Abstract
Mycobacterium tuberculosis is the causative agent of human tuberculosis (TB). Mycobacterial secretory protein ESAT-6 induces MMP-9 in epithelial cells neighboring infected macrophages. MMP-9 then enhances recruitment of uninfected macrophages, which contribute to nascent granuloma maturation and bacterial growth. Disruption of MMP-9 function attenuates granuloma formation and bacterial growth. The abundant mycobacterial HSP65 chaperone is the major target for immune response and a critical component in M. tuberculosis adhesion to macrophages. We hypothesized that HSP65 is susceptible to MMP-9 proteolysis and that the resulting HSP65 immunogenic peptides affect host adaptive immunity. To identify MMPs which cleave HSP65, we used the MMP-2 and MMP-9 gelatinases, the simple hemopexin domain MMP-8, the membrane associated MMP-14, MMP-15, MMP-16 and MMP-24, and the glycosylphosphatidylinositol-linked MMP-17 and MMP-25 in our studies. We determined both the relative cleavage efficiency of MMPs against the HSP65 substrate and the peptide sequence of the cleavage sites. Cleavage of the unstructured PAGHG474L C-terminal region initiates the degradation of HSP65 by MMPs. This initial cleavage destroys the substrate-binding capacity of the HSP65 chaperone. Multiple additional cleavages of the unfolded HSP65 then follows. MMP-2, MMP-8, MMP-14, MMP-15 and MMP-16, in addition to MMP-9, generate the known highly immunogenic N-terminal peptide of HSP65. Based on our biochemical data, we now suspect that MMP proteolysis of HSP65 in vivo, including MMP-9 proteolysis, also results in the abundant generation of the N-terminal immunogenic peptide and that this peptide, in addition to intact HSP65, contributes to the complex immunomodulatory interplay in the course of TB infection.
Introduction
M. tuberculosis is an obligate pathogenic bacterial species of the genus Mycobacterium and the causative agent of human tuberculosis (TB). The host immune response to M. tuberculosis antigens plays a key role in determining defense mechanisms against infection [1]. Lung granulomas, organized aggregates of lung epithelial and immune cells, are a hallmark of M. tuberculosis pathogenic process. By encasing mycobacteria, granulomas have been thought to curtail pulmonary and extrapulmonary infection. The latest findings, however, challenge this paradigm [2]. Thus, infected macrophages migrate from granulomas to the lung surface to face the pleural cavity and recruit uninfected macrophages which, in turn, migrate back to granulomas [3]. As a result, infected macrophages induce granuloma formation by promoting recruitment of additional macrophages. Following the death of infected macrophages, newly arriving macrophages phagocytose the debris and become infected. Iteration of these processes transforms an early granuloma into a bacterial expansion site.
Recent evidence suggests that the mycobacterial 6 kDa early secreted protein ESAT-6 induces matrix metalloproteinase–9 (MMP-9) in epithelial cells neighboring infected macrophages and that MMP-9 then contributes to the further recruitment of uninfected macrophages, granuloma maturation and bacterial growth [4, 5]. Because of the unique parameters of its extended promoter region, the MMP-9 gene is readily transcribed in inflammatory diseases [6]. Conversely, silencing of MMP-9 function reduces granuloma and mycobacterial burden suggesting that a better understanding of the MMP-9’s role in M. tuberculosis pathogenesis may contribute to the development of novel diagnostic methods and the identification of novel drug targets in TB [7-11].
MMP-9 is a member of the matrix metalloproteinase (MMP) family. This family consists of 24 zinc proteinases in humans [12-14]. Because of their ability to proteolyze multiple components of the extracellular matrix, cell signaling adhesion receptors, and soluble cytokines and growth factors, MMPs play an important role in normal development and disease. MMPs share common structural motifs including a pro-peptide that maintains the latency of the respective zymogen, a catalytic domain with the zinc-containing active site, a hinge region, and a hemopexin-like domain. Synthesized as pro-enzymes, most MMPs are secreted before conversion to their active form. MMP activities are modulated on several levels including transcription, pro-enzyme activation, or by their endogenous inhibitors, tissue inhibitors of metalloproteinases (TIMPs) [15].
Because of the presence of additional substrate-binding sites in the non-catalytic protein domains, there is a level of diversity in the substrate specificity of MMPs. As a consequence, MMPs have different, albeit significantly overlapping and frequently redundant, biological functions. In contrast with other MMPs and similar with matrix metalloproteinase-2 (MMP-2; Gelatinase A), the catalytic domain of MMP-9 (Gelatinase B) exhibits three inserted fibronectin repeats. Because of the presence of these repeats, MMP-2 and MMP-9 efficiently bind cleave collagen and gelatin [6].
The mycobacterial 65 kDa heat shock protein (HSP65) has been identified as the major clinically important antigen of M. tuberculosis. HSP65 is structurally and functionally similar to the eukaryotic HSP70 chaperon [16, 17]. HSP65 is one of the major cellular proteins produced by mycobacteria. Naturally, the HSP65 antigen is considered to be highly relevant to a subunit vaccine design against mycobacteria. Based on the importance of HSP65 and MMP-9 to the TB pathogenesis, we hypothesized that HSP65 is susceptible to MMP-9 proteolysis and that the resulting HSP65 peptides, especially if they represent the potent antigenic epitopes, play a role in either disease progression or latency or both. Because of the functional redundancy among MMPs and because MMPs distinct and additional to MMP-9 are also likely to play a potential role in the TB immunopathology [9, 11, 18-20], we evaluated multiple MMPs in our studies rather than MMP-9 alone.
