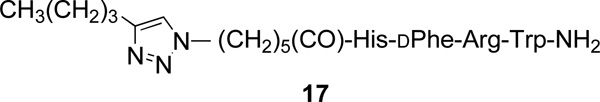Abstract
A spherical molecular scaffold bearing eight terminal alkyne groups was synthesized in one step from sucrose. One or more copies of a tetrapeptide azide, either N3(CH2)5(C=O)-His-dPhe-Arg-Trp-NH2 (MSH4) or N3(CH2)5(C=O)-Trp-Met-Asp-Phe-NH2 (CCK4), were attached to the scaffold via the copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) reaction. Competitive binding assays using Eu-labeled probes based on the superpotent ligands Ser-Tyr-Ser-Nle-Glu-His-dPhe-Arg-Trp-Gly-Lys-Pro-Val-NH2 (NDP-α-MSH) and Asp-Tyr-Met-Gly-Trp-Met-Asp-Phe-NH2 (CCK8) were used to study the interactions of monovalent and multivalent MSH4 and CCK4 constructs with Hek293 cells engineered to overexpress MC4R and CCK2R. All of the monovalent and multivalent MSH4 constructs exhibited binding comparable to that of the parental ligand, suggesting that either the ligand spacing was inappropriate for multivalent binding, or MSH4 is too weak a binder for a second “anchoring” binding event to occur before the monovalently-bound construct is released from the cell surface. In contrast with this behavior, monovalent CCK4 constructs were significantly less potent than the parental ligand, while multivalent CCK4 constructs were as or more potent than the parental ligand. These results are suggestive of multivalent binding, which may be due to increased residence times for monovalently bound CCK4 constructs on the cell surface relative to MSH4 constructs, the greater residence time being necessary for the establishment of multivalent binding.
Keywords: Multimeric ligands, Melanocortin 4 receptor ligands, Cholecystokinin 2 receptor ligands
1. Introduction
Early detection and diagnosis of many human cancers could be aided by reagents that seek out and selectively bind to cancer cells and report their existence and location by non-invasive molecular imaging.1–4 One strategy for development of such reagents involves attaching imaging agents to molecular scaffolds that bear multiple copies of ligands to receptors present on the surface of cancer cells.5–11 Such multivalent constructs could display enhanced affinity and selectivity for cancer cells based on cooperative binding.12–16
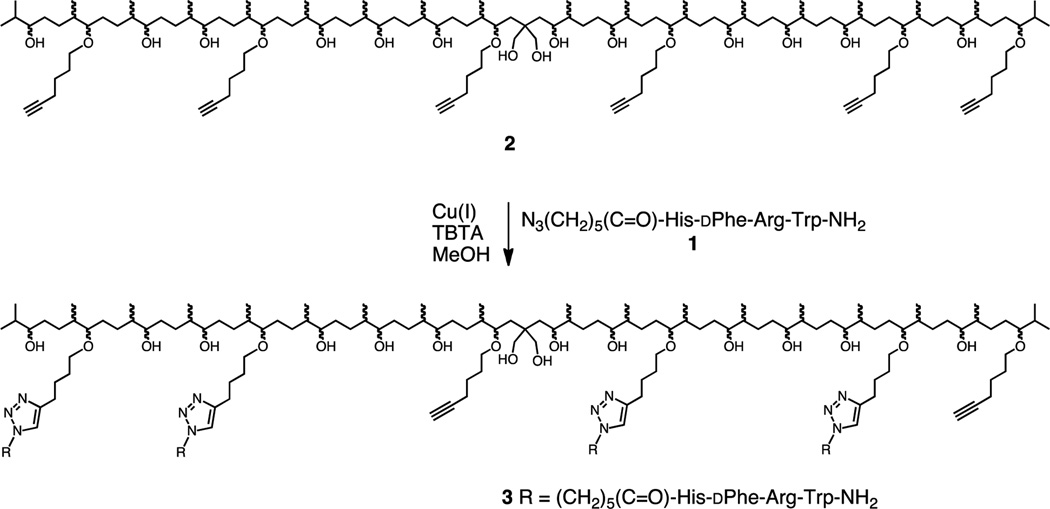
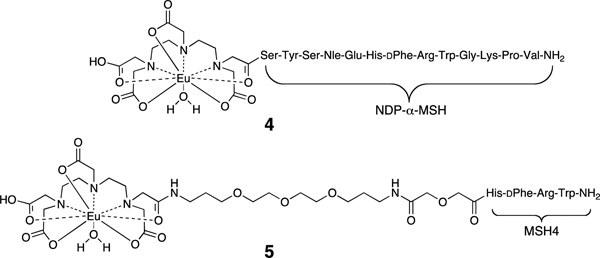
Previously we described the preparation and testing of multivalent constructs derived from squalene17 and solanesol.18 These constructs bore sidechains based on the ligand Ac-His-dPhe-Arg-Trp-NH2 (MSH4FN1)19–21 which has a low micromolar affinity for binding to the melanocortin 4 receptor (MC4R).22 In our prior work, the azide N3(CH2)5(C=O)-His-dPhe-Arg-Trp-NH2 (1, Scheme 1) was attached to a linear scaffold (e.g., 2) bearing terminal alkyne groups using the copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC),23–26 producing triazole-containing multivalent constructs (e.g., 3).18 The abilities of 3 and of similar monovalent and multivalent constructs based on MSH4 to bind to MC4R were tested in competitive binding assays against probes 411 and 527 using Hek293 cells engineered to overexpress this receptor. Probe 4 is based on the superpotent ligand Ser-Tyr-Ser-Nle-Glu-His-dPhe-Arg-Trp-Gly-Lys-Pro-Val-NH2 (NDP-α-MSH),28,29 while the probe 5 is based on MSH4. Interestingly, all of the solanesol-derived monovalent and multivalent constructs bearing MSH4 bound monovalently to MC4R when competed against either probe.18 To determine whether or not the solanesol-derived linear scaffold was responsible for this unexpected binding behavior, we have prepared and tested monovalent and multivalent constructs using azide 1 and a spherical scaffold derived from sucrose (vide infra). To determine whether or not this binding behavior was ligand and/or receptor dependent, we have also prepared and tested monovalent and multivalent constructs derived from this spherical scaffold and an azide that incorporates the ligand Trp-Met-Asp-Phe-NH2 (CCK4) which has a low nanomolar affinity for binding to the cholecystokinin 2 receptor (CCK2R).30
Scheme 1.
Synthesis of Multivalent Constructs 3.
