Abstract
Cellular replicases include three subassemblies: a DNA polymerase, a sliding clamp processivity factor, and a clamp loader complex. The E. coli clamp loader is the DnaX complex (DnaX3δδ'χψ), where DnaX occurs either as τ or as the shorter γ that arises by translational frameshifting. Complexes comprised of either form of DnaX are fully active clamp loaders, but τ confers important replicase functions including chaperoning the polymerase to the newly loaded clamp to form an initiation complex for processive replication. The kinetics of initiation complex formation were explored for DnaX complexes reconstituted with varying τ and γ stoichiometries, revealing that τ-mediated polymerase chaperoning accelerates initiation complex formation 100-fold. Analyzing DnaX complexes containing one or more K51E variant DnaX subunits demonstrated that only one active ATP-binding site is required to form initiation complexes, but the two additional sites increase the rate ca.1000-fold. For τ-containing complexes, the ATP analogue ATPγS was found to support initiation complex formation at 1/1000th the rate with ATP. In contrast to previous models that ATPγS drives hydrolysis-independent initiation complex formation by τ-containing complexes, the rate and stoichiometry of ATPγS hydrolysis coincide with those for initiation complex formation. These results show that although one ATPase site is sufficient for initiation complex formation, the combination of polymerase chaperoning and the binding and hydrolysis of three ATPs dramatically accelerates initiation complex formation to a rate constant (25–50 s−1) compatible with double-stranded DNA replication.
Introduction
Cellular DNA replicases are composed of three subassemblies of distinct function: a replicative polymerase, a sliding clamp processivity factor, and a clamp loader 1. The E. coli cellular replicase is the Pol III holoenzyme. The trimeric core polymerase (Pol III) contains the active site for nucleotide addition. The ring-shaped homodimeric sliding clamp (β2) encircles the DNA, tethering the polymerase to the template and conferring the high processivity necessary for replicating chromosomal DNA 2, 3. The 7-subunit clamp loader complex (DnaX complex) is a specialized AAA+ ATPase that opens the β2 ring and places it onto the DNA 4, 5. Once loaded onto the DNA, the sliding clamp binds the polymerase to form an initiation complex capable of processive elongation in the presence of dNTPs 6, 7.
The DnaX complex consists of the products of five genes with stoichiometry of DnaX3δδ′ψχ, and DnaX3δδ′ is the core pentameric ring for clamp loading8, 9. The ψ and χ subunits function peripherally to stabilize oligimerization of the complex, stabilize conformational intermediates associated with clamp loading, and to interact with single-stranded DNA binding protein (SSB4; the E. coli SSB binds DNA as tetrameric unit) 10–12. The core ring is asymmetric, with the DnaX subunits functioning as ATPases. The δ and δ′ subunits have folds similar to DnaX but lack ATP binding sites 13, 14.
The DnaX subunits can be either the full length product of the dnaX gene (τ) or a truncated product (γ). The γ subunit arises by a programmed translational frameshift that eliminates from the C-terminus two of the five domains found in τ 15–17. Both γ and τ contain the domains that contact the other subunits in the DnaX complex and are involved in the clamp loading mechanism 13. Indeed, active clamp-loading complexes can be reconstituted with any combination of τ and γ subunits 5, and the γ-only DnaX complex (γ3 complex) has been used extensively as a model system to study the clamp loading mechanism 18. τ is essential to replisome function. The two C-terminal domains of τ absent from τ bind the replicative helicase DnaB6 19 and the α catalytic subunit of Pol III20. By this contact to Pol III, two τ subunits within the DnaX complex serve to dimerize leading and lagging strand polymerases within the replicase 21–23. Thus, the τ subunits function as organizers of the replisome, linking the polymerases and clamp loader of the Pol III holoenzyme with the DnaB6 helicase and DnaG primase (via its interaction with DnaB6) activities of the replisome 24, 25. The subunit composition of the DnaX complex has long been thought to be τ2γδδ′ψχ (τ2γ complex) 9. All published preparations of DnaX complex purified intact from cells have been mixed τ/γ complexes 26, and γ occupies a unique position within the complex 10.
In addition to its role as replisome organizer, τ enables the DnaX complex to chaperone Pol III to β2 as it is loaded onto the DNA, facilitating efficient formation of the Pol III/β2/DNA initiation complex 27. This chaperoning effect was shown to accelerate initiation complex formation and to greatly reduce the required Pol III concentration. τ serves as a link between Pol III and the SSB-contacting χ subunit, aiding in polymerase progression on SSB4-coated templates 28. The τ subunit also promotes an interaction with SSB independently of the χ subunit that is favorable for initiation complex formation 27.
The clamp loading cycle is driven by both ATP binding and hydrolysis by the clamp loader. Binding of ATP stabilizes a clamp loader conformation with high affinity for both the clamp and primed DNA, facilitating ternary complex formation 29, 30. In this complex the β2 ring is bound in an open conformation, allowing DNA to pass into the ring. Subsequent ATP hydrolysis results in a conformational change that may close the ring around the DNA and decreases the affinity of the clamp loader for the ternary complex, resulting in clamp loader dissociation from the DNA-bound β2 30. Based primarily on detailed mechanistic studies utilizing the γ3 clamp loader complex, clamp loading has been thought to require binding of ATP by each of the three DnaX subunits and hydrolysis of all three of these ATPs 31. Recently, we prepared and analyzed reconstituted DnaX complexes containing all possible combinations of wild-type and non-ATP-binding K51E variant τ or γ subunits, and we discovered that complexes containing a single wild-type ATPase subunit could facilitate Pol III holoenzyme function in vitro 32. It is also well established that the poorly hydrolyzed ATP analogue ATPγS can support initiation complex formation with τ-containing holoenzyme 27, 33–35. These results suggest that τ-mediated chaperoning of Pol III to the β2 clamp provides alternative clamp loading pathways with different requirements for ATP binding and hydrolysis. Therefore, to understand the molecular mechanism of initiation complex formation it is critical to examine the complete process, with τ-containing DnaX complexes (τ-complexes) bound to Pol III throughout the β2 loading and subsequent Pol III-β2 association reaction steps.
In this work, we explored the pre-steady state kinetics for initiation complex formation by the E. coli Pol III holoenzyme. The results reveal the profound effects that chaperoning of Pol III has on the kinetics of initiation complex formation, accelerating the process 100-fold over an unchaperoned reaction and altering the fundamental requirements for ATP binding and hydrolysis. Probing variant DnaX complexes reconstituted with one to three of the DnaX subunits replaced by non-ATP-binding variants demonstrated that only one ATPase site is essential, but the additional sites dramatically enhance initiation complex formation to rates compatible with replication fork progression. Probing the ATP hydrolysis requirements of the system yielded a surprising observation that the DnaX complex slowly hydrolyzes the analogue ATPγS at a rate similar to initiation complex formation. This finding leads us to reexamine previous models for how ATPγS participates in initiation complex formation.