We focused our studies on MMPs which are representatives of the major MMP groups including gelatinases (MMP-2 and MMP-9), the simple hemopexin domain MMPs (matrix metalloproteinase-8; MMP-8), the membrane type-MMPs (MT-MMPs), including membrane type-1 matrix metalloproteinase/matrix metalloproteinase-14 (MT1-MMP/MMP-14), membrane type-2 matrix metalloproteinase/matrix metalloproteinase-15 (MT2-MMP/MMP-15), membrane type-3 matrix metalloproteinase/matrix metalloproteinase-16 (MT3-MMP/MMP-16) and membrane type-5 matrix metalloproteinase/matrix metalloproteinase-24 (MT5-MMP/MMP-24), and the glycosylphosphatidylinositol-linked MMPs including membrane type-4 matrix metalloproteinase/matrix metalloproteinase-17 (MT4-MMP/MMP-17) and membrane type-6 matrix metalloproteinase/matrix metalloproteinase-25 (MT6-MMP/MMP-25) [21]. We determined both the efficiency of each MMP in the cleavage of HSP65 and the peptide sequence of the cleavage sites. We demonstrated that, in addition to MMP-9, MMP-2, MMP-8, MMP-14, MMP-15 and MMP-16 proteolysis of the HSP65 antigen generates the highly immunogenic N-terminal peptide. Based on our data, we now suspect that the in vivo cleavage of HSP65 by MMPs, including MMP-9, contributes to the complex immunomodulatory interplay in the course of TB infection.
Results
In vitro cleavage of HSP65 by MMPs
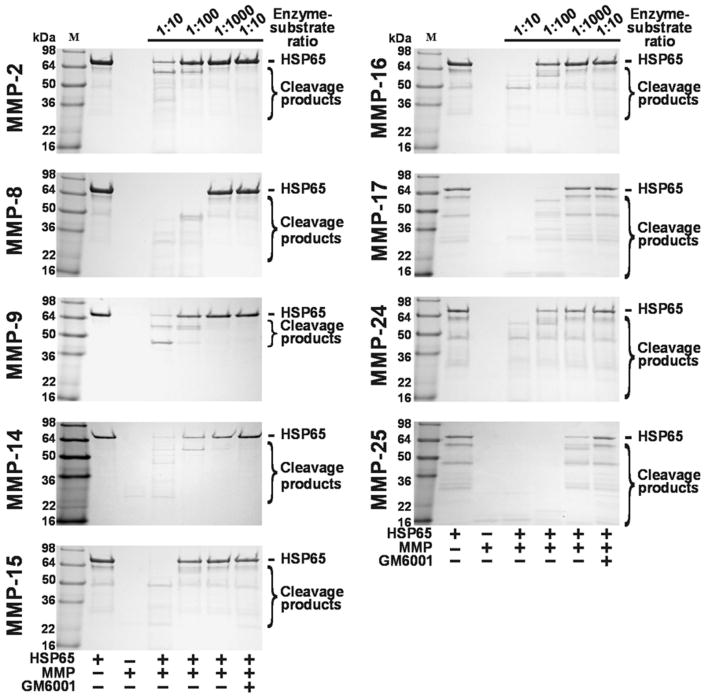
HSP65 (an apparent molecular mass 65-66 kDa) was co-incubated for 1 h at 37°C with the individual MMPs. The digests were separated by SDS-polyacrylamide gel electrophoresis (Fig. 1). Where indicated, the samples included GM6001 (a potent, broad-range inhibitor of MMPs). Our result clearly indicated that HSP65 was sensitive to MMP proteolysis. As expected, GM6001 fully blocked MMP proteolysis of HSP65. MMP-8, MMP-17 and, especially MMP-25, appeared the most efficient in cleaving HSP25 while MMP-2 was least efficient. The proteolysis of HSP65 by MMP-9 and MMP-25 inactivated the HSP65 immunoreactivity and, as a result, only the major high molecular mass digest products were recognized by the HSP65 monoclonal antibody (clone 9L497) (Supplemental Fig. S1).
Figure 1.
In vitro cleavage of HSP65 by MMPs. HSP65 was co-incubated with the indicated amounts of MMPs. The digested samples were analyzed by SDS-polyacrylamide gel electrophoresis. Were indicated, GM6001 (50 μM) was added to the reactions to block MMP activity.
MS analysis of the digest reactions
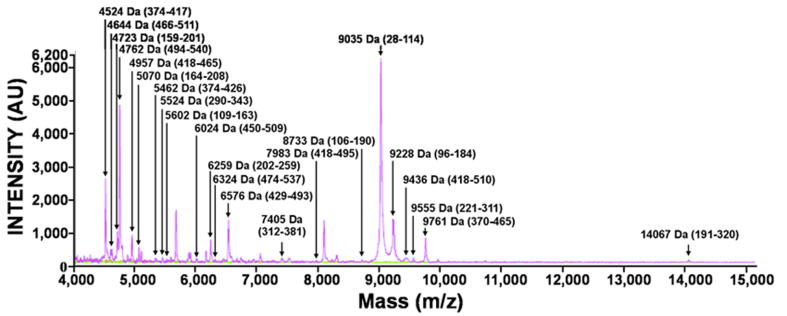
HSP65 was subjected to proteolysis by the individual MMPs. The mass of the digest peptides was then determined using MALDI-TOF MS. The results of a representative MS analysis of MMP-9 proteolysis of HSP65 are shown in Fig. 2. In our analyses, we tried to identify the most representative cleavage fragments rather than to determine the identity of each peptide in the samples. Generally, because of the cleavage preference redundancy among MMPs, multiple individual MMPs are capable of generating, albeit with widely varying kinetics, similar cleavage peptides as a result of the HSP65 proteolysis. To map the experimental cleavage sites we used a cleavage prediction program that predicted both the MMP cleavage sites and the size of the digest fragments [22-24]. These predictions aided in the identification of the size and the sequence of the cleavage fragments. The number of the cleavage sites in HSP65 and, consequently, of the HSP65 peptides we identified varied from 8 for MMP-24 to 37 for MMP-8. These high numbers allowed us to develop a detailed map of the HSP65 proteolysis by the individual MMPs. The digest peptide sequences are presented in Supplemental Table S1. The cleavage maps are shown in Fig. 3.