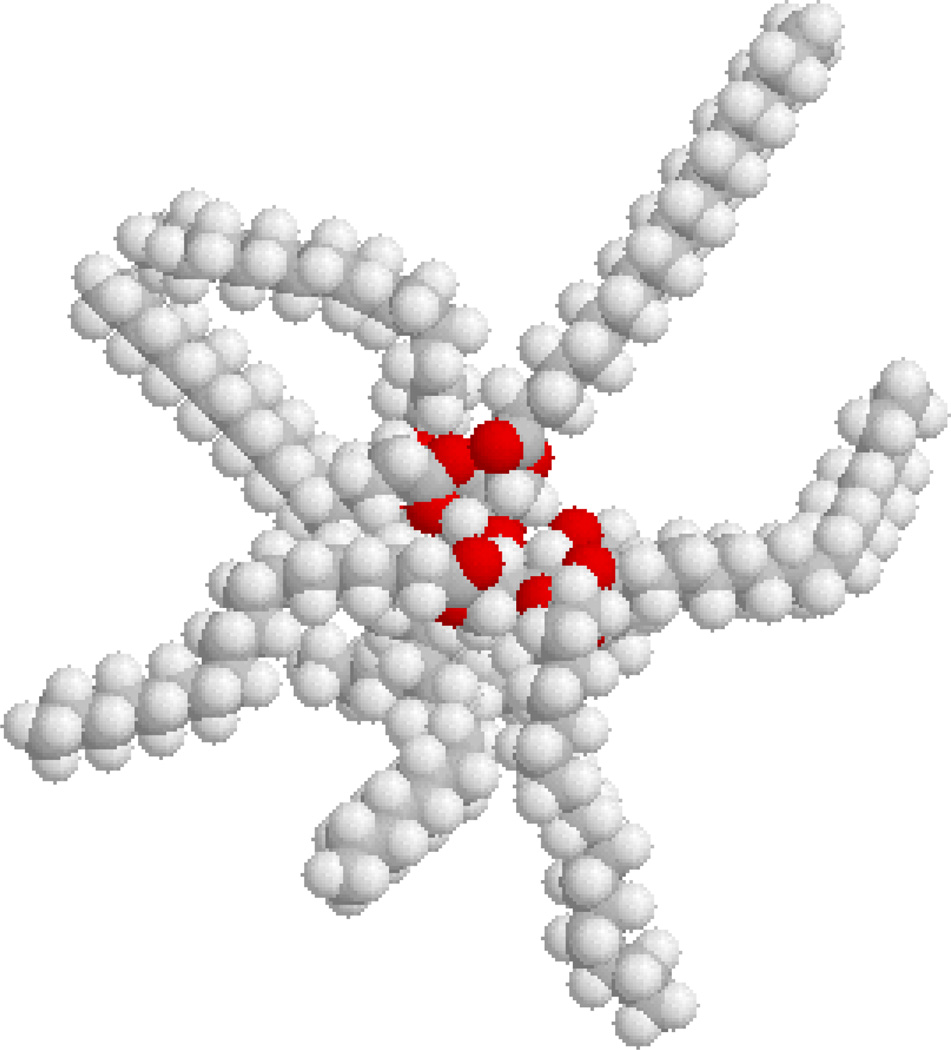
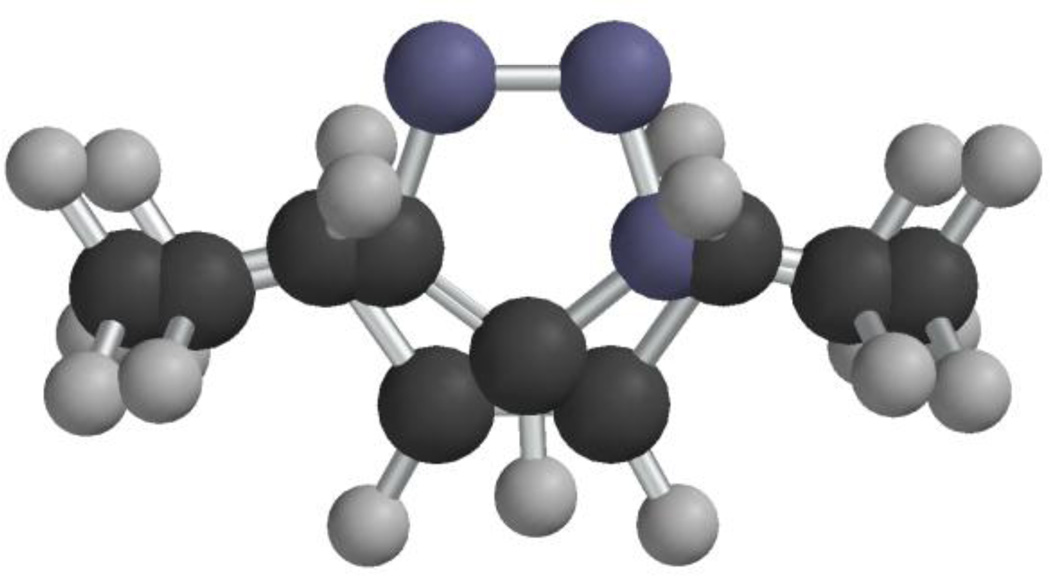
While searching for alternatives to linear scaffolds for ligand display, our attention was drawn by Olestra, a non-digestible fat substitute (e.g. 6, Figure 1).31,32 We reasoned that sucrose-derived molecules might make useful scaffolds for multimerization if ligands and other moieties of interest, such as imaging and therapeutic agents, could be attached to the termini of the fatty acid chains. Additionally, we determined that the 1,4-disubstituted-1H-1,2,3-triazole groups that are produced via CuAAC map reasonably well onto the cis-1,2-disubstituted alkene groups of unsaturated fatty acid components of Olestra (compare 7 and 8, Figure 2). These considerations led to the synthesis and biological testing of sucrose-derived multivalent constructs that display one or more copies of MSH4 or CCK4 as described herein.
Figure 1.
A space-filling representation of one component of Olestra (6). This representation © 1997 by Daniel J. Berger and may be copied without limit if its use is for non-profit educational purposes.
Figure 2.
Superposition of cis-3-pentene (7) and 1,4-dimethyl-1H-1,2,3-triazole (8). The methyl carbon-to-methyl carbon distances are 6.04 Å and 5.00 Å for 7 and 8, respectively. The conformers of 7 and 8 used in this comparison were generated using Spartan '02 v1.0.5 for Macintosh.
2. Materials and methods
2.1. Chemical synthesis
2.1.1. General experimental
Dichloromethane (DCM), diethyl ether, and tetrahydrofuran (THF) were dried by passage through activated alumina. Other solvents and commercial reagents were used as supplied. For moisture sensitive reactions, glassware was flame-dried under argon. Analytical thin-layer chromatography (TLC) was carried out on pre-coated silica gel 60 F-254 plates with visualization by UV exposure, by exposure to I2 vapor, or by staining with 10% phosphomolybdic acid solution in ethanol or with 5% H2SO4 in ethanol and heat. Gravity-driven column chromatography was accomplished using silica gel 60 (63–210 µm). 1H NMR and 13C NMR spectra were recorded at 300 MHz or 500 MHz for 1H NMR and at 75 MHz or 125 MHz for 13C NMR. Chemical shifts (δ) are expressed in ppm and are internally referenced (7.24 ppm for CDCl3 and 3.31 ppm for CD3OD for 1H NMR and 77.0 ppm for CDCl3 and 49.15 ppm for CD3OD for 13C NMR). Analytical HPLC was performed on a 4.6 × 75 mm Waters Symmetry® C18 column and preparative HPLC was performed on a 19 × 256 mm Waters X-Bridge Preparative C18 column. The mobile phase was 10–90% acetonitrile and water containing 0.1% trifluoroacetic acid (TFA) within 50 min. The flow rates were 1 mL/min and 15 mL/min for analytical and preparative runs, respectively. The dual UV detector system operated at 230 and 280 nm. Matrix-assisted desorption/ionization time-of-flight (MALDI-TOF) experiments were carried out on a Bruker Ultraflex III MALDI TOF-TOF instrument. Both the reflectron and linear techniques were used for positive ion detection. The matrix, sinapic acid, and the analyte were dissolved in water:acetonitrile 1:1 containing 0.1% formic acid and the solutions mixed in a ratio of 100:1. ESI was also used to ionize some of the samples. These experiments were performed on an ESI Bruker Apex Qh 9.4 T FT-ICR instrument.
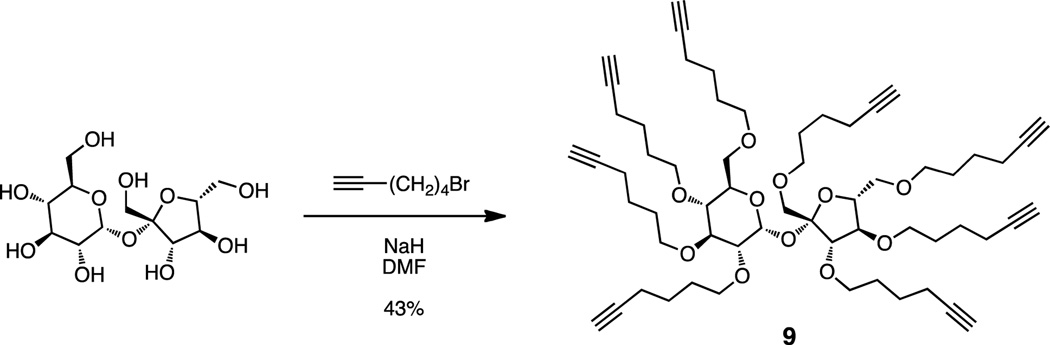
2.1.2. Scaffold synthesis (Scheme 2)
Scheme 2.
Synthesis of sucrose derivative 9.