Results
Chaperoning of Pol III by τ-complexes accelerates initiation complex formation 100 fold
The role of the E. coli DnaX complex in loading the β2 sliding clamp processivity factor onto DNA has been well established. We have recently shown that the polymerase Pol III, when tightly bound to τ–containing DnaX complexes, is preferentially attached to newly loaded β2 sliding clamps 27. We hypothesized that this chaperoning of Pol III by τ-complexes enhances the kinetics for formation of initiation complexes over an unchaperoned reaction in the absence of the τ subunit. To test this possibility directly, we measured and compared the kinetics for initiation complex formation with τ3 and γ3 forms of the DnaX complex. We adapted a primer extension assay for Pol III holoenzyme initiation complex formation for rapid mixing experiments in a quench-flow instrument. This assay separates initiation complex formation from subsequent DNA synthesis by mixing, in the absence dNTPs, the Pol III holoenzyme components (Pol III, DnaX complex, and β2) and ATP with 32P-labeled primed DNA template, quenching the initiation complex formation reaction with excess unlabeled primer termini and with ATP-depleting hexokinase, and then extending the 32P-labeled initiation complexes that had formed by adding dNTPs.
Quench-flow kinetics experiments with τ3 complex, sampled in the absence of SSB4 with Pol III concentrations ranging from 3 to 40 nM, showed significant initiation complex formation within the first 125 ms reaction time (Fig. 1A). The data sets were each fit to an observed first order rate constant, kobs, yielding values between 34 and 55 s−1 (Table 1). Under identical conditions, the γ3 complex, which lacks the Pol III-chaperoning function conferred by the τ subunit, showed kobs values for initiation complex formation ranging from 0.20 to 0.56 s−1 (Fig. 1B). Thus, the association between Pol III and the τ subunit accelerates initiation complex formation by approximately100-fold.
Fig. 1. The τ subunit accelerates initiation complex formation ca. 100-fold.
(a) The kinetics for initiation complex formation for the τ3 complex from 0 to 0.125 s, with Pol III concentrations of 3.0 nM (blue), 5.0 nM (cyan), 10 nM (green), 20 nM (red) and 40 nM (black). (b) Kinetics for initiation complex formation for the γ3 complex from 0 to 32 s, with Pol III concentrations of 3.0 nM (blue), 5.0 nM (cyan), 10 nM (green), 20 nM (red) and 40 nM (black). The data sets are each fit to a single exponential rate constant (solid line), yielding the values summarized in Table 1. All of these reactions were conducted without SSB4.
Table 1.
Rate constants for initiation complex formation with τ3 and γ3 complexes without SSB4 at varying Pol III concentrations
| [Pol III] (nM) | γ3 complex kobs (s−1) |
τ3 complex kobs (s−1) |
|---|---|---|
| 3 | 0.20 | 55 |
| 5 | 0.26 | 54 |
| 10 | 0.34 | 35 |
| 20 | 0.43 | 34 |
| 40 | 0.56 | 47 |
If the rate-limiting step for initiation complex formation with the γ3 complex is diffusion of Pol III to the newly loaded β2, then kobs would be predicted to scale proportionally with Pol III concentration. Although the rate constant for initiation complex formation does increase measurably with increasing Pol III, the effect is less than 3-fold over a more than 10-fold range of Pol III concentrations (Table 1). Thus, the diffusion of unbound Pol III to β2-loaded primer/template DNA does not appear to be the sole rate-limiting step for unchaperoned initiation complex formation. For τ3 complex-catalyzed reactions, we observe no dependence on Pol III concentration, consistent with a unimolecular reaction (Table 1).
The τ3 complex data deviate from simple single exponential behavior when time points longer than 0.125 s are sampled (Supplementary Fig. S1). Data sets sampled out to 4 s are well described by double exponential curve fits, yielding the rate constant values summarized in Supplementary Table S1. Double exponential kinetics is indicative of two subpopulations within the sample exhibiting different initiation kinetics. In this case, each data set showed a fast phase with rate constants similar to the single exponential fits for the 0 to 0.125 s data in Fig. 1 and Table 1 and a slow phase with a rate constant of ca. 1 s−1 (Supplementary Table S1). Biphasic kinetic behavior has been reported for the eukaryotic clamp loading and unloading reactions, and the data were interpreted as reflecting two distinct populations of clamp/clamp loader/DNA ternary complexes, one of which can go on to form active initiation complexes 36. The double exponential behavior observed here with the E. coli holoenzyme could reflect a similar competition between reaction pathways that form incomplete or inactive sub-assemblies of holoenzyme components on the DNA and pathways that form initiation complexes active for DNA synthesis. The faster sub-population represented more than half of the total initiation complexes formed in all of the data sets and, because the slower phase is too slow to be biologically relevant, is the focus of data analysis in the main text of this report.
SSB4 increases the maximum percentage of primers extended but does not alter the kinetics of initiation complex formation
We have shown previously that SSB4 stimulates initiation complex formation by the τ3 complex while inhibiting the γ3 complex-catalyzed reaction 27. Here, we tested whether SSB4 affects the rate of initiation complex formation for the τ3 complex by measuring the kinetics for initiation complex formation in the presence and absence of 250 nM SSB4. The results show that SSB4 increases the total fraction of primers extended (Fig. 2). Fitting the curves yields similar rate constants with and without SSB4 (kobs of 6.7 and 5.2 s−1, respectively). Thus, the advantages conferred by SSB4 do not arise from an acceleration of initiation kinetics. Since SSB4 does not impact the initiation kinetics but does increase the overall yield of the assay and therefore its sensitivity, the experiments in the subsequent sections were conducted in the presence of SSB4 unless otherwise noted.
Fig. 2. The τ3 complex forms initiation complexes with similar kinetics with and without SSB4.
The kinetics for initiation complex formation with 250 nM SSB4 and without SSB4 are shown. The solid lines represent fits to single exponential, yielding kobs = 6.7 s−1 for the 250 nM SSB4 data and kobs = 5.2 s−1 for the no SSB4 data.