Figure 2.
Representative MALDI-TOF MS spectra of the cleavage peptides of HSP65. HSP65 was co-incubated with MMP-9. The digests were analyzed using a Bruker Daltonics Autoflex II MALDI TOF TOF mass spectrometer to determine the molecular mass of the resulting peptides. The intact and the digested Hsp65 samples are in red and magenta, respectively. The buffer alone, blue line; MMP-9 alone, green line. The numbers in the parentheses show the HSP65 sequence numbering. Only the low molecular mass fragments are shown.
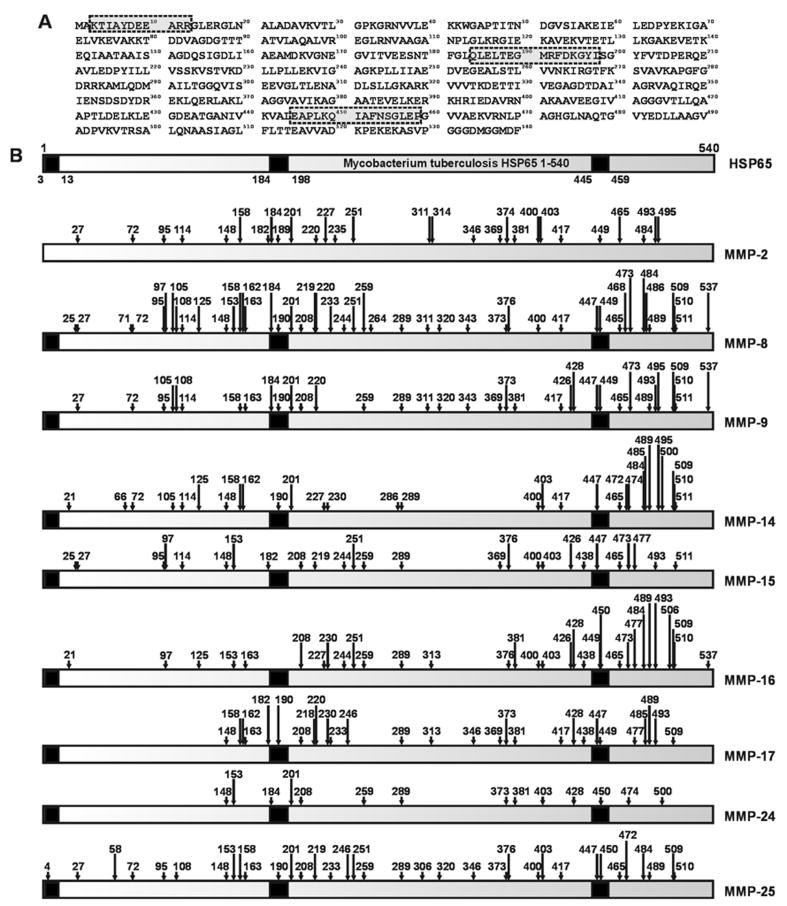
Figure 3.
The cleavage map of HSP65. A. The HSP65 1-540 sequence (M. tuberculosis H37Rv; GenBank accession number CAA17397). The antigenic regions are boxed and shaded in grey. B. The cleavages by MMPs in the HSP65 sequence. The numbers above arrows indicate positions of the cleavage sites. The immunogenic regions (3-13, 184-198 and 445-459) are shown in black.
The P1′–S1′ interaction has been identified as the major determinant of the cleavage position of MMPs in peptide substrates [22, 25, 26]. The study results in agreement with previous works suggesting that MMPs accept a variety of amino acids with hydrophobic and/or aliphatic residues at the P1’ postion. Val and Leu, however, were predominant at the P1′ position − 60% of the peptide’s exhibited either Val or Leu at the P1’. There are a similar number of Leu and Val residues in the peptide sequence of HSP65 (55 and 54, respectively). The Val-Leu ratio at the P1’ was roughly equal for gelatinases (MMP-2 and MMP-9). In turn, the P1’ Leu was preferred over Val by membrane-tethered MMPs (MMP-14, MMP-15, MMP-16 and MMP-24) and GPI-linked MT1-MMPs (MMP-17 and MMP-25). Leu was clearly preferred by MMPs over Ile at the P1’ residue position (Supplemental Table S2). As expected, Pro was frequently detected at the P3 position of the cleavage sites preferred by many MMPs we tested. Multiple amino acid types including Ala, Gly, Glu and Leu were observed at the P2 and P3 positions (Supplemental Tables S3-S5).
We next determined the identity of the major cleavage products generated as a result of MMP-9 proteolysis of HSP65. For this purpose, HSP65 was co-incubated with MMP-9 and, following separation of the digest products using SDS-polyacrylamide gel electrophoresis, the 57-58 kDa and 45 kDa protein bands were excised from the gel, reduced, oxidized and in-gel digested with trypsin. The resulting peptides were analyzed using LC/MS. The sequence data are summarized in Fig. 4. Overall, the sequence coverage exceeded 35%. The identified peptides covered the whole HSP65 peptide sequence except its C-terminal, substrate binding region. From the results of this analysis it becomes clear that the high molecular mass digest products represent the N-terminal portion of HSP65. These results suggest that the initial cleavages of HSP65 by MMPs take place in the C-terminal substrate-binding domain of HSP65, inactivating, as a result, the substrate-binding capacity of the chaperone.
Figure 4.