2.1.2.1. Octa-O-(5-hexyn-1-yl)-β-d-fructofuranosyl-α-d-glucopyranoside (9)
To a suspension of NaH (330 mg, 13.7 mmol) in dry dimethylformamide (DMF, 10 mL) under argon were added sucrose (200 mg, 0.58 mmol), 6-bromo-1-hexyne33 (1.13 g, 6.97 mmol), and tetrabutylammonium bromide (50 mg, 0.15 mmol). The mixture was stirred at room temperature for 48 h. The reaction was quenched with sat NH4Cl and the mixture extracted with ethyl acetate (3 × 15 mL). The combined organic extracts were washed with water, brine, dried over Na2SO4, and filtered. Removal of volatiles in vacuo afforded a residue that was subjected to silica gel chromatography (63–210 µm) using ethyl acetate/hexanes (2:8) as elutant. This afforded 250 mg (0.25 mmol, 43%) of 9 as a colorless oil, Rf 0.6 (ethyl acetate/hexanes, 3:7), [α]D25 17.0 (c 0.5, CHCl3); IR (cm−1) 3308, 2928, 2854, 1213, 1152, 1094; 1H NMR (500 MHz, CDCl3) δ 1.57–1.71 (m, 32H), 1.95 (m, 8H), 2.18–2.23 (m, 16H), 3.17 (dd, J = 9.5 Hz, J = 3.4 Hz, 1H), 3.28 (t, J = 9.5 Hz, 1H), 3.35–3.71 (m, 20H), 3.77–3.92 (m, 5H), 4.07 (d, J = 7.2 Hz, 1H), 5.50 (d, J = 3.8 Hz, 1H); 13C NMR (125 MHz, CDCl3) δ 18.3, 24.7, 25.3, 28.6, 28.7, 29.1, 29.2, 29.6, 31.7, 62.3, 68.4, 68.5, 69.5, 70.2, 70.5, 70.6, 70.8, 71.0, 71.1, 71.7, 72.3, 72.4, 72.7, 79.5, 80.5, 81.6, 82.9, 84.0, 84.2, 84.3, 89.9, 104.3; HRMS (MALDI-TOF) calculated for C60H86NaO11 [M+Na]+ 1005.6068, observed 1005.6068.
2.1.3. Solid phase synthesis (Scheme 3)
Scheme 3.
Solid Phase Synthesisa
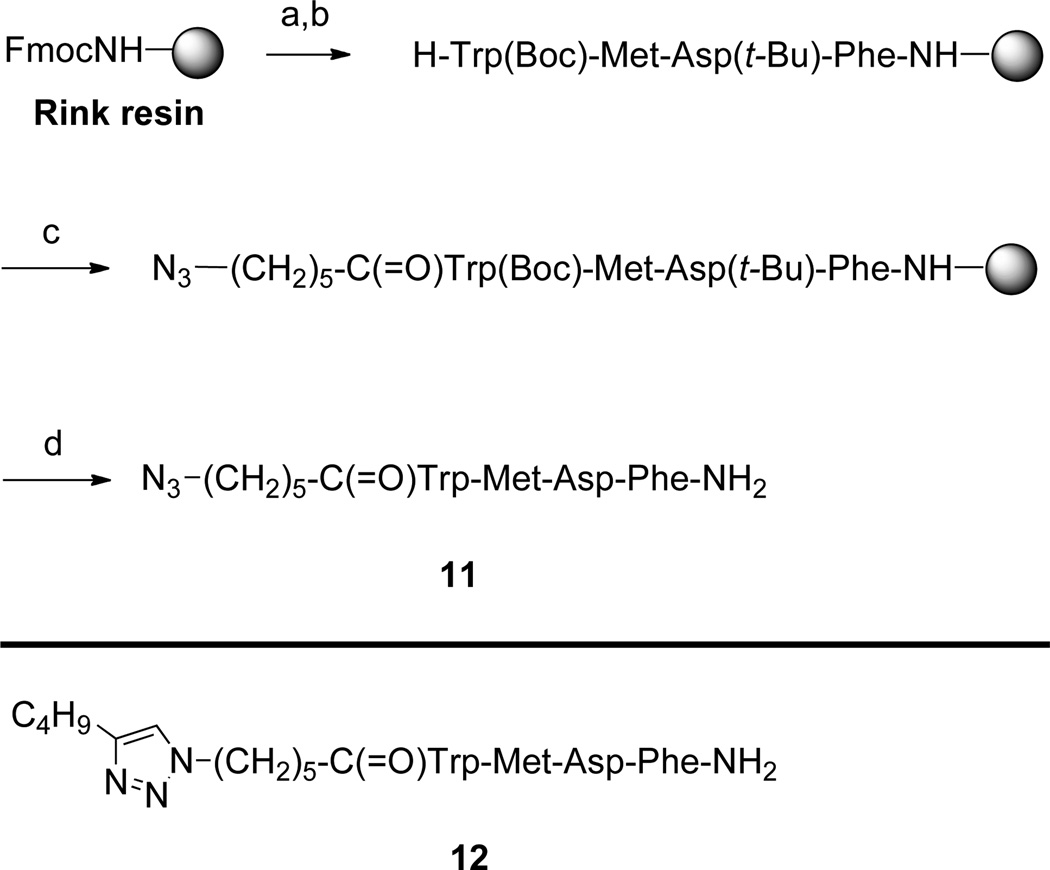
aReagents: (a) piperidine. (b) Fmoc/tBu solid phase synthesis. (c) N3(CH2)5CO2H, Cl-HOBt, DIC. (d) trifluoroacetic acid/triisopropylsilane/thioanisole/water (91/3/3/3).
In a syringe (polypropylene reaction tube equipped with a polypropylene frit) Rink amide resin (1 g, 0.68 mmol) was allowed to swell in THF for 1 h. THF was removed, and 20% piperidine in DMF (15 mL) was added for 2 min to cleave the 9-fluorenylmethyoxycarbonyl (Fmoc) group. The liquid phase was removed, and 20% piperidine solution in DMF (15 mL) was again added and the mixture shaken for 18 min. The liquid phase was removed, and the resin was washed with DMF (3 × 15 mL), DCM (3 × 15 mL), DMF (3 × 15 mL), 0.5 M 1-hydroxybenzotriazole (HOBt) in DMF (15 mL), 0.5 M HOBt in DMF (15 mL) plus a drop of bromophenol blue, DMF (2 × 15 mL), and DCM (15 mL), in that order. A solution of the first Fmoc-amino acid, Fmoc-Phe (1.05 g, 2.04 mmol), 6-chloro-1-hydroxybenzotriazole (Cl-HOBt, 345 mg, 2.04 mmol), and diisopropylcarbodiimide (DIC, 512 mg, 4.08 mmol) in DMF (15 mL) was allowed to react for 2 min, then added to the resin and the mixture shaken for 1 h, at which time the blue color had disappeared. The resin was then washed with DMF (3 × 15 mL), DCM (3 × 15 mL), and DMF (3 × 15 mL). Free NH2 groups were capped by addition of a 1:1 mixture of acetic anhydride and pyridine (6 mL). After the mixture was shaken for 20 min, the resin was washed with DMF (3 × 15 mL), DCM (3 × 15 mL), and DMF (3 × 15 mL). The absence of free amine groups was confirmed by the Kaiser test. The above cycle of procedures was repeated for coupling of each of the amino acids in the sequence, and finally for attachment of the N-terminal 6-azidohexanoic acid34 residue or the N-terminal 6-(4-butyl-1H-1,2,3-triazol-1-yl)hexanoic acid18 residue, thus producing the resin-bound peptide derivatives related to compounds 11 and 12. Cleavage and deprotection were achieved using a 91:3:3:3 mixture of trifluoroacetic acid, triisopropylsilane, thioanisole, and water (10 mL). The mixture of cleavage cocktail and resin was shaken for overnight, the solution was separated from the resin, volatiles were evaporated, the residue triturated with diethyl ether, and the crude product collected by centrifugation. Purification of the tetrapeptide products 11 and 12 was accomplished by preparative reversed phase HPLC. Yields ranged from 39–46% over several batches. The purity of compounds 11 and 12 was checked by analytical reversed phase HPLC. Compounds 11 and 12 were recovered from solution by lyophilzation and were further characterized by ESI mass spectrometry (see Table 1).
Table 1.
Mass spectral and HPLC characterization of compounds 11 and 12.
| Compound | Formula [M] |
Calc Mass [Ion] |
Mass Found (error) |
tRa |
|---|---|---|---|---|
| 11 | C36H47N9O7 | 718.3671 [M+1]+ | 718.3669 (0.3 ppm) | 23.52 |
| 12 | C44H60N14O5 | 800.4454 [M+1]+ | 800.4450 (0.5 ppm) | 24.16 |
Linear gradient of from 10→90% acetonitrile in water containing 0.1% TFA over 50 min.
2.1.4. Multimer synthesis (Scheme 4)
Scheme 4.