Only one τsubunit is required to confer enhanced initiation complex formation kinetics
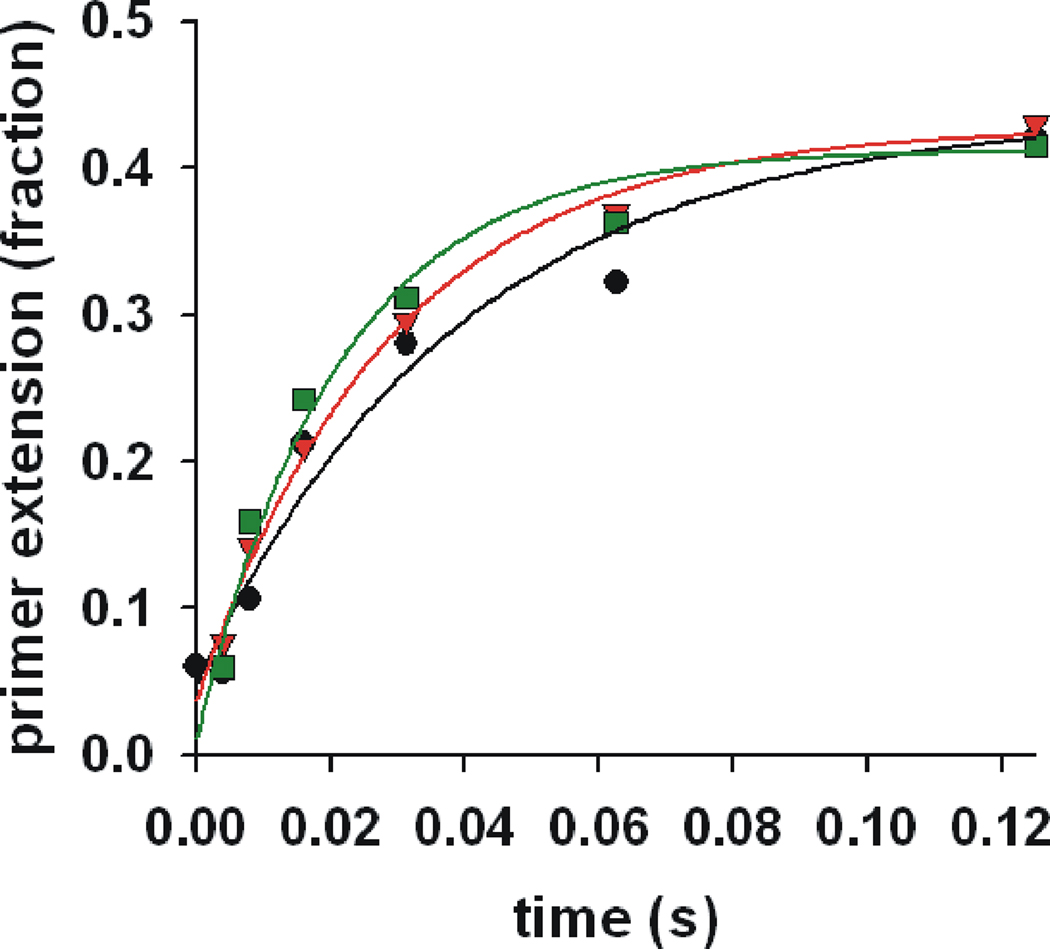
Because the τ3 complex has clear advantages over the γ3 complex, both in the speed of initiation complex formation and in the ability to interact favorably with SSB4, we determined how many τ subunits are required to confer these advantages. The kinetics for initiation complex formation were measured for the τ3, τ2γ, and τγ2 DnaX complexes in the presence of SSB4. All of the τ-containing complexes showed similar kinetics for initiation complex formation (Fig. 3), with kobs values ranging from 26 to 48 s−1 (Table 2). These rate constants for the τ-containing complexes with SSB4 are approximately 100 fold faster than γ3 complex in the absence of SSB4 (Table 1). Thus, one τ subunit in the DnaX complex is required to accelerate initiation kinetics. As has been observed previously for the τ3 complex, both the τ2γ and τγ2 complexes show robust activity with SSB4, demonstrating that a single τ subunit is sufficient to overcome the SSB4 inhibition associated with the γ3 complex. Similar to the τ3 complex in the absence of SSB4, sampling longer time points revealed double exponential behavior for the mixed τ/γ complexes (Supplementary Fig. S2).
Fig. 3. A single τ subunit is sufficient for rapid initiation complex formation.
The kinetics for initiation complex formation were measured for the τ3 (black), τ2γ (red), and τγ2 (green) complexes. The data sets are each fit to a single exponential (solid lines), with the results shown in Table 2. The experiments included 250 nM SSB4.
Table 2.
kobs for initiation complex formation for various τ-complexes with 250 nM SSB4 and ATP or ATPγS.
| DnaX complex | kobs (s−1) |
|---|---|
| τ3-ATP | 26 |
| τ2γ-ATP | 35 |
| τγ2-ATP | 48 |
| τ2γm-ATP | 1.2 |
| τγm2-ATP | 0.010 |
| τ3-ATPγS | 0.020 |
| τ2γ-ATPγS | 0.021 |
| τγ2-ATPγS | 0.027 |
Initiation complex formation requires binding of only one ATP, but binding of the second and third ATPs enhance the kinetics 3500-fold
It is generally thought that clamp loading by the DnaX complex is associated with binding of three ATPs to promote enhanced affinity for β2 and the primer/template and that these ATPs are hydrolyzed upon closing of the β2 ring and release onto the DNA 31. Recently, we prepared an assortment of DnaX complexes with one or more of the DnaX subunits containing a K51E mutation in the Walker A motif that eliminates ATP binding, and we showed complexes containing only one wild-type DnaX subunit could support processive DNA synthesis in a Pol III holoenzyme reconstitution assay 32. Here, we investigated the kinetics for initiation complex formation for DnaX complexes that can bind two, one, or no ATPs, using complexes with two wild-type τ subunits and one γ-K51E subunit (τ2γm complex, where the m signifies “mutant”), one wild-type τ and two γ-K51E subunits (τγm2 complex), and three τ-K51E subunits (τm3 complex), respectively. Since we have shown above that complexes with one, two or three τs have the same kinetics for initiation complex formation, the differences between these complexes should arise primarily from their differences in ATP binding and hydrolysis.
The kinetics for initiation complex formation were examined for these complexes under the same conditions as the all wild-type complexes in the previous section. The results show that τ2γm complex forms initiation complexes with a kobs of 1.2 s−1 (Fig. 4A, Table 2). Thus, the loss of one ATP binding site slows initiation 30-fold relative to the all wild-type τ2γ complex. However, the τ2γm complex remains an effective facilitator of initiation complex formation, forming initiation complexes two-fold faster than wild-type γ3 complex (in the absence of SSB4). This result illustrates the significance of the contribution to the initiation mechanism made by the τ subunit: a clamp loader weakened as an ATPase that chaperones Pol III to the initiation complex is more effective than a fully active clamp loader lacking a τ subunit.
Fig. 4. Multiple ATPase sites in the DnaX complex accelerate initiation complex formation, but only one site is required for activity.
(a) Initiation kinetics with the τ2γm complex (two wild-type and one mutant ATP binding site). (b) Initiation kinetics with the τγ2m complex (one wild-type and two mutant ATP binding sites). Solid lines represent fits to single exponentials, with the results shown in Table 2. The experiments included 250 nM SSB4.
The loss of a second ATP binding site does not eliminate initiation complex formation, but the kobs of 0.010 s−1 for the τγm2 complex is two orders of magnitude slower than that for the τ2γm complex (Fig. 4B, Table 2). The relatively slow initiation complex formation with the τγm2 complex (half-life of 69 s) is sufficient to support the full processive DNA synthesis activity observed previously for this complex in assays employing 5 minute reaction times 32. A DnaX complex that does not bind ATP, τm3 complex, did not show significant initiation complex formation in our assay after up to 20 min of initiation time, even at elevated β2 concentrations. The results for these complexes with K51E mutants confirm that only one active ATPase site is necessary and sufficient for initiation complex formation. The two additional ATP sites are not absolutely necessary to form initiation complexes, but they do accelerate initiation by 3500-fold, pushing the kinetics to a level compatible with replication fork progression.
A single τ subunit is required for initiation complex formation supported by the ATP analogue ATPγS
To further explore the role of ATP binding and hydrolysis in initiation complex formation, we used the ATP analogue ATPγS. It is thought that binding of ATPγS to the DnaX complex mimics the positive allosteric effects of ATP binding in increasing the affinity of the complex for β2 and primed DNA, but ATPγS does not provide the benefits of ATP hydrolysis in promoting closing of the β2 ring around and release onto the DNA 29, 37. ATPγS promotes initiation complex formation with τ-complexes but not with γ3 complexes 27, 34.