MMP-9 proteolysis of HSP65. A, In vitro cleavage of HSP65 by MMP-9. HSP65 was co-incubated with the indicated amounts of MMP-9. The digested samples were analyzed by SDS-polyacrylamide gel electrophoresis followed by Coomassie staining. The high molecular mass bands (marked by * and **) were excised from the gel and subjected to in-gel trypsin digestion. B, The tryptic peptides were analyzed by LC/MS/MS. MS/MS spectra were searched against the Swiss-Prot data base using SEQUEST Sorcerer software. The peptides with a probability score of 0.95 and a cross-correlation (Xcorr) value of 2.0 were further analyzed and annotated. The sequence and molecular mass of the annotated peptides are shown. Top and bottom panels, the peptide coverage in the top (*) and bottom (**) HSP65 fragments, respectively. C, Peptide sequence coverage of the 1-540 HSP65 (M. tuberculosis H37Rv; GenBank accession number CAA17397). The regions covered by the annotated peptides are framed and shaded in grey.
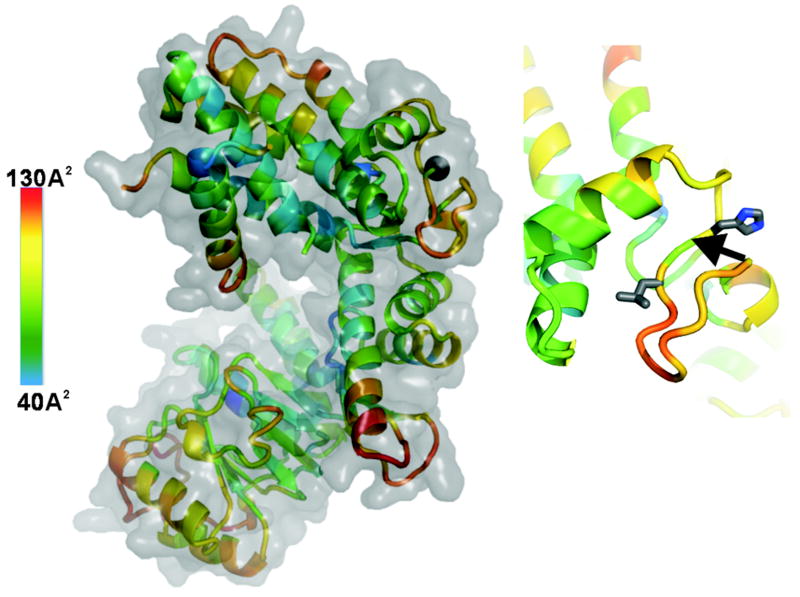
Based on the available structure of HSP65 (PDB entry 1SJP) [16], the peptide sequence and the size of the 57-58 and 45 kDa cleavage fragments, we suggest that VAEK↓V466R and PAGH↓G474L are the first potential sites cleaved by MMP-9 in the HSP65 protein (Fig. 5). The VAEK↓V466R region, however, represents a portion of an α-helix which, normally, is not readily accessible to proteolysis. In turn, the PAGH↓G474L sequence is a fragment of an extended and flexible region which is localized at the surface of the HSP65 molecule. As a result, we believe that the PAGH↓G474L sequence in the flexible region of HSP65 is the primary cleavage site of MMP-9. This initial cleavage site is then followed by additional cleavages leading to the extensive fragmentation of HSP65. Because the cleavage peptides derived from this region were identified for all MMPs we analyzed (except MMP-14), it is also likely that the PAGHG474L region is the primary cleavage site for multiple MMPs rather than for MMP-9 alone.
Figure 5.
Structure of HSP65 (PDB 1SJP) and position of the potential primary cleavage. Left, HSP65. The cartoon is colored according the B-factor distribution. The B-factor scale is on the left: low flexibility, blue; high flexibily, red. The B-factors of protein crystal structures reflect the fluctuation of atoms about their average positions and provide important information about protein dynamics. Black sphere shows the position of the H↓G474 scissile bond. Right, close-up of the GH↓G474L region. The side-chains of the P1’ His and P2 Leu are shown as sticks. Arrow points to the H↓G474 scissile bond. The structure of HSP65 was visualized using PyMol.
Digest peptides with the known antigenic determinants
There are several highly immunogenic regions in the HSP65 peptide sequence including the 3-13, 184-198 and 445-459 peptide sequences from the N-terminal, the central and the C-terminal regions of HSP65. According to our cleavage data, multiple MMPs, including MMP-9, are capable of readily generating the digest peptides which exhibit of the HSP65 antigenic determinants. The 184-198 and 445-459 sequence regions could be generated by MMP proteolysis but they, however, can next be inactivated by the further proteolysis. In contrast, the N-terminal 1-21, 1-25 and 1-27 peptides that included known 3-13 immunogenic sequence region of HSP65 could readily arise as a result of MMP proteolysis including MMP-9 proteolysis.
Discussion
Immune system acts as an effective barrier to infection. Understanding the mechanisms which bacterial pathogens employ to circumvent innate immune systems improves our ability to control the disease. At the initial stage of lung infection, M. tuberculosis adheres to the host alveolar type II epithelial cells. These cells subsequently engulf mycobacteria and support their intracellular replication [27]. The capsule (an outer layer of the cell wall) modulates interactions of M. tuberculosis with host macrophages. Mycobacterial HSP65, a portion of which is associated with the capsule [28], plays an important, however incompletely understood, role in these interactions [28-32]. It is not clear if an intracapsular pool of HSP65 results because of the re-binding of the released cellular HSP65 or because of the specific trafficking of HSP65 to the bacterial surface. Regardless of the delivery mechanisms, both the extracellular and intracapsular HSP65 pools represent critical components in the adhesion and phagocytosis processes of M. tuberculosis. There is also a body of evidence that HSP65 provides other essential functions in M. tuberculosis pathogenesis, including its chaperone function. HSP65 is also the main mycobacterial antigen associated with establishing the immune response by the host.