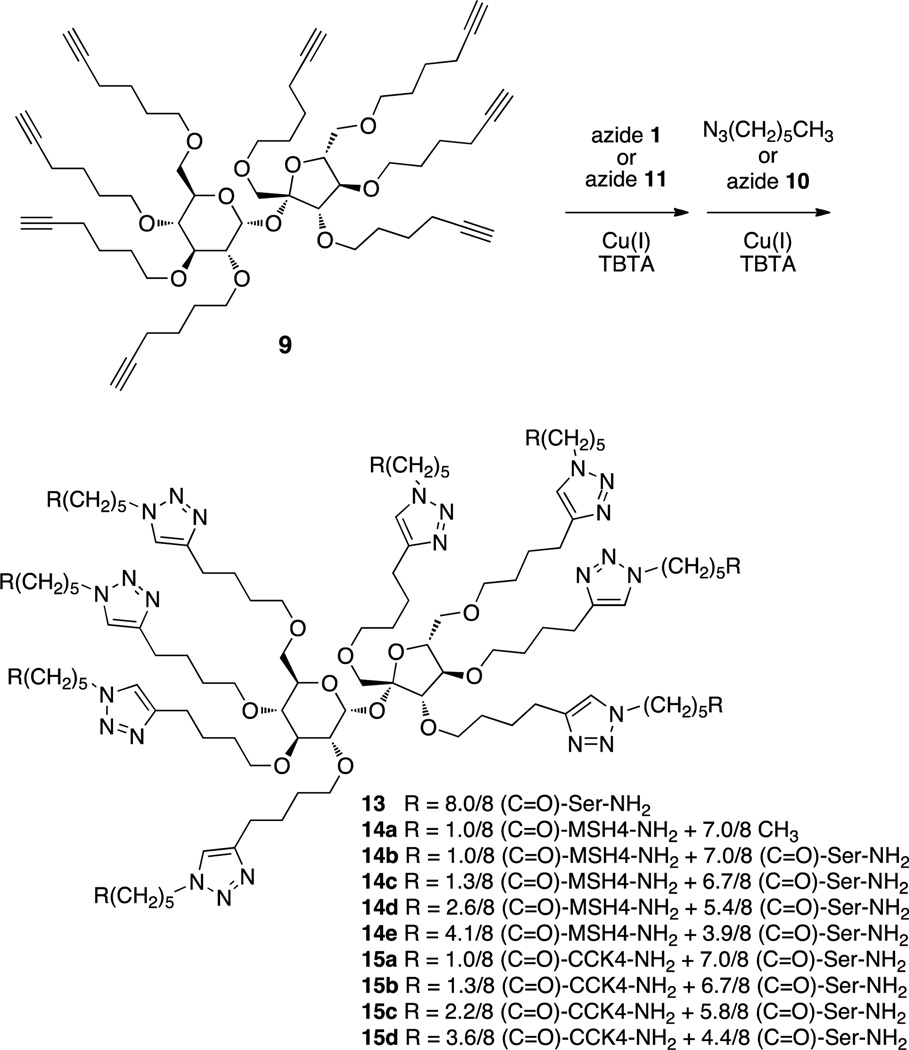
Synthesis of multivalent compounds 13, 14a–14e, and 15a–d via CuAAC.a
aThe positions of ligand attachment are presumed to be random.
2.1.4.1. Serine Amide Multimer 13
A mixture of 9 (10 mg, 10 µmol), azide 1018 (39 mg, 162 µmol), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, 4.3 mg, 8 µmol), and tetrakis(acetonitrile)copper(I) hexafluorophosphate (3 mg, 8 µmol) in dry methanol (1.5 mL) was irradiated for 4 h in a Biotage microwave reactor (100 °C). After the reaction was complete, water (25 mL) was added and the mixture was extracted with CHCl3 containing dithizone (20 mg/L, 3 × 15 mL) to remove copper.35 The water layer was then washed with DCM (2 × 15 mL) to remove any remaining 10 and TBTA. After lyophilization, fractionation of the residue by preparative HPLC (10→90% acetonitrile in water containing 0.1% TFA within 50 min, tR 11.1 min) and recovery by lyophilization afforded 4.5 mg (1.5 µmol, 15% yield) of 13 as a white solid, mp 72–73 °C, [α]D25 6.5 (c 0.55, CHCl3); IR (cm−1) 3283, 2926, 2852, 1635, 1212, 1152; 1H NMR (500 MHz, CD3OD) δ 1.31–1.36 (m, 16H), 1.57–1.70 (m, 48H), 1.88–191 (m, 16H), 2.27 (t, J = 7.0 Hz, 16H), 2.68–2.72 (m, 16H), 3.06–3.09 (m, 1H), 3.15–3.20 (m, 1H), 3.33–3.95 (m, 43H), 4.05 (d, J = 7.5 Hz, 1H), 4.33–4.42 (m, 23H), 5.50 (d, J = 3.5 Hz, 1H), 7.81 (br s, 8H); 13C NMR (125 MHz, CD3OD) δ 24.6, 25.6, 25.7, 25.8, 28.8, 29.2, 29.5, 35.1, 49.9, 55.1, 61.8, 66.2, 68.0, 69.6, 70.0, 70.2, 70.4, 70.6, 70.9, 71.0, 71.7, 71.9, 72.2, 72.5, 77.9, 79.2, 80.3, 81.3, 82.0, 83.7, 89.3, 92.5, 104.1, 109.4, 111.5, 113.4, 115.8, 122.1, 122.2, 147.3, 173.7, 174.5; HRMS (MALDI-TOF) calculated for C132H222N40NaO35 [M+Na]+ 2950.6719, observed 2950.6423.
2.1.4.2. Procedure for Reaction of Azide-functionalized MSH4 Derivative 1 and 1-Azidohexane with 9 to Produce Multivalent Constructs 14a
A mixture of 9 (56 mg, 56 µmol), azide 118 (15 mg, 19 µmol), TBTA (2.0 mg, 3.8 µmol), and tetrakis(acetonitrile)copper(I) hexafluorophosphate (1.4 mg, 3.8 µmol) in dry methanol (1.5 mL) was irradiated for 2 h in a Biotage microwave reactor (100 °C). After the reaction was complete, water (25 mL) was added and the mixture extracted with CHCl3 containing dithizone (20 mg/L, 3 × 15 mL) and with DCM (3 × 15 mL) to remove copper,35 TBTA, and excess 9. After lyophilization, the residue (30 mg) was taken up in dry methanol (1.5 mL), 1-azidohexane (97 mg, 763 µmol), TBTA (6.3 mg, 11.8 µmol), and tetrakis(acetonitrile)copper(I) hexafluorophosphate (4.4 mg, 11.8 µmol) were added, and the reaction mixture was irradiated for 4 h in a Biotage microwave reactor (100 °C). Following workup as above and lyophilization, the residue was fractionated by preparative HPLC (10→90% acetonitrile in water containing 0.1% TFA within 50 min, tR 29–33 min) to afford 14a as an oil; yield 13 mg (4.9 µmol, 26%). Product 14a was analyzed by MALDI-TOF (see Figure S2 in the Supplementary Data) and by UV spectroscopy.18
2.1.4.3. Procedure for Reaction of Azide-functionalized MSH4 Derivative 1 and Azide-functionalized Serine Amide Derivative 10 with 9 to Produce Multivalent Constructs 14b
A mixture of 9 (56 mg, 56 µmol), azide 118 (15 mg, 19 µmol), TBTA (2.0 mg, 3.8 µmol), and tetrakis(acetonitrile)copper(I) hexafluorophosphate (1.4 mg, 3.8 µmol) in dry methanol (1.5 mL) was irradiated for 2 h in a Biotage microwave reactor (100 °C). After the reaction was complete, water (25 mL) was added and the mixture extracted with CHCl3 containing dithizone (20 mg/L, 3 × 15 mL) and with DCM (3 × 15 mL) to remove copper,35 TBTA, and excess 9. After lyophilization, the residue (29 mg) was taken up in dry methanol (1.5 mL), azide 1018 (54 mg, 224 µmol), TBTA (6.9 mg, 3.8 µmol), and tetrakis(acetonitrile)copper(I) hexafluorophosphate (4.8 mg, 3.8 µmol) were added, and the reaction mixture was irradiated for 4 h in a Biotage microwave reactor (100 °C). Following workup as above and lyophilization, the residue was fractionated by preparative HPLC (10→90% acetonitrile in water containing 0.1% TFA within 50 min, tR 12.5–16.5 min) to afford 14b as a white powder; yield 3.0 mg (0.87 µmol, 5%). Product 14b was analyzed by MALDI-TOF (see Figure S3 in the Supplementary Data) and by UV spectroscopy.18
2.1.4.4. General Procedure for Reaction of Azide-functionalized MSH4 Derivative 1 and Azide-functionalized Serine Amide Derivative 10 with 9 to Produce Multivalent constructs 14c–e
Mixtures of 9 (variable amount, see Table 2), azide 118 (variable amount, see Table 2), TBTA (0.43 mg/µmol of 9), and tetrakis(acetonitrile)copper(I) hexafluorophosphate (0.3 mg/µmol of 9) in dry methanol (100 µL/µmol of 9) were irradiated for 2–4 h in a Biotage microwave reactor (100 °C). Azide 1018 (variable amount, see Table 2) was then added to the reaction mixtures and irradiation was resumed for another 4 h. After the reactions were complete, water (2.5 mL/µmol of 9) was added and the mixtures were extracted with CHCl3 containing dithizone (20 mg/L, 3 × 15 mL) and with DCM (3 × 15 mL) to remove copper,35 TBTA, and excess 10. After lyophilization, the residues were fractionated by preparative HPLC (10→90% acetonitrile in water containing 0.1% TFA within 50 min, tR 12.5–16.5 min) to afford products 14c–14e as white powders; yields are given in Table 2. Products were analyzed by MALDI-TOF (see Figure 3 and Figures S4–S6 in the Supplementary Data) and by UV spectroscopy.18
Table 2.