Here, we examined how many τ subunits are required for this ATPγS-driven initiation complex formation. As an initial test, single data points were measured for 60 s initiation times with the τ3, τ2γ, τγ2, and γ3 complexes. Since we have shown previously that τ3 complex initiation with ATPγS is greatly enhanced by SSB4 whereas γ3 complex is inhibited by SSB4 27, we tested the complexes both with and without SSB4. We found that, like the τ3 complex, the τ2γ and τγ2 complexes show low levels of initiation activity with ATPγS in the absence of SSB4, and this activity is stimulated by SSB4 (Fig. 5). As we observed previously, γ3 complex initiates poorly with ATPγS in the absence of SSB4 and is inhibited by SSB4. These findings demonstrate that a single τ subunit enables initiation complex formation in the absence of normal ATP hydrolysis and confers a beneficial interaction with SSB4. The results are consistent with a previous observation that complexes with one or more τs support Pol III holoenzyme activity in a replication reconstitution assay 38.
Fig. 5. One τ subunit is sufficient for SSB4-enhanced initiation complex formation with ATPγS.
Polyacrylamide gel electrophoresis analysis of initiation complex formation for the τ3, τ2γ, and τγ2, and γ3 complexes with ATPγS substituted for ATP. The complexes were assayed both without SSB4 (lanes 1–4 from left) and with 250 nM SSB4 (lanes 5–8). The right-most lane shows a control experiment with no polymerase added. The lower bands represent the 32P-5′ end-labeled 30 nt primer, and the upper bands represent the 52 nt primer extension product. The fraction of the primer population extended for each reaction is indicated above its respective lane.
The rate of initiation complex formation is more than 1000-fold slower with ATPγS than with ATP
We next measured the kinetics for initiation complex formation for the various τ/γ complexes with ATPγS to determine how the rates compare to those with ATP. The results show that ATPγS supports initiation complex formation for the τ3, τ2γ, and τγ2 complexes, with kobs in the range of 0.02 to 0.03 s−1 (Fig. 6, Table 2). All three τ-complexes had similar rate constants, but the maximum fraction of primers extended was significantly lower with τγ2 complex than with the other two τ-complexes. This difference was not observed with ATP-driven initiation (Fig. 3), suggesting that the effect arises from the system being challenged by ATPγS.
Fig. 6. τ-containing complexes slowly form initiation complexes in the presence of ATPγS.
The kinetics for initiation complex formation were measured for the τ3 (black), τ2γ (red), τγ2 (green), and γ3 (blue) complexes. The data are fit to single exponentials (solid lines), yielding the kobs values listed in Table 2. The activity was too low with the γ3 complex data for a meaningful curve fit.
Initiation complex formation with each of these DnaX complexes is nearly three orders of magnitude slower with ATPγS than with ATP. If ATPγS is an effective substitute for promoting the positive allosteric effects of ATP binding, then this result illustrates the importance of ATP hydrolysis in accelerating the kinetics of initiation complex formation to a biologically useful rate.
Initiation complex formation coincides with slow hydrolysis of ATPγS
The difference in the rates of initiation complex formation between ATP- and ATPγS-driven reactions was so large that it raised the question of whether slow hydrolysis of ATPγS was contributing to initiation complex formation. The γ3 complex has been shown previously to hydrolyze ATPγS, albeit with a kcat of 1 × 10−4 s−1 that is likely too slow to support initiation complex formation at the rates observed here 35. Since, unlike γ3 complexes, τ-complexes do form initiation complexes with ATPγS, we speculated that the mechanism that enables this to occur might enhance the rate of hydrolysis. We tested for ATPγS hydrolysis associated with initiation complex formation, using thin layer chromatography (TLC) to separate the [35S]thiophosphate hydrolysis product from [γ-35S]ATPγS.
We quantified ATPγS hydrolysis by the τ3, τγ2, and γ3 complexes in the presence of all components used in the initiation assay (Pol III, β2, primer/template, and SSB4). Surprisingly, all three DnaX complexes showed significant ATPγS hydrolysis (Fig. 7, Table 3). The effects of omitting each of the reaction components individually were examined to determine the factors on which ATPγS hydrolysis depends. For each DnaX complex tested, the hydrolysis absolutely required primer/template and was stimulated by β2. These results are consistent with ATPγS hydrolysis arising from DnaX complex activity. Hydrolysis was observed in the presence of Pol III, but the activity was higher when Pol III was omitted. Pol III at the concentration used in this assay does not inhibit initiation complex formation (see Supplementary Fig. S3), so the Pol III effect does not arise from high Pol III levels interfering with normal holoenzyme function. These results are consistent with Pol III forming initiation complexes and sequestering the primer/template (which is the limiting reagent) from serving as substrates for multiple turnovers of ATPγS hydrolysis by the DnaX complex. Surprisingly, the γ3 complex, which does not form productive initiation complexes in the presence of ATPγS, also showed reduced hydrolysis in the presence of Pol III. This finding could indicate that γ3 complex catalyzes formation of an initiation complex, but the complex is rapidly disassembled by ATPγS bound γ3 complex (see Discussion). Alternatively, a stable initiation complex-like assembly could be formed with ATPγS that is incompetent for DNA synthesis and is therefore not detected by our primer extension assay.
Fig. 7. DnaX complexes hydrolyze ATPγS with β2 and DNA dependence.
TLC analysis of 15 min ATPγS hydrolysis reactions with τ3, τγ2, and γ3 complexes. The effects of Pol III, β2, primer/template (labeled “DNA”), and SSB4 were tested by omitting each component individually, as indicated. The fraction of ATPγS hydrolyzed to thiophosphate (labeled PO3S3−) for each lane is reported in Table 3.
Table 3.
Fraction of 1.0 µM ATPγS hydrolyzed by 0.10 µM of various DnaX complexes in 15 min1.
| DnaX complex | all components |
−Pol III | −β2 | −DNA | −SSB4 |
|---|---|---|---|---|---|
| τ3 | 0.10 | 0.66 | 0.02 | 0.0 | 0.23 |
| τγ2 | 0.055 | 0.64 | 0.039 | 0.0 | 0.12 |
| γ3 | 0.15 | 0.70 | 0.011 | 0.0 | 0.094 |
The 1% background observed in the –DnaX lane in Fig. 7 is subtracted from these values.
SSB4 does not prevent ATPγS hydrolysis by the DnaX complex, but omitting SSB4 results in higher levels of hydrolysis. SSB4 has little effect on hydrolysis by the τγ2 complex in the absence of Pol III, whereas increasing levels of SSB4 caused a reduction in hydrolysis in the presence of Pol III (Supplementary Fig. S4). This result is consistent with SSB4 stimulating initiation complex formation and thus preventing multiple turnovers of ATPγS hydrolysis by the clamp loader complex.
To estimate the rate of ATPγS hydrolysis, a time course was run with τγ2 complex in a reaction including Pol III, β2, primer/template, and SSB4. Theτγ2 complex was selected so that the τ subunits would be saturated with Pol III under the conditions of 2-fold excess of Pol III over DnaX complex. After 32 min, 10% of the 1.0 µM ATPγS had been hydrolyzed (Fig. 8A). Fitting the data from the first 8 min to a straight line yields an estimate of 0.09 nM s−1 for the initial rate of hydrolysis. A parallel primer extension assay was run to determine the rate of initiation complex formation with the same component concentrations as the hydrolysis assay (Fig. 8B). A linear fit of the 0 to 8 min data for initiation complex formation yields a rate of 0.02 nM s−1. Taking the ratio of these values gives an estimate 4.5 ATPγS molecules hydrolyzed per initiation complex formed. This value is in reasonable agreement with the hydrolysis of three ATPγS that would be expected if the hydrolysis is associated with one turnover of clamp loading and initiation.