MMPs are important regulators in the inflammatory response and tissue destruction in multiple diseases, including TB. MMPs and, especially MMP-9, are frequently up-regulated in inflammatory conditions [6]. This up-regulation then contributes to recruitment of the blood cells including leukocytes, macrophages and neutrophils which bring their own endogenous MMPs into the affected tissue. It becomes increasingly clear now that disruption of the host MMP-9 function attenuates granuloma formation [5, 8, 10, 11]. Based on these previous data, we hypothesized that MMP-9 cleaves HSP65 and that the resulting peptides play a role in either the granuloma maintenance or the disease spread outside of granulomas or both. Because there is a level of functional redundancy among MMPs, we evaluated several individual MMPs rather than MMP-9 alone in our HSP65 cleavage studies.
The use of mass spectrometry allowed us to identify the molecular mass and, consequently, the sequence of the resulting HSP65 digest peptides and the affected scissile bonds. As a result, we now know that HSP65 can be efficiently fragmented by multiple MMPs. Because MMP-9 is specifically up-regulated in granulomas, this proteinase, most probably, initiates the proteolysis of the cellular HSP65 pools that is released by the cells into the extracellular milieu. It is likely that the proteolysis of HSP65 ensues from the initial cleavage of the flexible PAGHG474L C-terminal region in the substrate-binding domain of HSP65. This initial cleavage unavoidably inactivates the substrate-binding capacity of the chaperone. Multiple additional cleavages of the partially unfolded HSP65 molecule then follow.
Based on our cleavage data and the results of others [28], it also likely that the exposed substrate-binding C-terminal region of the intracapsular HSP65 is accessible by MMPs, while the N-terminal portion of HSP65 is buried in the capsular compartment of the bacteria. In its reverse orientation, the capsular HSP65 would not be capable of exhibiting its recorded adhesin functionality [28].
HSP65 is obviously a prominent mycobacterial antigen. It exhibits several highly immunogenic regions [30, 31]. According to our results, multiple MMPs seem to be capable of generating the peptides which span the highly immunogenic 3-13 region. It may be suggested that the MMP-9 activity in granulomas [5, 8, 11] also readily generates significant amounts of this immunogenic peptide.
If the results of our studies are combined with the pre-existing knowledge about mycobacterial HSP65, it is tempting to hypothesize that either intact secreted HSP65 or its digest fragments or both function as a decoy in the extracellular milieu to attract immune cells to the infected site. In principle, this suggestion is consistent with the observations by others including the studies of the host adaptive immune response towards the M. tuberculosis Ag85B antigen in mice [33]. Hence, to evade the detection by the antigen-specific CD4+ T cells, the pathogen transfers its antigens from the infected cells to the neighboring uninfected cells in granulomas. As a result, the uninfected cells act as a decoy to rescue the infected host cells. Our suggestion that soluble HSP65 and/or its digest fragments perform a decoy function in order to misdirect the host immune system from attacking mycobacteria warrants additional studies in model systems in vivo.
Materials and Methods
Reagents
All reagents unless otherwise indicated were from Sigma. A hydroxamate inhibitor of MMPs (GM6001) was from Chemicon (Temecula, CA, USA). Another hydroxamate inhibitor of MMPs (AG3340) was a kind gift of Dr. Peter Baciu (Allergan, Irvine, CA, USA). The recombinant purified HSP65 protein that was N-terminally tagged with a Hisx6 tag was purchased from ProSpec-Tany Technogene (East Brunswick, NJ, USA). A monoclonal HSP65 antibody 9L497 was from United States Biological (Swampscott, MA, USA). Trypsin Gold mass-spectrometry grade was from Promega (Madison, WI, USA).
Expression and purification of MMPs
The individual catalytic domains of MMP-8, MMP-14, MMP-15, MMP-16, MMP-17, MMP-24 and MMP-25 were expressed in E. coli, purified from the inclusion bodies in 8 M urea using metal-chelating chromatography [24, 34]. The purified samples were then refolded to restore their native conformation and proteolytic activity [35]. The recombinant pro-forms of the catalytic domains of MMP-2 and MMP-9 were purified from the serum-free medium conditioned by the stably transfected HEK293 cells using the gelatin-column chromatography [36]. Pro-MMP-2 and pro-MMP-9 were activated using 4-aminophenylmercuric acetate as described earlier [36]. The purity of the isolated MMPs was confirmed by SDS-polyacrylamide gel electrophoresis followed by Coomassie staining of the gels. Only the samples the level of purity of which exceeded 95% were used in our subsequent studies. Both the refolded and activated MMPs were used immediately in activity assays.
To standardize the activities of the individual MMPs, the concentrations of catalytically active MMPs in the purified samples were always quantified by active site titration prior to kinetic studies. Active site titration of MMPs was performed with AG3340 (ki = 0.5 - 100 nM for different MMPs) [37]. Briefly, 10 nM MMP was incubated with increasing concentration of AG3340. Residual activity of MMPs was then measured by determining the rate of cleavage of a fluorescent peptide substrate {(7-methoxycoumarin-4-yl)acetyl-Pro-Leu-Gly-Leu-(3-[2,4-dinitrophenyl]-L-2,3-diaminopropionyl)-Ala-Arg-NH2} (Bachem Americas, Torrance, CA, USA). The steady-state rate of the substrate cleavage by MMPs was plotted as a function of inhibitor concentration and fitted with the equation V = SA(E0 - 0.5{(E0 + I + Ki) - [(E0 + I + Ki)2 - 4E0I]0.5}), where V is the steady-state rate of substrate hydrolysis, SA is specific activity (rate per unit of enzyme concentration), E0 is enzyme concentration. I is inhibitor concentration, and Ki is the dissociation constant of the enzyme-inhibitor complex [38]. The data were plotted vs. the amounts of AG3340 and a line was fitted through the data points. The intercept on the x-axis equals to the concentration of active enzyme. The concentrations of active MMPs were in a 20-80% range when compared to the respective protein concentration.