Synthesis of 13 and 14a–e.
| Product | mg 9 | mg 1 (equiv) | mg 10 (equiv) | Yield, mg (%) | MALDI-TOF |
|---|---|---|---|---|---|
| 13 | 10 | 0 | 39 (16) | 4.5 (15) | Figure S1 |
| 14a | 56 | 15 (0.34) | 97 (45)a | 13 (26) | Figure S2 |
| 14b | 56 | 15 (0.34) | 63 (16) | 3.0 (5) | Figure S3 |
| 14c | 18 | 29 (2) | 44 (10) | 44 (61) | Figure S4 |
| 14d | 7.0 | 17 (3) | 17 (10) | 14 (42) | Figure S5 |
| 14e | 8.0 | 31 (5) | 20 (10) | 22 (49) | Figure S6 |
1-Azidohexane used in place of 10.
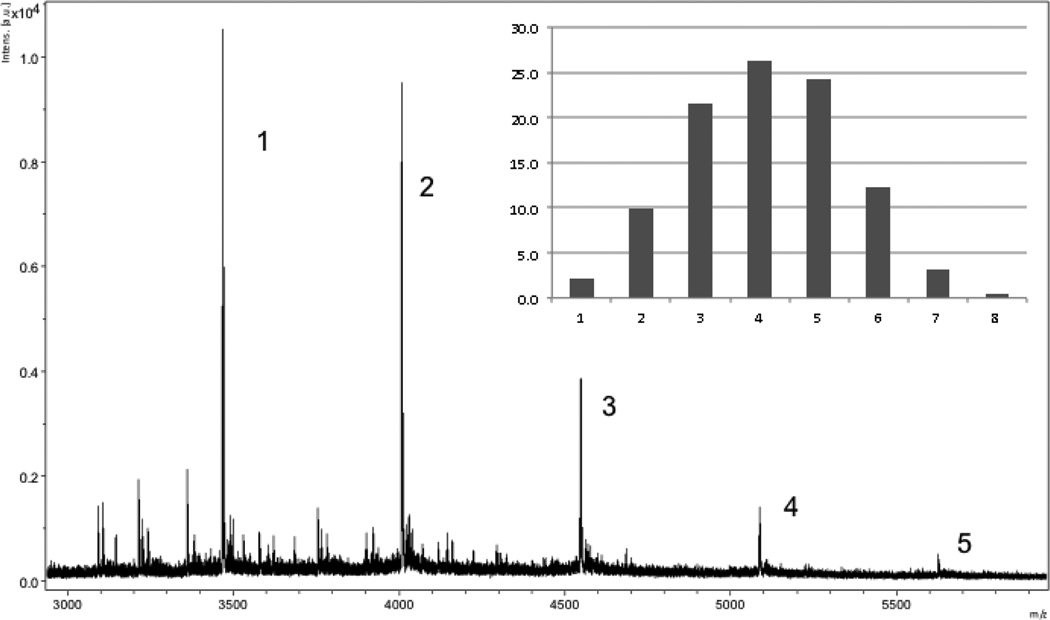
Figure 3.
MALDI-TOF mass spectrum of 14e showing the distribution of products. The numbers indicate the number of attached MSH4 ligands. The insert shows the anticipated statistical product distribution assuming attachment of 4.1 ligands per scaffold. For peak identification and for spectra of 13, 14a–14e, and 15a see the Supplementary Data.
2.1.4.5. Procedure for Reaction of Azide-functionalized CCK4 Derivative 11 and Azide-functionalized Serine Amide Derivative 10 with 9 to Produce Multivalent Constructs 15a
A mixture of 9 (205 mg, 209 µmol), azide 11 (30 mg, 41 µmol), TBTA (4.3 mg, 8.2 µmol), and tetrakis(acetonitrile)copper(I) hexafluorophosphate (3 mg, 8.2 µmol) in dry DMF (1.5 mL) was irradiated for 2 h in a Biotage microwave reactor (100 °C). After the reaction was complete, the DMF was evaporated under reduced pressure to afford a residue that was subjected to silica gel chromatography (63–210 µm) using DCM:methanol (9:1). After evaporation of the volatile materials, the residue (40 mg) was taken up in dry DMF (1.5 mL), azide 1018 (57 mg, 230 µmol), TBTA (8 mg, 16 µmol), and tetrakis(acetonitrile)copper(I) hexafluorophosphate (6 mg, 16 µmol) were added, and the reaction mixture was irradiated for 6 h in a Biotage microwave reactor (100 °C). Following workup as described above for compounds 14c–e and lyophilization, the residue was fractionated by preparative HPLC (10→90% acetonitrile in water containing 0.1% TFA within 50 min, tR 29 min) to afford 15a as white solid; yield 14 mg (4.1 µmol, 17%). Product 15a was analyzed by MALDI-TOF (see Figure S7 in the Supplementary Data) and by UV spectroscopy.18
2.1.4.6. Procedure for Reaction of Azide-functionalized CCK4 Derivative 11 and Azide-functionalized Serine Amide Derivative 10 with 9 to Produce Multivalent Constructs 15b–d
A mixture of 9 (variable amounts, see Table 3), azide 11 (variable amounts, see Table 3), TBTA (0.43 mg/µmol of 9), and tetrakis(acetonitrile)copper(I) hexafluorophosphate (0.3mg/µmol of 9) in dry DMF (1.5 mL) was irradiated for 2 h in a Biotage microwave reactor (100 °C). Azide 1018 (variable amounts, see Table 3) was then added to the reaction mixtures and irradiation was resumed for another 4 h. After the reactions were complete, water (2.5 mL/µmol of 9) was added and the mixtures were extracted with CHCl3 containing dithizone (20 mg/L, 2 × 15 mL) to remove copper.35 The water layers were then washed with DCM (2 × 15 mL) to remove any remaining 10 and TBTA. After lyophilization, the residues were fractionated by preparative HPLC (10→90% acetonitrile in water containing 0.1% TFA within 50 min, tR 22–25 min) to afford products 15b–d as white solids; yields are given in Table 3. Products were analyzed by UV spectroscopy.18
Table 3.
Synthesis of 15a–d.
| Product | mg 9 | mg 11 (equiv) | mg 10 (equiv) | Yield, mg (%) | MALDI-TOF |
|---|---|---|---|---|---|
| 15a | 205 | 30 (0.2) | 57 (10)a | 14 (17)a | Figure S7 |
| 15b | 17 | 25 (2) | 50 (12) | 15 (23) | nrb |
| 15c | 19 | 41 (3) | 47 (10) | 11 (13) | nrb |
| 15d | 11 | 40 (5) | 27 (10) | 15 (26) | nrb |
Equivalents and yield based on 40 mg (23 µmol) of purified mono-CCK4 intermediate; see Experimental Section for details.
MALDI-TOF spectra showed no ions that could be identified as belonging to product. Yield determined by sample weight and UV spectroscopy.