Fig. 8. ATPγS is hydrolyzed by the τγ2 complex with kinetics and stoichiometry consistent with initiation complex formation.
(a) The rate of ATPγS hydrolysis from the TLC assay. The data are plotted in terms of the fraction of [γ-35S]ATPγS counts in the product band (left axis) and of the fraction multiplied by the 1000 nM initial ATPγS concentration (right axis). The solid line represents a fit of the 0 to 8 min time points to a straight line, yielding an initial rate of 0.09 nM s−1. (b) The rate of initiation complex formation from the primer extension assay under the same conditions as (a). The data are plotted in terms of the fraction of the total 32P counts in the product band (left axis) and of the initiation complex concentration determined from the fraction multiplied by the 50 nM initial primer/template concentration (right axis). The solid line represents a fit of the 0 to 8 min time points to a straight line, yielding an initial rate of 0.02 nM s−1.
The component omission results suggest that DnaX complexes, including the γ3 complex, are primer/template- and β2-dependent ATPγSases, and the kinetic data show that this hydrolysis occurs on a timescale and with stoichiometry consistent with initiation complex formation. Thus, it is likely that initiation complex formation by τ-complexes with ATPγS does not reflect a mechanism for initiation in the absence of hydrolysis, as we have suggested previously 27, but is instead associated with slow ATPγS hydrolysis leading to productive initiation by a τ-mediated mechanism.
Discussion
Processivity of a DNA replicase is conferred by tethering the polymerase to a bracelet-like sliding clamp, which is loaded onto the primed template DNA by AAA+ clamp loader complexes. In the E. coli Pol III holoenzyme model system, the loading of the β2 sliding clamp and subsequent binding of β2 by Pol III are tightly coupled by tethering the Pol III to the DnaX clamp loader complex via the τ subunit 27. In this work, we show that this chaperoning by Pol III to the loaded β2 requires only one τ subunit and accelerates formation of initiation complexes by ca. 100-fold over the unchaperoned reaction supported by the γ3 complex.
It has been proposed that the slowest step of the clamp loading reaction with the γ3 complex is release of the β2 from the DnaX complex onto the DNA, with a rate constant of 3 s−1 31. The γ3 complex data observed here show that complete initiation complexes form with a rate constant that increases with Pol III concentration up to 0.6 s−1 at the highest Pol III level tested (40 nM). Thus, as Pol III increases the initiation rate approaches a level similar to the rates of β2 release, suggesting that Pol III can quickly bind to β2 once the γ3 complex releases it onto the DNA. This mechanism is analogous to the T4 bacteriophage replicase, which has been shown to form initiation complexes with a kobs of 1 – 2 s−1 that is similar to the rate-limiting step for the clamp loading stage 39, 40. In striking contrast, our data for Pol III-bound τ complexes show rate constants for initiation complex formation of 25 – 50 s−1 (at 20 nM DnaX complex) that are much faster than the rate of β2 release measured in the absence of polymerase and are on the order of that for the rate for DNA binding to the β2-bound DnaX complex (466 µM−1 s−1 × 0.020 µM DNA = 9 s−1) 31. This enhanced rate suggests the chaperoned Pol III plays an active role in accelerating the clamp loading process, for example by signaling the clamp loader to release the β2.
The semi-discontinuous nature of double-stranded DNA replication requires that the lagging strand polymerase dissociate from the sliding clamp upon completion of an Okazaki fragment and form a new initiation complex with a sliding clamp at the downstream primer. The rate of fork progression, ca. 600 nt s−1 41, is similar to that of Pol III holoenzyme progression on a single-stranded template 1. Thus, most of the reaction time in double-stranded DNA replication is spent on nucleotide addition, and release of the lagging strand polymerase from a completed fragment and formation of an initiation complex on the next primer must be relatively rapid to avoid slowing the replicase. At 20 nM DnaX complex and Pol III, values similar to the total concentrations in cells estimated from a cell volume of 1 fL 42, 43, the γ3 complex forms initiation complexes with a half-life of 1.6 s. An Okazaki fragment is completed within 2 s, so the unchaperoned reaction is too slow to be compatible with fork progression. Under the same conditions, the half-life measured for τ-complexes is in the range of 15 – 30 ms, enabling initiation complex formation in a small fraction of the total time needed to complete Okazaki fragment synthesis.
This work shows that a major function of multiple ATPase sites within the DnaX complex is to impart a rapid rate of initiation complex formation necessary for fork progression. In contrast to earlier models where clamp loading is driven by the binding and subsequent hydrolysis of three ATPs by the DnaX complex, we demonstrate that the τγ2m complex, with only one ATP binding site, catalyzes initiation complex formation. The τ-mediated chaperoning of Pol III at least partially compensates for the loss of ATPase activity. Indeed, the τ2γ1m complex with only two active ATPases forms initiation complexes faster than a three ATPase γ3 complex. Given that the τ1γ2m complex already requires several minutes to form initiation complexes, it is likely that a τ-less ATPase variant would be even more impaired. The kinetic results are consistent with our previous report that K51E-variant DnaX complexes with at least one active ATPase site support processive DNA synthesis 32. These findings show that the binding and hydrolysis of three ATPs is not mechanistically essential to form an initiation complex. Our kinetic data dramatically illustrate that hydrolysis of multiple ATPs is vital to formation initiation complexes on a functionally useful timeframe. The 2nd and 3rd ATPase sites accelerate the process from a minutes timescale for the one ATPase complex to a 10 ms timescale for the three ATPase complex that is compatible with replication fork progression. Together, the results for the ATPase variant complexes and those for the all wild type complexes demonstrate that by using multiple ATPases and by chaperoning Pol III to β2, the Pol III holoenzyme achieves initiation complex formation rapid enough to support re-initiation on the lagging strand during replication fork progression.
One of the most surprising results of this study was that the DnaX complex hydrolyzes the nucleotide analogue ATPγS on a timescale similar to the rate of ATPγS-driven initiation complex formation. It has been known for more than 25 years that when τ is present the Pol III holoenzyme can form initiation complexes with ATPγS substituted for ATP 33, and we have shown previously that the Pol III chaperoning mechanism functions with ATPγS 27. While initial tests of Pol III holoenzyme purified from cells showed possible hydrolysis of ATPγS 44, subsequent studies have not observed hydrolysis 29, and the initial positive result was ascribed to a contaminant in the Pol III holoenzyme preparation 26. It has therefore been assumed that the analogue only serves to mimic the allosteric effects of ATP binding in the clamp loading cycle but not the effects arising from hydrolysis.
Here, we present several lines of evidence that strongly suggest that hydrolysis of ATPγS contributes to initiation complex formation by τ-containing Pol III holoenzyme. 1) DnaX complexes hydrolyze ATPγS. Thiophosphate forms on a timescale of minutes when ATPγS is incubated with DnaX complex (Fig. 7). It was crucial that excesses of unlabeled ATPγS and thiophosphate were used as a carrier in the TLC assay. Omitting the carrier lead to retention of thiophosphate near the origin, unresolved from ATPγS. Our experiment was conducted under conditions where a single turnover of ATP hydrolysis would result in a significant level of thiophosphate formation. An earlier study also used a TLC assay but found ATPγS hydrolysis to be less significant 29. In that study, no use of a TLC carrier is reported, and ATPγSase activity was measured in a steady state experiment with ATPγS in vast molar excess over the DnaX complex, making detection of a single turnover of hydrolysis difficult. 2) The ATPγS hydrolysis observed here requires primer/template DNA and is strongly stimulated by β2. These requirements mirror that for ATP hydrolysis by DnaX complex. 3) ATPγS hydrolysis occurs with similar kinetics to initiation complex formation. The initial rates of initiation complex formation and of ATPγS hydrolysis are similar under similar conditions (Fig. 8). 4) When Pol III is present, the molar ratio of ATPγS hydrolyzed to initiation complex formed is 4.5:1. Initiation complexes are likely be dead ends for the τ-complex that prevent enzymatic turnover, and this ratio is consistent with one turnover of clamp loading initiation complex formation.
