleoceanography. 2005;20:1–9. [Google Scholar]
- 51.Ravelo CA, Fairbanks RG. Oxygen isotopic composition of multiple species of planktonic foraminifera: recorders of the modern photic zone temperature gradient. Paleoceanography. 1992;7:815–831. [Google Scholar]
- 52.Chiessi CM, Ulrich S, Mulitza S, Pätzold J, Wefer G. Signature of the Brazil-Malvinas Confluence (Argentine Basin) in the isotopic composition of planktonic foraminifera from surface sediments. Marine Micropaleontology. 2007;64:52–66. [Google Scholar]
- 53.King AL, Howard WR. δ18O seasonality of planktonic foraminifera from Southern Ocean sediment traps: Latitudinal gradients and implications for paleoclimate reconstructions. Marine Micropaleontology. 2005;56:1–24. [Google Scholar]
- 54.Martínez-Garcia A, Rosell-Melé A, Geibert W, Gersonde R, Masqué P, et al. Links between iron supply, marine productivity, sea surface temperature, and CO2 over the last 1.1 Ma. Paleoceanography. 2009;24:PA1207. [Google Scholar]
- 55.Martinez-Garcia A, Rosell-Mele A, McClymont EL, Gersonde R, Haug GH. Subpolar link to the emergence of the modern equatorial Pacific cold tongue. Science. 2010;328:1550–1553. doi: 10.1126/science.1184480. [DOI] [PubMed] [Google Scholar]
- 56.Posada D. jModelTest: Phylogenetic Model Averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 57.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 58.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 59.Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. New York: Freeman; 1995. 887 [Google Scholar]
- 60.Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:1–9. [Google Scholar]
- 61.Carter L, McCave IN, Williams MJM, Fabio F, Martin S. Chapter 4 circulation and water masses of the southern ocean: a review. Developments in Earth and Environmental Sciences: Elsevier; 2008. pp. 85–114. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SSU rDNA based phylogenetic tree of Globoconella inflata . Evolutionary relationships between 50 SSU rDNA clones of G. inflata from 13 localities in the Atlantic, Pacific and Indian Oceans (see Table 1 and Figure 1 for station names and locations). This Maximum Likelihood inference shows the relationships between the two phylotypes (Type I in red and Type II in blue). The bootstrap scores (500 replicates) greater than 80% are given next to branches. The scale and branch lengths are given in % of nucleotide substitution per site. The colors associated to leaf labels indicate geographic area of collection: blue = subpolar Indian Ocean; light blue = subpolar South Atlantic; Pink = South Atlantic north of the Subpolar Front; Yellow: Indian Ocean north of the Subpolar Front; red = North Atlantic; green = South Pacific; Orange = North Pacific. Circles associated to specific colors indicate clones sequenced from the same individuals.
(TIF)
Principal components correlation coefficients. PCA loading histograms, showing the correlations between the 13 depth-temperatures and the two first resulting Principal Components, representing 82.2% and 13.5% of the total variance, respectively. PC1 is a mean thermal state of the upper 500 m of the water column, whereas PC2 contrasts the temperatures of the first 125 m (negative weight) with the temperatures recorded between 150 and 500 m (positive weight) (weak contrast correspond to high PC2-value; high contrast correspond to low PC2-value).
(TIF)