2.2. Biological studies
2.2.1. Formulation of Solutions
Solutions of MSH4 and CCK4 constructs for binding assays were made up in water and dimethylsulfoxide (HYBRI-MAX), respectively, based on the expected incorporations of MSH4 or CCK4. Concentrations of ligand in solution were then determined by measurement of the UV absorbance at 280 nm using a standard calibration plot.18
2.2.2. Binding Assays
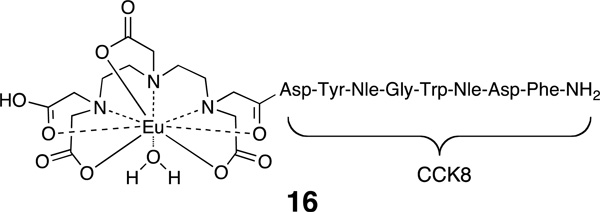
Quantitative receptor-binding assays were carried out following a previously described method.11 Hek293 cells engineered to express both CCK2R and MC4R were used to assess ligand binding. Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum. For MC4R assays, cells were seeded in PerkinElmer, tissue culture treated, black-frame with white well, 96-well plates (part # 6005060) at a density of 1.5 × 104 cells per well and were allowed to reach 80–90% confluence. For CCK2R assays, 2.5 × 105 cells were plated in each well of SigmaScreen poly-o-lysine coated 96-well, black/clear bottom plates (catalog # M5307-SEA) and incubated for 48 h at 37 °C. On the day of the experiment, media was removed from all wells. Ligands were diluted in binding buffer (MEM, 25 mM Hepes [pH 7.4], 0.3% BSA, 1 mM 1,10-phenanthroline, 0.5 mg/L leupeptin, and 200 mg/L bacitracin). Test compound solutions (50 µL), in a range of dilution concentrations, were added to 50 µL of a 10 nM solution of probe 4 (MC4R assay) or a 2 nM solution of probe 16 (CCK2R assay) were added to each well and each concentration was tested in quadruplicate. Cells were incubated in the presence of unlabeled and labeled ligands at 37 °C and 5% CO2 for 1 h. Following incubation, media was removed and the wells washed 3× with wash buffer (50 mM tris(hydroxymethyl)aminomethane hydrochloride, 0.2% BSA, 30 mM NaCl). Enhancement solution (PerkinElmer 1244-105) was added (100 µL/well) and plates were incubated for 30 minutes at 37 °C before measuring fluorescence using a VICTOR™ X4 2030 Multilabel Reader (PerkinElmer) instrument and the standard Eu time-resolved fluorescence (TRF) measurement settings (340 nm excitation, 400 µs delay, and emission collection for 400 µs at 615 nm).
Competitive binding data were analyzed with GraphPad Prism software using nonlinear regression analysis and fitted to a classic one site binding competition equation. Each EC50 value was generated from individual competitive binding assays and converted to a Ki value using the equation Ki = EC50/(1 + ([ligand]/KD)) where [ligand] refers to the concentration of the probe used as the labeled competed ligand. For probe 4, [ligand] = 10 nM and KD = 8.3 nM. For probe 16, [ligand] = 2 nM and KD = 34.6 nM. Results are given in Table 4. The value given represents the average of n independent competition binding experiments.
Table 4.
Competitive binding of MSH4, CCK4, 12, 13, 14a–14e, 15a–15d, and 17 to MC4R or CCK2R.
| Compound | MC4R | CCK2R | ||
|---|---|---|---|---|
| Kia (µM) | nb | Kia (nM) | nb | |
| MSH4 | 1.3 ± 0.38 | 5 | ndc | |
| 17 | 1.9 ± 0.14 | 5 | nd | |
| 14a | 7.3 ± 1.1 | 4 | nd | |
| 14b | 1.6 ± 0.16 | 4 | nd | |
| 14c | 0.54 ± 0.04 | 4 | nd | |
| 14d | 0.23 ± 0.02 | 4 | nd | |
| 14e | 0.17 ± 0.02 | 4 | nd | |
| CCK4 | nd | 3.1d | ||
| 12 | nd | 18 ± 5.7 | 3 | |
| 15a | nd | 67 ± 9.4 | 3 | |
| 15b | nd | 1.5 ± 0.7 | 3 | |
| 15c | nd | 2.0 ± 0.3 | 3 | |
| 15d | nd | 0.80 ± 0.2 | 3 | |
| 13 | nbe | 3 | nbe | 2 |
Ki values were calculated using the equation Ki = EC50/(1 + ([ligand]/KD)) where [ligand] = 10 nM and KD = 8.3 nM for probe 4 and [ligand] = 2 nM and KD = 34.6 nM for probe 16.
The value given represents the average of n independent competition binding experiments, each done in quadruplicate.
Not determined.
Taken from Reference 30.
Compound 13 was unable to inhibit the binding of probe 4 or probe 16 in the concentration range tested (10−5–10−12 M in serine amide).
3. Results
3.1. Chemistry
Reaction of sucrose with 24 equivalents of sodium hydride in DMF, followed by addition of 12 equivalents of 6-bromo-1-hexyne33 afforded octaalkyne 9 in 43% yield after chromatography (Scheme 2). Azides 1 and 10 were prepared as previously described.18 Azide 11 was prepared by solid phase synthesis36,37 on Rink amide Tentagel S resin as depicted in Scheme 3. 6-Azidohexanoic acid34 was coupled to the N-terminus of the resin-bound tetrapeptide. Simultaneous side chain deprotection and cleavage of the tetrapeptide from the resin was effected using a mixture of trifluoroacetic acid, triisopropylsilane, thioanisole, and water (91:3:3:3), producing the desired azideterminated ligand, N3(CH2)5(C=O)-Trp-Met-Asp-Phe-NH2 (11). The triazole-containing ligand CH3(CH2)3(C2N3)(CH2)5(C=O)-Trp-Met-Asp-Phe-NH2 (12) was prepared by N-terminal acylation of the resin-bound tetrapeptide with 6-(4-butyl-1H-1,2,3-triazol-1-yl)hexanoic acid18 in place of 6-azidohexanoic acid. Compounds 11 and 12 were purified by reversed phase C18 preparative HPLC (yields ranged from 40–45%) and were characterized by analytical HPLC and ESI-MS. Details appear in Table 1.
Reaction of 9 with an excess of 1018 in the presence of tetrakis(acetonitrile)copper(I) hexafluorophosphate and tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) in methanol in a Biotage microwave reactor at 100 °C gave the corresponding serine amide octamer 13 (Scheme 4). Copper ions were removed from this mixture by complexation with dithizone and removal of the complex by extraction with CHCl3.35 Other small organic-soluble molecules (TBTA, excess 10) were also removed during this extraction. The water-soluble product 13 was purified by preparative reversed phase HPLC, recovered by lyophilization, and characterized by MALDI-TOF mass spectrometry (see Figure S1 in the Supplementary Data).
Reaction of 9 with varying amounts of 1 in the presence of copper(I) and TBTA in methanol in a Biotage microwave reactor at 100 °C, followed by reaction either with an excess of 1-azidohexane or with the serinamide-derived azide 10 under the same conditions, gave the corresponding multimeric mixtures 14a–14e. Copper ions and small organic molecules were removed as previously described. The crude product mixtures were further purified by preparative reversed phase HPLC and were characterized by MALDI-TOF mass spectrometry (see Figure 3 and Figures S2–S6 in the Supplementary Data). The average numbers of R(C=O)-MSH4-NH2 ligands per scaffold were determined to be 1.0, 1.0, 1.3, 2.6, and 4.1 for the multimeric mixtures 14a–14e, respectively, by UV spectroscopy (see the Discussion Section and the Supplementary Data for details).18
Similarly, reaction of 9 with varying amounts of azide 11 in the presence of copper(I) and TBTA in DMF in a Biotage microwave reactor at 100 °C, followed by reaction with an excess of azide 10 under the same conditions, gave the corresponding multimeric mixtures 15a–15d. Copper ions and small organic molecules were removed as previously described. The crude product mixtures were further purified by preparative reversed phase HPLC and were characterized, in the case of 15a, by MALDI-TOF mass spectrometry (see Figure S7 in the Supplementary Data). Satisfactory mass spectra of 15b–15d could not be obtained. The average numbers of R(C=O)-CCK4-NH2 ligands per scaffold were determined to be 1.0, 1.3, 2.2, and 3.6 for the multimeric mixtures 15a–15d, respectively, by UV spectroscopy.18
3.2. Bioassays
Hek293 cells overexpressing both MC4R22,38 and 30,39 were used to assess ligand binding using previously described europium-based competitive binding assays that employed Eu-DTPA-NDP-α-MSH-NH2 (4) or Eu-DTPA-CCK8-NH2 (16) as the labeled probe.11 The Ki values for the parental ligands CH3C(C=O)-His-dPhe-Arg-Trp-NH2 (listed as MSH4) and Trp-Met-Asp-Phe-NH2 (listed as CCK4), for the corresponding triazole-containing control compounds 12 and 1718, and for the constructs 13, 14a–14e, and 15a–15d are listed in Table 4.