In vitro cleavage reactions and mass-spectrometry analysis
HSP65 (1.5 μg; 1 μM) was co-incubated for 1 h at 37°C with the individual MMPs (1-100 nM; an enzyme-substrate ratio 1:10 – 1:1,000) in 20 μl reactions containing 50 mM HEPES, pH 6.8, supplemented with 10 mM CaCl2 and 50 μM ZnCl2. Where indicated, GM6001 (2.5 mM) was added to the reactions to inhibit MMPs. The cleavage was stopped using a 5xSDS sample buffer. The digest samples were analyzed by SDS-polyacrylamide gel electrophoresis using 4-20% gradient gels, by Western blotting with the HSP65 antibody and by mass-spectrometry using a Bruker Daltonics Autoflex II MALDI-TOF TOF mass spectrometer. For mass spectrometry analysis, the reactions were cooled on ice and equal volumes of a sample and of a sinapic acid (20 mg/ml) in 50% acetonitrile-0.1% trifluoroacetic acid solution were co-crystallized directly on the MALDI target plate and allowed to dry for 5 min. Mass spectra were processed with FlexAnalysis 2.4. The singly charged cleavage products, which were observed only in the cleavage reactions but not in the controls, were recorded and processed further.
To determine the peptide sequence of the major high molecular mass digest products, HSP65 (1.5 μg; 1 μM) was co-incubated with MMP-9 (1-100 nM; an enzyme-substrate ratio 1:10 – 1:1,000). The digested samples were separated by SDS-polyacrylamide gel electrophoresis using 4-20% gradient gels. The gels were stained with SimplyBlue SafeStain (Invitrogen, Carlsbad, CA, USA). The samples were then processed by the Proteomics Core facility of the SBMRI. The stained protein bands which corresponded to the high molecular weight digest products were excised were reduced (10 mM Tris(2-carboxyethyl)phosphine, 37°C 30 min), alkylated (20 mM iodoacetamide, 37°C 40 min in dark), and digested with Trypsin Gold, Mass Spectrometry Grade (1:100 w/w ratio; 37°C 16-18 h). The samples were desalted using a SepPack cartridge, dried using a SpeedVac and re-suspended in 0.1 ml 5% formic acid. The resulting peptides were separated into 24 fractions using HPLC with a Strong Cation Exchange column. The 1/10 aliquot of each peptide fraction was analyzed by LC/MS using an LTQ Orbitrap L mass-spectrometer (Thermo Scientific, Waltham, MA, USA) coupled with a C18 column. MS/MS spectra were searched against the Swiss-Prot database using SEQUEST Sorcerer software and the respective tryptic peptide Mycobacterium databases. The peptides with a probability score >0.95 and a cross-correlation (Xcorr) value >2.0 were further analyzed and annotated.
Prediction and ranking of the cleavage sites
To predict the cleavage sites of MMPs in the HSP65 sequence we used a specialized computer program we had developed [22, 23]. The program determines the contribution of each amino acid residue at each of the P3-P2’ positions to the efficiency of the protein proteolysis by a proteinase and assigns a numerical score to every peptide bond in the protein sequence. The score is based on the Positional Weight Matrix (PWM) approach we developed for the individual MMPs using the high volume data from the substrate phage library cleavage. The elements of the PWM define the probability of the presence of each amino acid type at the P3 to P2’ sub-site position of the substrate relative to the cleavage-resistant substrates. The PWM score is a sum of the log2 elements (log-odds) of the P3 to P2’ positions (eq.1-2).
| (1) |
| (2) |
where j is the j-th position near the cleavage site and iAA is the amino acid type. The offset and threshold values are specific for each MMP and they were determined using the 10-fold cross-validation test. A peptide bond is considered to be cleavable if the score value exceeds the threshold value. The offset is the log2 value of the score when the PWM element equals to zero. The data were filtered using the Jnet secondary structure prediction software [39] and the Disopred2 disordered region prediction software [40]. Only those potential cleavage sites which were predicted by both programs to be at the high, 5-9, confidence levels and which are localized in the unstructured and disordered regions were considered further.
Supplementary Material
Supplemental Figure S1. In vitro cleavage of HSP65 by MMP-9 and MMP-25. HSP65 was co-incubated with the indicated amounts of MMPs. The digested samples were analyzed Western blotting with the HSP65 antibody. Were indicated, GM6001 (50 μM) was added to the reactions to block MMP activity.
Supplemental Table S1. MMP proteolysis of HSP65 and a MALDI-TOF MS analysis of the digest fragments. The arrows indicate the scissile bonds.
Supplemental Table S2. The P1’ residue positions in the HSP65 cleavage peptides. PEX, hemopexin.
Supplemental Table S3. The P1 residue positions in the HSP65 cleavage peptides. PEX, hemopexin.
Supplemental Table S4. The P2 residue positions in the HSP65 cleavage peptides. PEX, hemopexin.
Supplemental Table S5. The P3 residue positions in the HSP65 cleavage peptides. PEX, hemopexin.
Acknowledgments
This work was supported by Public Health Service grants CA83017 and CA77470 (to AYS) from the National Cancer Institute and by University of Massachusetts Medical School (to AS).