The revelation that ATPγS-driven initiation complex formation by τ-complexes coincides with ATPγS hydrolysis required reexamination of earlier models for initiation complex formation. In a previous study, we proposed that ATPγS mimics the allosteric effects of ATP in promoting the DnaX complex to bind DNA and open β2, and the τ subunit positions Pol III to bind and capture a transiently populated closed β2 intermediate, bypassing the requirement for hydrolysis to form an initiation complex 27. While this could still be a valid alternate pathway, the simplest mechanism consistent with the data presented here is that β2 is loaded through the slow hydrolysis of ATPγS and then bound by the chaperoned Pol III. The resulting initiation complex is protected from subsequent unloading by associated τ-complex.
The γ3 complex shows little ability to form initiation complexes with ATPγS. However, the γ3 complex was as effective as the τ-complexes at hydrolyzing ATPγS. If any DnaX complex can hydrolyze ATPγS, then why do only τ-complexes form active initiation complex? A possible explanation is that ATPγS may promote disassembly of Pol III holoenzyme from the DNA, but the τ subunit protects against this process. A previous study from our lab showed that Pol III holoenzyme exclusively forms leading strand initiation complexes in the presence of ATPγS, and addition of ATPγS to existing dimeric leading/ lagging strand initiation complexes (formed in the presence of ATP) causes lagging strand complexes to dissociate from the DNA 34. This finding coupled with our observations suggests that ATPγS causes initiation complex disassembly unless the complex is configured to be the leading strand replicase and bound to a τ-complex. Marians and colleagues have demonstrated that holoenzyme assembled by γ3 complex on a rolling circle template is disassembled by γ3 complex during active elongation, but the elongating complex is protected by the addition of the τ subunit 45. Thus, the τ subunit protects initiation and elongation complexes from disassembly by the DnaX complex for holoenzyme-DNA complexes configured as leading strands. These facts support a model that τ-complexes can form initiation complexes in the presence of ATPγS by binding and hydrolyzing ATPγS to load β2, chaperoning in Pol III, and remaining bound to the initiation complex to protect against removal by exogenous ATPγS-bound DnaX complex (Fig. 9a). A leading strand τ-initiation complex is able to accumulate from the slow hydrolysis of ATPγS. The mechanism of initiation would be similar with ATP or ATPγS, the latter would just occur 1000-fold more slowly. By contrast, the γ3 complex might be able to slowly hydrolyze ATPγS and load the β2 clamp as effectively as the τ-complex, perhaps even transiently forming an initiation complex, but the γ3 complex does not remain associated with these complexes to protect against subsequent ATPγS-driven disassociation (Fig. 9b). Any β2 loading or initiation complex formation associated with slow γ3 complex-catalyzed ATPγS hydrolysis cannot compensate for ATPγS-driven disassembly. With ATP, dissociation by exogenous γ3 complex is presumably offset by rapid re-initiation supported by ATP hydrolysis, enabling γ3 complex to form initiation complex with the natural nucleotide.
Fig. 9. Models for ATPγS-driven initiation complex formation and disassembly.
The reaction steps other than ATPγS hydrolysis are discussed and cited in the text. (a) Model for initiation complex formation by the τ-complex. Step a represents ATP (or ATPγS)-bound τ-complex binding the primer/template DNA and the β2 in an open conformation. Step b represents nucleotide hydrolysis and closing of the β2 ring to form the closed conformation. Steps a through b are 1000-fold faster with ATP than with ATPγS. Step c represents the chaperoned binding of Pol III to the loaded β2 to form an initiation complex. This initiation complex (when configured as a leading strand complex) is protected from unloading by the τ-complex, which remains associated. (b) Model for initiation complex formation and disassembly by the γ3 complex. Steps a′ and b′ are the same as the analogous steps for τ-complex. Step c′ represents γ3 complex releasing β2 and dissociating, leaving a β2/DNA complex. Step d′ represents binding of Pol III to complete initiation complex formation. Steps e′ and f′ represent unloading of β2 and disassembly of the unprotected initiation complex, respectively, by ATPγS-bound γ3 complex. ATP-bound γ3 complex could dissociate the complexes by steps analogous to e′ and f′, but they would be rapidly replaced by ATP hydrolysis-driven initiation complex formation (steps a′ through d′).
This work shows that a single τ subunit is required to sustain a rate of initiation complex formation compatible with the physiological rate of fork progression and to benefit from the presence of SSB, both functions of the lagging strand replicase. The cellular form of DnaX complex is thought to be τ2γδδ’χψ, leading to a (Pol III)2τ2γδδ’χψ form of the holoenzyme 26. The presence of two τ subunits enables binding of two Pol IIIs to dimerize the replicase 21–23, and this process is thought to couple leading and lagging strand synthesis 24, 25. A dimeric replicase would tether a dissociated lagging strand polymerase to the still-elongating leading strand assembly, increasing the local concentration of the dissociated replicase and potentially contributing to its rate of initiation complex formation. The presence of a DnaX complex bound as part of the lagging strand replicase may also be required to sense the presence of a new primer at the replication fork and facilitate polymerase release prior to chaperoning formation of a new initiation complex, a possibility that has not been addressed experimentally. Two τ subunits may also be required to protect both the leading and lagging strand assemblies from premature dissociation by exogenous DnaX complex 45, an issue that also remains to be experimentally addressed.
In many organisms, the replicative polymerase is not tightly bound by its allied clamp loader like in E. coli, raising the question of how the polymerase reaches the loaded sliding clamp in these systems. The Gram-positive bacterium B. subtilis forms only weak interactions between its polymerases and the τ subunits of its clamp loader 46, making it unclear whether the clamp loader can chaperone the polymerases to initiation complexes. The T4 replicase has been shown to form transient interactions between the gp43 polymerase and the gp44/62 clamp loader, and the polymerase binds rapidly upon completion of clamp loading but does not appear to accelerate clamp loading 40, 47. Perhaps the B. subtilis holoenzyme could also utilize a relatively transient polymerase/clamp loader interaction to rapidly complete initiation complex formation after clamp loading. The eukaryotic clamp loading process is rapid (rate constant as fast as ca. 10 s−1 36), but no binding interactions have been identified between the eukaryotic clamp loader and polymerases, and the mechanism by which the polymerases associate with the loaded clamp is not well understood. While it is clearly important that clamp loading be fast, what ultimately matters for replication fork progression is the rate for completing initiation complex formation. We have shown that the E. coli replicase maximizes this rate by tightly coupling the clamp loading and polymerase-binding-to-loaded-clamp reaction stages, and it will be interesting to see how other replicases meet the challenge of forming initiation complexes on a timescale compatible with Okazaki fragment synthesis and re-initiation.