4. Discussion
The one-step synthesis of octaalkyne 9 starting from sucrose was reasonably efficient and can provide access to multigram quantities of this product. Presumably, the identities of the sugar starting material and the alkylating agent can be varied to produce many such scaffolds. Microwave-driven CuAAC was used to attach zero, one, or an average of 1.3, 2.6, or 4.1 copies of the MSH4 azide 1 to scaffold 9. The remaining alkyne residues of the scaffold were then reacted with 1-azidohexane to produce 14a, or with serinamide-derived azide 10 to produce 13 and 14b–e. The constructs so produced were purified from copper and from small molecules by extraction, further purified by preparative reversed phase HPLC, and characterized by MALDI-TOF mass spectrometry and by UV spectroscopy. While MALDI-TOF analysis was reasonably consistent with statistical attachment of ligands in the case of 14c, the mass spectra for 14d and 14e suggested that ligand attachment was not statistical (see Figure 3 and Figures S4–S6 in the Supplementary Data). MALDI-TOF analysis of a 1:1 mixture of monovalent and divalent MSH4 constructs on a squalene scaffold17,27 demonstrated a highly attenuated sensitivity for detection of the divalent construct. Thus, MALDI-TOF analysis of multimers 14c–14e is qualitative, with actual distributions of MSH4 ligands somewhere between the MALDI-TOF limit of analysis and the expected statistical distributions. For this reason, concentrations of multivalent species for bioassays were determined by UV analysis (see calculations in the Supplementary Data).
Constructs 15a–15d bearing one or an average of 1.3, 2.2, or 3.6 CCK4 ligands were similarly prepared from scaffold 9, CCK4 azide 11, and azide 10. In principle, other ligands, imaging agents, and/or therapeutic agents might be attached to scaffold 9.
Constructs 13, 14a–e, and 15a–d were subjected to biological testing using previously described competitive binding assays.11 Serinamide derivative 13 was ineffective at blocking probes 4 and 16 from binding to cells displaying MC4R and CCK2R over the range of concentrations tested (Table 4). The Ki for the monovalent MSH4 control compound 17 was 1.5 times the value for the parental ligand, indicating that attachment of the triazole-containing “spacer” to the N-terminus of MSH4 has a modest detrimental effect on ligand binding to MC4R.18 The Ki values differed among the various sucrose-derived constructs 14a–e. Comparison of the Ki values for 14a and 14b suggests that termination of the non-MSH4-bearing sidechains with the more hydrophilic serinamide residues, as opposed to hydrophobic alkyl groups, enhances construct binding. That this enhancement is not due to specific binding by the serinamide residues is supported by the inactivity of compound 13. While the Ki values decrease with higher levels of MSH4 incorporation (compare 14b–e), the observed changes can be attributed to statistics and proximity effects and suggest effective, but monovalent binding of 14b–e at available MC4R. These results are consistent with results from solanesol-derived multivalent MSH4 constructs18 and divalent MSH4 constructs derived from a flexible linker.8 One might suppose that 14b–e are competent binders as monovalent species, but are incompetent binders as multivalent species due to improper ligand spacing and/or presentation for binding.40 However, as to ligand spacing, estimates of the distance between ligand binding sites for adjacent receptors range from 20–50 Å.9 If a spherical distribution of ligands is assumed for these sucrose-derived multivalent constructs, the maximum distance between ligands is approximately 40 Å.41 If a linear array of ligands is assumed for the solanesol-derived multivalent constructs, the distances between ligands are variable with a maximum separation of approximately 90 Å.42 As to ligand presentation, it seems unlikely that all of the MSH4 multivalent and bivalent constructs studied would improperly present a second ligand for binding, given the wide range of permitted N-terminal and C-terminal modifications of this ligand43 and the fact that the observed binding affinities for the first ligand are comparable to the parental ligand.
Monovalent binding of multivalent MSH4 constructs would be expected if the off rates of monovalently bound constructs are faster than the binding of a second ligand arm to MC4R on the cell surface. The NDP-α-MSH ligand, upon which 4 is based, is known to have a slow off-rate (t½ ~ 8 h).44 The off rates of MSH4 and related constructs, such as 14a–b and 17, are unknown, but are expected to be much larger than the rates for NDP-α-MSH and presumably for 4. For rigid linkers that are too short to bridge between adjacent receptors, small increases in avidity have been attributed to statistically- and proximity-enhanced rebinding.7 In these cases, the shorter the linker the greater the avidity.6,7 These observations are consistent with fast off-rates for monovalently bound MSH4 constructs.
Another factor that may inhibit multivalent binding of 14c–e is reduction in the number of MC4R receptors at the cell surface due to receptor cycling. In support of this point, internalization of probe 4 contributes significantly to the fluorescence measured in these assays, which may more properly be termed “binding and uptake” assays. In a preliminary study that compared measured fluorescence at 37 °C and 4 °C, as much as 90% of the fluorescence at 37 °C was attributable to internalized probe.45
Results from assays with CCK4 constructs 15a–d stand in contrast with results from assays with MSH4 constructs 14b–e. The Ki for the monovalent CCK4 control compound 12 was six times the value for the parental ligand, indicating that attachment of the triazole-containing “spacer” to the N-terminus of CCK4 has a significant detrimental effect on ligand binding to CCK2R. The Ki values differed among the various sucrose-derived constructs 15a–d. The Ki value for 15a which bears one copy of the CCK ligand was 22 times the value for the parental ligand and 3.7 times the value for the control compound 12. Apparently, attachment of the CCK4 ligand to the sucrose-derived scaffold has a much greater negative effect on binding than was observed for MSH4. This observation is in keeping with the more restrictive limits on N-terminal substitution for this ligand.46 The Ki values decrease with higher levels of CCK4 incorporation, with 15b–d of comparable potency and each 30–80 times as potent as 15a, suggestive of multivalent binding. Since the affinity of the parental CCK4 ligand for its receptor is 420 times that of the parental MSH4 ligand for its receptor, it seems reasonable to assume that the off-rates of 15a–d are lower than the off-rates for 14b–e, perhaps providing sufficient time for a second binding event to occur before dissociation of the monovalently bound construct. The fact that the greater multivalency of 15d does not more considerably enhance the avidity observed for 15c or 15b may be a consequence of the spatial distribution of the CCK4 ligands attached to this spherical scaffold, which necessitates that on average half of the ligands must be oriented away from the cell surface. This fact limits the number of simultaneous attachments that can be made through ligand binding. For a fully loaded sucrose-derived scaffold bearing eight copies of ligand, there can be at most four simultaneous attachments. For steric and/or electrostatic reasons, it is unlikely that full scaffold loading with ligand could commonly be achieved. Assuming half loading (four ligands per scaffold) or less, as is the case for 14c–e and 15b–d, the analysis above suggests that bivalent attachment is a more realistic expectation.
Supplementary Material
Acknowledgments
This work was supported by grants R33 CA 95944, RO1 CA 97360, RO1 CA 123547, and P30 CA 23074 from the National Cancer Institute.
Abbreviations
- Boc
tert-butoxycarbonyl
- BSA
bovine serum albumin
- CCK2R
cholecystokinin 2 receptor
- CCK4
Trp-Met-Asp-Phe-NH2
- Cl-HOBt
6-chloro-1-hydroxybenzotriazole
- CuAAC
copper(I)-catalyzed azide-alkyne cycloaddition
- DCM
dichloromethane
- DIC
diisopropyl carbodiimide
- DMEM
Dulbecco’s Modified Eagle Medium
- DMF
N,N-dimethylformamide
- DTPA
diethylenetriaminepentaacetic acid
- EC50
effective concentration, 50%
- ESI
electrospray ionization
- Fmoc
9-fluorenylmethyoxycarbonyl
- HOBt
1-hydroxybenzotriazole
- HRMS
high resolution mass spectroscopy
- MALDI-TOF
matrix-assisted desorption/ionization time-of-flight
- MC4R
melanocortin 4 receptor
- MEM
Minimum Essential Medium
- MSH4
Ac-His-dPhe-Arg-Trp-NH2
- NDP-α-MSH
Ser-Tyr-Ser-Nle-Glu-His-dPhe-Arg-Trp-Gly-Lys-Pro-Val-NH2
- TBTA
tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine
- t-Bu
tert-butyl
- TFA
trifluoroacetic acid
- THF
tetrahydrofuran
- TLC
thin-layer chromatography
- TRF
time-resolved fluorescence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data associated with this article (mass spectra of multivalent constructs 13, 14a–e, and 15a, analysis of ligand distribution for 14c–e and 15b–d, and 1H and 13C NMR spectra of compounds 9 and 13) can be found, in the online version, at XXX.