Abbreviations
- HSP65
65 kDa heat shock protein
- MMP
matrix metalloproteinase
- MS
mass-spectrometry
- MT-MMP
membrane type MMP
- PMW
Positional Weight Matrix
- TB
tuberculosis
References
- 1.Dorhoi A, Reece ST, Kaufmann SH. For better or for worse: the immune response against Mycobacterium tuberculosis balances pathology and protection. Immunol Rev. 2011;240:235–251. doi: 10.1111/j.1600-065X.2010.00994.x. [DOI] [PubMed] [Google Scholar]
- 2.Lesley R, Ramakrishnan L. Insights into early mycobacterial pathogenesis from the zebrafish. Curr Opin Microbiol. 2008;11:277–283. doi: 10.1016/j.mib.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flynn JL, Chan J, Lin PL. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunol. 2011;4:271–278. doi: 10.1038/mi.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aagaard C, Hoang T, Dietrich J, Cardona PJ, Izzo A, Dolganov G, Schoolnik GK, Cassidy JP, Billeskov R, Andersen P. A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat Med. 2011;17:189–194. doi: 10.1038/nm.2285. [DOI] [PubMed] [Google Scholar]
- 5.Volkman HE, Pozos TC, Zheng J, Davis JM, Rawls JF, Ramakrishnan L. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science. 2010;327:466–469. doi: 10.1126/science.1179663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Steen PE, Dubois B, Nelissen I, Rudd PM, Dwek RA, Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9) Crit Rev Biochem Mol Biol. 2002;37:375–536. doi: 10.1080/10409230290771546. [DOI] [PubMed] [Google Scholar]
- 7.Harris JE, Nuttall RK, Elkington PT, Green JA, Horncastle DE, Graeber MB, Edwards DR, Friedland JS. Monocyte-astrocyte networks regulate matrix metalloproteinase gene expression and secretion in central nervous system tuberculosis in vitro and in vivo. J Immunol. 2007;178:1199–1207. doi: 10.4049/jimmunol.178.2.1199. [DOI] [PubMed] [Google Scholar]
- 8.Price NM, Gilman RH, Uddin J, Recavarren S, Friedland JS. Unopposed matrix metalloproteinase-9 expression in human tuberculous granuloma and the role of TNF-alpha-dependent monocyte networks. J Immunol. 2003;171:5579–5586. doi: 10.4049/jimmunol.171.10.5579. [DOI] [PubMed] [Google Scholar]
- 9.Rivera-Marrero CA, Schuyler W, Roser S, Ritzenthaler JD, Newburn SA, Roman J. M. tuberculosis induction of matrix metalloproteinase-9: the role of mannose and receptor-mediated mechanisms. Am J Physiol Lung Cell Mol Physiol. 2002;282:L546–555. doi: 10.1152/ajplung.00175.2001. [DOI] [PubMed] [Google Scholar]
- 10.Sheen P, O’Kane CM, Chaudhary K, Tovar M, Santillan C, Sosa J, Caviedes L, Gilman RH, Stamp G, Friedland JS. High MMP-9 activity characterises pleural tuberculosis correlating with granuloma formation. Eur Respir J. 2009;33:134–141. doi: 10.1183/09031936.00127807. [DOI] [PubMed] [Google Scholar]
- 11.Taylor JL, Hattle JM, Dreitz SA, Troudt JM, Izzo LS, Basaraba RJ, Orme IM, Matrisian LM, Izzo AA. Role for matrix metalloproteinase 9 in granuloma formation during pulmonary Mycobacterium tuberculosis infection. Infect Immun. 2006;74:6135–6144. doi: 10.1128/IAI.02048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez D, Morrison CJ, Overall CM. Matrix metalloproteinases: what do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim Biophys Acta. 2010;1803:39–54. doi: 10.1016/j.bbamcr.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Hadler-Olsen E, Fadnes B, Sylte I, Uhlin-Hansen L, Winberg JO. Regulation of matrix metalloproteinase activity in health and disease. FEBS J. 2011;278:28–45. doi: 10.1111/j.1742-4658.2010.07920.x. [DOI] [PubMed] [Google Scholar]
- 15.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta. 2010;1803:55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qamra R, Mande SC. Crystal structure of the 65-kilodalton heat shock protein, chaperonin 60.2, of Mycobacterium tuberculosis. J Bacteriol. 2004;186:8105–8113. doi: 10.1128/JB.186.23.8105-8113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewthwaite JC, Coates AR, Tormay P, Singh M, Mascagni P, Poole S, Roberts M, Sharp L, Henderson B. Mycobacterium tuberculosis chaperonin 60.1 is a more potent cytokine stimulator than chaperonin 60.2 (Hsp 65) and contains a CD14-binding domain. Infect Immun. 2001;69:7349–7355. doi: 10.1128/IAI.69.12.7349-7355.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elkington P, Shiomi T, Breen R, Nuttall RK, Ugarte-Gil CA, Walker NF, Saraiva L, Pedersen B, Mauri F, Lipman M, et al. MMP-1 drives immunopathology in human tuberculosis and transgenic mice. J Clin Invest. 2011;121:1827–1833. doi: 10.1172/JCI45666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green JA, Elkington PT, Pennington CJ, Roncaroli F, Dholakia S, Moores RC, Bullen A, Porter JC, Agranoff D, Edwards DR, et al. Mycobacterium tuberculosis upregulates microglial matrix metalloproteinase-1 and -3 expression and secretion via NF-kappaB- and Activator Protein-1-dependent monocyte networks. J Immunol. 2010;184:6492–6503. doi: 10.4049/jimmunol.0903811. [DOI] [PubMed] [Google Scholar]
- 20.Rand L, Green JA, Saraiva L, Friedland JS, Elkington PT. Matrix metalloproteinase-1 is regulated in tuberculosis by a p38 MAPK-dependent, p-aminosalicylic acid-sensitive signaling cascade. J Immunol. 2009;182:5865–5872. doi: 10.4049/jimmunol.0801935. [DOI] [PubMed] [Google Scholar]