Materials and Methods
Proteins and nucleic acids
DNA polymerase III holoenzyme and SSB proteins were expressed and purified as previously described: Pol III 21;β;48; reconstituted τ3 and γ3 DnaX complexes 9; τ2γ and τγ2 complexes and mixed τ/γ complexes containing K51E point mutations in τ or γ 32; and SSB 49. Hexokinase was purchased from Sigma-Aldrich. Protein concentrations were determined by the method of Bradford a Bio-Rad protein assay kit using bovine serum albumin as a standard.
M13Gori single-stranded DNA was prepared as described 48 and annealed as described previously 27 to a 30 nt RNA oligonucleotide primer purchased from Thermo Fisher Scientific/Dharmacon with the sequence: 5′UGAGCUCGGGGAAUGCGGCGGCGAGAUAGU. Where applicable, the primer was 5′-32P end-labeled with T4 polynucleotide kinase (New England Biolabs). Calf thymus DNA “activated” by partial digestion with DNase I (USB) to produce free 3′ termini was prepared as previously described 21.
Buffers
All reactions herein were conducted with the following buffer: 50 mM HEPES-KOH (pH 7.5), 100 mM potassium glutamate, 10 mM magnesium acetate, 0.20 mg/mL bovine serum albumin, 10 mM dithiothreitol, 2.5% (v/v) glycerol, and 0.02% (v/v) Nonidet-P40 detergent.
Primer extension assay for initiation complex formation
All reactions were conducted with initiation complex formation and primer extension as separate reaction steps, under single turnover conditions where all Pol III holoenzyme components were present in molar excess over the primer/template substrate. Unless otherwise noted, all assays for initiate complex formation were conducted with 20 nM Pol III, 20 nM DnaX complex, 50 nM β2, 0.20 mM ATP or ATPγS, 1.0 nM 32P-5′ end-labeled primer/template, and 0 or 250 nM SSB4 as noted. This activity assay for initiation complex formation has been shown previously to require ATP and β2 for primer extension, showing that the assay reports only fully assembled processive initiation complexes 27.
Data sets with time points of less than 5 s were collected by rapid mixing and quenching using a KinTek Model RQF-3 rapid quench-flow device at 25 °C. Initiation complex formation was initiated by rapidly mixing 27 µL of Pol III, DnaX complex, β2, hexokinase, and ATP with 27 µL 32P-5′ end-labeled primer/template solution and (where applicable) SSB4. The SSB4 dependence data in Fig. 2 were measured with 10 nM Pol III and 7.8 nM DnaX complex, with the remaining component concentrations the same as above. After a reaction time programmed by the instrument, initiation complex formation was quenched with ca. 100 µL 2 µg/µL activated calf thymus DNA (to provide excess 3′ termini) and 20 mM glucose (to facilitate the rapid hydrolysis of ATP by hexokinase). The quench solution also contained 80 µM each dTTP, dCTP, and dGTP and 4.0 µM ddATP to facilitate primer extension. The primer elongates until incorporating the ddA nucleotide and terminating, yielding a 53 nt extension product. The quenched samples were held for a 5 s primer extension time in the instrument “exit line” prior to ejecting into tubes containing a stop solution of 150 µL 96% formamide/25 mM EDTA denaturing gel loading solution. Reaction products were separated by 16% (w/v) polyacrylamide gel electrophoresis with 8 M urea denaturant. The radioactivity in the primer and product bands was quantified by phosphorimaging using a Storm 840 imager and ImageQuant 5.2 software (Amersham Biosciences). The fraction of primer elongated for each reaction was determined as the counts for the extension product band divided by the sum of the primer and product bands.
Data sets with all points ≥ 5 s were sampled manually, with initiation complex formation initiated by combining 12.5 µL Pol III, DnaX complex, β2, ATP (or ATPγS where applicable), and hexokinase with 12.5 µL 32P-5′ end-labeled primer/template solution and SSB4 (where applicable). These reactions were quenched and primer-extended by pipetting in 5.0 µL 2 µg/µL calf thymus DNA; 60 mM glucose; 240 µM each dTTP, dCTP, and dGTP; and 12 µM ddATP. After 5 s primer extension, 30 µL 96% formamide/25 mM EDTA denaturing gel loading solution was added to stop the reactions. The samples were analyzed by gel electrophoresis as above.
The initiation experiments paralleling the conditions of the ATPγS hydrolysis assays were conducted similarly to the other manually sampled experiments, except that the reactions were conducted with the concentrations described in the next section and initiated by combining 7.5 µL of Pol III, DnaX complex, β2, 32P-5′ end-labeled primer/template, SSB4 (where applicable), and 12 mM glucose with 2.5 µL ATPγS. These reactions were quenched and primer-extended for 5 s with 2.0 µL of the same solution as the other manually sampled experiments but also containing 10 µM hexokinase.
ATPγS hydrolysis assayed by thin layer chromatography
ATPγS hydrolysis was initiated by combining 4.0 µL 200 nM Pol III, 100 nM DnaX complex, 200 nM β2, 50 nM unlabeled primer/template, and 3.0 µM SSB4 with 1.0 µL 1.0 µM [γ-35S]ATPγS (ca. 0.2 µCi/pmol). After the reaction times described, the reactions were quenched with 2.0 µL 100 mM EDTA, 5.0 mM thiophosphate, and 1.0 mM unlabeled ATPγS. The latter two reagents function both as “cold traps” for the hydrolysis reaction and as carriers in thin layer chromatography (TLC). In the absence of these carriers, the [35S]thiophosphate remained the origin and did not migrate. For each sample, 5 µL was spotted 1 µL at a time on a polyethylenimine-TLC plate. The plates were developed in a solution of 1.0 M formic acid and 0.50 M lithium chloride until the solvent front reached ca. 80% to the top of the plate. The plates were dried and analyzed by phosphorimaging. The fraction of ATPγS hydrolyzed was quantified as the counts in the faster-migrating thiophosphate product band divided by the sum of the thiophosphate and slower-migrating ATPγS bands.
Data analysis
Kinetic data were fit to a simple exponential function: f = f0 + Δfmax * {1 − exp(−kobst)}, where f is the fraction of primers elongated, f0 is the fraction of primers elongated at zero reaction time, Δfmax is the maximum change in fraction of primers elongated, kobs is the observed first order rate constant, and t is the reaction time elapsed. The rapid kinetic data for τ-containing complexes deviated from a simple exponential when time points longer than 0.125 s were sampled. The longer time courses were fit to a sum of two exponential function: f = f0 + Δf1 * {1 − exp(−k1t)} + Δf2 * {1 − exp(−k2t)}, where Δf1 is the maximum change in fraction of primers elongated for one subpopulation, k1 is the observed first order rate constant for this subpopulation,Δ f2 is the maximum change in fraction of primers elongated for a second subpopulation, k2 is the observed first order rate constant for the second subpopulation. The double exponential curve fits are shown in the Supplementary Data and yielded qualitatively similar rate constants for the faster-initiating population to those derived from single exponential fits to time points ≤ 0.125 s.
Highlights.