References and notes
- 1.Weissleder R, Mahmood U. Radiology. 2001;219:316. doi: 10.1148/radiology.219.2.r01ma19316. [DOI] [PubMed] [Google Scholar]
- 2.Gillies RJ, Hruby VJ. Expert Opin. Ther. Targets. 2003;7:137. doi: 10.1517/14728222.7.2.137. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy PJ, Radda GK. J. R. Soc. Interface. 2005;2:133. doi: 10.1098/rsif.2005.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morse DL, Gillies RJ. Biochem. Pharmacol. 2010;80:731. doi: 10.1016/j.bcp.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Handl HL, Vagner J, Han H, Mash E, Hruby VJ, Gillies RJ. Expert Opin. Ther. Targets. 2004;8:565. doi: 10.1517/14728222.8.6.565. [DOI] [PubMed] [Google Scholar]
- 6.Vagner J, Handl HL, Gillies RJ, Hruby VJ. Bioorg. Med. Chem. Lett. 2004;14:211. doi: 10.1016/j.bmcl.2003.09.079. [DOI] [PubMed] [Google Scholar]
- 7.Vagner J, Handl HL, Monguchi Y, Jana U, Begay LJ, Mash EA, Hruby VJ, Gillies RJ. Bioconjugate Chem. 2006;17:1545. doi: 10.1021/bc060154p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowen ME, Monguchi Y, Sankaranarayanan R, Vagner J, Begay LJ, Xu L, Jagadish B, Hruby VJ, Gillies RJ, Mash EA. J. Org. Chem. 2007;72:1675. doi: 10.1021/jo062276g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handl HL, Sankaranarayanan R, Josan JS, Vagner J, Mash EA, Gillies RJ, Hruby VJ. Bioconjugate Chem. 2007;18:1101. doi: 10.1021/bc0603642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vagner J, Xu L, Handl HL, Josan JS, Morse DL, Mash EA, Gillies RJ, Hruby VJ. Angew. Chem. Int. Ed. 2008;47:1685. doi: 10.1002/anie.200702770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu L, Vagner J, Josan JS, Lynch RM, Morse DL, Baggett B, Han H, Mash EA, Hruby VJ, Gillies RJ. Mol. Cancer Ther. 2009;8:2356. doi: 10.1158/1535-7163.MCT-08-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mammen M, Choi S-K, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:2754. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Kiessling LL, Gestwicki JE, Strong LE. Angew. Chem. Int. Ed. 2006;45:2348. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnamurthy VM, Estroff LA, Whitesides GM. In: Fragment-based Approaches in Drug Discovery. Jahnke W, Erlanson DA, editors. Weinheim: Wiley-VCH; 2006. pp. 11–53. [Google Scholar]
- 15.Carlson CB, Mowery P, Owen RM, Dykhuizen EC, Kiessling LL. ACS Chemical Biology. 2007;2:119. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]
- 16.Choi S-K. Synthetic Multivalent Molecules. Hoboken, NJ: Wiley-Interscience; 2004. [Google Scholar]
- 17.Jagadish B, Sankaranarayanan R, Xu L, Richards R, Vagner J, Hruby VJ, Gillies RJ, Mash EA. Bioorg. Med. Chem. Lett. 2007;17:3310. doi: 10.1016/j.bmcl.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alleti R, Rao V, Xu L, Gillies RJ, Mash EA. J. Org. Chem. 2010;75:5895. doi: 10.1021/jo101043m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The “low-affinity” ligand MSH4, Ac-His-dPhe-Arg-Trp-NH2, was based on the minimal active sequence for full agonist activity of α-MSH; see Hruby VJ, Wilkes BC, Hadley ME, Al-Obeidi F, Sawyer TK, Staples DJ, de Vaux AE, Dym O, de Lauro Castrucci AM, Hintz MF, Riehm JP, Rao KR. J. Med. Chem. 1987;30:2126. doi: 10.1021/jm00394a033.
- 20.Castrucci AML, Hadley ME, Sawyer TK, Wilkes BC, Al-Obiedi F, Staples DJ, de Vaux AE, Dym O, Hintz MF, Riehm JP, Rao KR, Hruby VJ. Gen. Comp. Endocrinol. 1989;73:157. doi: 10.1016/0016-6480(89)90066-x. [DOI] [PubMed] [Google Scholar]
- 21.Haskell-Luevano C, Hendrata S, North C, Sawyer TK, Hadley ME, Hruby VJ, Dickinson C, Gantz I. J. Med. Chem. 1997;40:2133. doi: 10.1021/jm960840h. [DOI] [PubMed] [Google Scholar]
- 22.The MC4R vector was originally received from Dr. Ira Gantz; see Gantz I, Miwa H, Konda Y, Shimoto Y, Tashiro T, Watson SJ, DelValle J, Yamada T. J. Biol. Chem. 1993;268:15174.
- 23.Kolb HC, Finn MG, Sharpless KB. Angew. Chem. Int. Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Gil MV, Arévalo MJ, López Ó. Synthesis. 2007:1589. [Google Scholar]
- 25.Angell YL, Burgess K. Chem. Soc. Rev. 2007;36:1674. doi: 10.1039/b701444a. [DOI] [PubMed] [Google Scholar]
- 26.Holub JM, Garabedian MJ, Kirshenbaum K. QSAR Comb. Sci. 2007;26:1175. [Google Scholar]
- 27.Xu L, Vagner J, Alleti R, Rao V, Jagadish B, Morse DL, Hruby VJ, Gillies RJ, Mash EA. Bioorg. Med. Chem. Lett. 2010;20:2489. doi: 10.1016/j.bmcl.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The “high-affinity” ligand, NDP-α-MSH, is Ac-Ser-Tyr-Ser-Nle-Glu-His-dPhe-Arg-Trp-Gly-Lys-Pro-Val-NH2; see Sawyer TK, Sanfilippo PJ, Hruby VJ, Engel MH, Heward CB, Burnett JB, Hadley ME. Proc. Natl. Acad. Sci. U.S.A. 1980;77:5754. doi: 10.1073/pnas.77.10.5754.
- 29.Hadley ME, Anderson B, Heward CB, Sawyer TK, Hruby VJ. Science. 1981;213:1025. doi: 10.1126/science.6973820. [DOI] [PubMed] [Google Scholar]
- 30.Horwell DC. Neuropeptides. 1991;19:57. doi: 10.1016/0143-4179(91)90083-u. [DOI] [PubMed] [Google Scholar]
- 31.Akoh CC. Food Sci. Tech. 2002;117:695. [Google Scholar]
- 32.Bimal C, Guonong Z. Food Rev. Int. 2006;22:245. [Google Scholar]
- 33.Sharma S, Oehlschlager AC. J. Org. Chem. 1989;54:5064. [Google Scholar]
- 34.Grandjean C, Boutonnier A, Guerreiro C, Fournier J-M, Mulard LA. J. Org. Chem. 2005;70:7123. doi: 10.1021/jo0505472. [DOI] [PubMed] [Google Scholar]
- 35.Caplin S. Tissue Culture Association Manual. 1976;2:439. [Google Scholar]
- 36.Merrifield RB. J. Am. Chem. Soc. 1963;85:2149. [Google Scholar]
- 37.Hruby VJ, Meyer J-P. In: Bioorganic Chemistry: Peptides and Proteins. Hecht SM, editor. New York: Oxford Univ. Press; 1998. pp. 27–64. [Google Scholar]
- 38.Yang X, Wang Z, Dong W, Ling L, Yang H, Chen R. J. Prot. Chem. 2003;22:335. doi: 10.1023/a:1025386022852. [DOI] [PubMed] [Google Scholar]
- 39.Noble F, Wank SA, Crawley JN, Bradwejn J, Seroogy KB, Hamon M, Roques BP. Pharmacol. Rev. 1999;51:745. [PubMed] [Google Scholar]
- 40.Shewmake TA, Solis FJ, Gillies RJ, Caplan MR. Biomacromol. 2008;9:3057. doi: 10.1021/bm800529b. [DOI] [PubMed] [Google Scholar]
- 41.This estimate is based on molecular dynamics studies of compound 13 in a water environment. Studies of components present in 15a–d are in progress.
- 42.This estimate is based on examination of Dreiding molecular models in which the chain is fully extended.
- 43.Holder JR, Haskell-Luevano C. Med. Res. Rev. 2004;24:325. doi: 10.1002/med.10064. [DOI] [PubMed] [Google Scholar]
- 44.Haskell-Luevano C, Miwa H, Dickinson C, Hadley ME, Hruby VJ, Yamada T, Gantz I. J. Med. Chem. 1996;39:432. doi: 10.1021/jm950407s. [DOI] [PubMed] [Google Scholar]
- 45.Xu L. Unpublished work. [Google Scholar]
- 46.de Tullio P, Delarge J, Pirotte B. Current Med. Chem. 1999;6:433. and references cited therein. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.