- 21.Sohail A, Sun Q, Zhao H, Bernardo MM, Cho JA, Fridman R. MT4-(MMP17) and MT6-MMP (MMP25), A unique set of membrane-anchored matrix metalloproteinases: properties and expression in cancer. Cancer Metastasis Rev. 2008;27:289–302. doi: 10.1007/s10555-008-9129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratnikov B, Cieplak P, Smith JW. High throughput substrate phage display for protease profiling. Methods Mol Biol. 2009;539:93–114. doi: 10.1007/978-1-60327-003-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Remacle AG, Shiryaev SA, Oh ES, Cieplak P, Srinivasan A, Wei G, Liddington RC, Ratnikov BI, Parent A, Desjardins R, et al. Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J Biol Chem. 2008;283:20897–20906. doi: 10.1074/jbc.M803762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiryaev SA, Savinov AY, Cieplak P, Ratnikov BI, Motamedchaboki K, Smith JW, Strongin AY. Matrix metalloproteinase proteolysis of the myelin basic protein isoforms is a source of immunogenic peptides in autoimmune multiple sclerosis. PLoS One. 2009;4:e4952. doi: 10.1371/journal.pone.0004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinz A, Jung MC, Duca L, Sippl W, Taddese S, Ihling C, Rusciani A, Jahreis G, Weiss AS, Neubert RH, et al. Degradation of tropoelastin by matrix metalloproteinases--cleavage site specificities and release of matrikines. FEBS J. 2010;277:1939–1956. doi: 10.1111/j.1742-4658.2010.07616.x. [DOI] [PubMed] [Google Scholar]
- 26.Heinz A, Taddese S, Sippl W, Neubert RH, Schmelzer CE. Insights into the degradation of human elastin by matrilysin-1. Biochimie. 2011;93:187–194. doi: 10.1016/j.biochi.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Bermudez LE, Goodman J. Mycobacterium tuberculosis invades and replicates within type II alveolar cells. Infect Immun. 1996;64:1400–1406. doi: 10.1128/iai.64.4.1400-1406.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hickey TB, Thorson LM, Speert DP, Daffe M, Stokes RW. Mycobacterium tuberculosis Cpn60.2 and DnaK are located on the bacterial surface, where Cpn60.2 facilitates efficient bacterial association with macrophages. Infect Immun. 2009;77:3389–3401. doi: 10.1128/IAI.00143-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geluk A, Taneja V, van Meijgaarden KE, Zanelli E, Abou-Zeid C, Thole JE, de Vries RR, David CS, Ottenhoff TH. Identification of HLA class II-restricted determinants of Mycobacterium tuberculosis-derived proteins by using HLA-transgenic, class II-deficient mice. Proc Natl Acad Sci U S A. 1998;95:10797–10802. doi: 10.1073/pnas.95.18.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mustafa AS, Lundin KE, Meloen RH, Shinnick TM, Oftung F. Identification of promiscuous epitopes from the Mycobacterial 65-kilodalton heat shock protein recognized by human CD4(+) T cells of the Mycobacterium leprae memory repertoire. Infect Immun. 1999;67:5683–5689. doi: 10.1128/iai.67.11.5683-5689.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagabhushanam V, Purcell AW, Mannering S, Germano S, Praszkier J, Cheers C. Identification of an I-Ad restricted peptide on the 65-kilodalton heat shock protein of Mycobacterium avium. Immunol Cell Biol. 2002;80:574–583. doi: 10.1046/j.1440-1711.2002.01128.x. [DOI] [PubMed] [Google Scholar]
- 32.Pelizon AC, Martins DR, Zorzella-Pezavento SF, Seger J, Justulin LA, Jr, da Fonseca DM, Santos RR, Jr, Masson AP, Silva CL, Sartori A. Neonatal BCG immunization followed by DNAhsp65 boosters: highly immunogenic but not protective against tuberculosis - a paradoxical effect of the vector? Scand J Immunol. 2010;71:63–69. doi: 10.1111/j.1365-3083.2009.02352.x. [DOI] [PubMed] [Google Scholar]
- 33.Urdahl KB, Shafiani S, Ernst JD. Initiation and regulation of T-cell responses in tuberculosis. Mucosal Immunol. 2011;4:288–293. doi: 10.1038/mi.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiryaev SA, Remacle AG, Savinov AY, Chernov AV, Cieplak P, Radichev IA, Williams R, Shiryaeva TN, Gawlik K, Postnova TI, et al. Inflammatory proprotein convertase-matrix metalloproteinase proteolytic pathway in antigen-presenting cells as a step to autoimmune multiple sclerosis. J Biol Chem. 2009;284:30615–30626. doi: 10.1074/jbc.M109.041244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kridel SJ, Sawai H, Ratnikov BI, Chen EI, Li W, Godzik A, Strongin AY, Smith JW. A unique substrate binding mode discriminates membrane type-1 matrix metalloproteinase from other matrix metalloproteinases. J Biol Chem. 2002;277:23788–23793. doi: 10.1074/jbc.M111574200. [DOI] [PubMed] [Google Scholar]
- 36.Chen EI, Li W, Godzik A, Howard EW, Smith JW. A residue in the S2 subsite controls substrate selectivity of matrix metalloproteinase-2 and matrix metalloproteinase-9. J Biol Chem. 2003;278:17158–17163. doi: 10.1074/jbc.M210324200. [DOI] [PubMed] [Google Scholar]
- 37.Fisher JF, Mobashery S. Recent advances in MMP inhibitor design. Cancer Metastasis Rev. 2006;25:115–136. doi: 10.1007/s10555-006-7894-9. [DOI] [PubMed] [Google Scholar]
- 38.Knight CG. Active-site titration of peptidases. Methods Enzymol. 1995;248:85–101. doi: 10.1016/0076-6879(95)48008-0. [DOI] [PubMed] [Google Scholar]
- 39.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 40.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. In vitro cleavage of HSP65 by MMP-9 and MMP-25. HSP65 was co-incubated with the indicated amounts of MMPs. The digested samples were analyzed Western blotting with the HSP65 antibody. Were indicated, GM6001 (50 μM) was added to the reactions to block MMP activity.
Supplemental Table S1. MMP proteolysis of HSP65 and a MALDI-TOF MS analysis of the digest fragments. The arrows indicate the scissile bonds.
Supplemental Table S2. The P1’ residue positions in the HSP65 cleavage peptides. PEX, hemopexin.
Supplemental Table S3. The P1 residue positions in the HSP65 cleavage peptides. PEX, hemopexin.
Supplemental Table S4. The P2 residue positions in the HSP65 cleavage peptides. PEX, hemopexin.
Supplemental Table S5. The P3 residue positions in the HSP65 cleavage peptides. PEX, hemopexin.