τ accelerates the rate of initiation complex formation ca. 100-fold
Multiple DnaX ATPases are required for maximal rate of initiation complex formation
τ complex drives initiation complexes with concomitant ATPγS hydrolysis
Only one τ is needed to confer stimulation by SSB in initiation complex formation
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 GM060273 and F32GM084697 (postdoctoral fellowship for C.D.D.) and in part by R01 GM49700. We thank Anna Wieczorek for preparation of the mixed τ/γ DnaX complexes and Nicholas Fingland and Rahul Saxena, PhD for insightful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg A, Baker TA. DNA replication. New York: WH Freeman and Company; 1992. [Google Scholar]
- 2.LaDuca RJ, Crute JJ, McHenry CS, Bambara RA. The β Subunit of the Escherichia coli DNA Polymerase III Holoenzyme Interacts Functionally with the Catalytic Core in the Absence of Other Subunits. J Biol Chem. 1986;261:7550–7557. [PubMed] [Google Scholar]
- 3.Burgers PMJ. Mammalian Cyclin/PCNA (DNA Polymerase δ Auxilliary Protein) Stimulates Processive DNA Synthesis by Yeast DNA Polymerase III. Nucleic Acids Res. 1988;16:6297–6307. doi: 10.1093/nar/16.14.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Indiani C, O'Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 5.McHenry CS. Chromosomal replicases as asymmetric dimers: Studies of subunit arrangement and functional consequences. Mol Microbiol. 2003;49:1157–1165. doi: 10.1046/j.1365-2958.2003.03645.x. [DOI] [PubMed] [Google Scholar]
- 6.Johanson KO, McHenry CS. The β subunit of the DNA polymerase III holoenzyme becomes inaccessible to antibody after formation of an initiation complex with primed DNA. J Biol Chem. 1982;257:12310–12315. [PubMed] [Google Scholar]
- 7.Stukenberg PT, Studwell-Vaughan PS, O'Donnell ME. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 8.Jeruzalmi D, O'Donnell ME, Kuriyan J. Crystal structure of the processivity clamp loader gamma complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 9.Pritchard AE, Dallmann HG, Glover BP, McHenry CS. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of δδ' with DnaX(4) forms DnaX(3)δδ'. EMBO J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glover BP, McHenry CS. The DnaX-binding Subunits δ' and ψ are bound to γ and not τ in the DNA Polymerase III Holoenzyme. J Biol Chem. 2000;275:3017–3020. doi: 10.1074/jbc.275.5.3017. [DOI] [PubMed] [Google Scholar]
- 11.Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O'Donnell M, Kuriyan J. The Mechanism of ATP-dependent Primer-template Recognition by a Clamp Loader Complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson SG, Williams CR, O'Donnell M, Bloom LB. A function for the ψ subunit in loading the Escherichia coli DNA polymerase sliding clamp. J Biol Chem. 2007;282:7035–7045. doi: 10.1074/jbc.M610136200. [DOI] [PubMed] [Google Scholar]
- 13.Jeruzalmi D, O'Donnell ME, Kuriyan J. Crystal Structure of the Processivity Clamp Loader Gamma Complex of E. coli DNA Polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 14.Bullard JM, Pritchard AE, Song MS, Glover BP, Wieczorek A, Chen J, Janjic N, McHenry CS. A Three-domain Structure for the δ Subunit of the DNA Polymerase III Holoenzyme δ Domain III Binds δ' and Assembles into the DnaX Complex. J Biol Chem. 2002;277:13246–13256. doi: 10.1074/jbc.M108708200. [DOI] [PubMed] [Google Scholar]
- 15.Flower AM, McHenry CS. The γ subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A. 1990;87:3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blinkowa AL, Walker JR. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuchihashi Z, Kornberg A. Translational frameshifting generates the γ subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloom LB. Dynamics of Loading the Escherichia coli DNA Polymerase Processivity Clamp. Crit Rev Biochem Mol Biol. 2006;41:179–208. doi: 10.1080/10409230600648751. [DOI] [PubMed] [Google Scholar]
- 19.Gao D, McHenry CS. τ Binds and Organizes Escherichia coli Replication Proteins through Distinct Domains. Domain IV, Located within the Unique C Terminus of τ, Binds the Replication Fork Helicase, DnaB. J Biol Chem. 2001;276:4441–4446. doi: 10.1074/jbc.M009830200. [DOI] [PubMed] [Google Scholar]
- 20.Gao D, McHenry CS. τ βinds and organizes Escherichia coli replication proteins through distinct domains: partial proteolysis of terminally tagged τ to determine candidate domains and to assign domain V as the α binding domain. J Biol Chem. 2001;276:4433–4440. doi: 10.1074/jbc.M009828200. [DOI] [PubMed] [Google Scholar]
- 21.Kim DR, McHenry CS. Biotin Tagging Deletion Analysis of Domain Limits Involved in Protein-Macromolecular Interactions: Mapping the τ Binding Domain of the DNA Polymerase III α Subunit. J Biol Chem. 1996;271:20690–20698. doi: 10.1074/jbc.271.34.20690. [DOI] [PubMed] [Google Scholar]
- 22.McHenry CS. Purfication and Characterization of DNA Polymerase III': Identification of τ as a Subunit of the DNA Polymerase III Holoenzyme. J Biol Chem. 1982;257:2657–2663. [PubMed] [Google Scholar]
- 23.Studwell-Vaughan PS, O'Donnell ME. Constitution of the Twin Polymerase of DNA Polymerase III Holoenzyme. J Biol Chem. 1991;266:19833–19841. [PubMed] [Google Scholar]
- 24.Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a Replicative Polymerase and Helicase: a τ-DnaB Interaction Mediates Rapid Replication Fork Movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Dallmann HG, McHenry CS, Marians KJ. τ Couples the Leading- and Lagging-strand Polymerases at the Escherichia coli DNA Replication Fork. J Biol Chem. 1996;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- 26.McHenry CS. DNA replicases from a bacterial perspective. Annu Rev Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- 27.Downey CD, McHenry CS. Chaperoning of a replicative polymerase onto a newly-assembled DNA-bound sliding clamp by the clamp loader. Mol Cell. 2010;37:481–491. doi: 10.1016/j.molcel.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glover BP, McHenry CS. The χψ Subunits of DNA Polymerase III Holoenzyme Bind to Single-stranded DNA-binding Protein (SSB) and Facilitate Replication of a SSB-coated Template. J Biol Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 29.Hingorani MM, O'Donnell ME. ATP Binding to the Escherichia coli Clamp Loader Powers Opening of the Ring-Shaped Clamp of DNA Polymerase III Holoenzyme. J Biol Chem. 1998;273:24550–24563. doi: 10.1074/jbc.273.38.24550. [DOI] [PubMed] [Google Scholar]
- 30.Bloom LB. Loading clamps for DNA replication and repair. DNA Repair (Amst) 2009;8:570–578. doi: 10.1016/j.dnarep.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson SG, Thompson JA, Paschall CO, O'Donnell M, Bloom LB. Temporal correlation of DNA binding, ATP hydrolysis, and clamp release in the clamp loading reaction catalyzed by the Escherichia coli gamma complex. Biochemistry. 2009;48:8516–8527. doi: 10.1021/bi900912a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wieczorek A, Downey CD, Dallmann HG, McHenry CS. Only one ATP-binding DnaX subunit is required for initiation complex formation by the E. coli DNA polymerase III holoenzyme. J Biol Chem. 2010;285:29049–29053. doi: 10.1074/jbc.C110.165076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johanson KO, McHenry CS. Adenosine 5'-O-(3-Thiotriphosphate) Can Support the Formation of an Initiation Complex between the DNA Polymerase III Holoenzyme and Primed DNA. J Biol Chem. 1984;259:4589–4595. [PubMed] [Google Scholar]
- 34.Glover BP, McHenry CS. The DNA Polymerase III Holoenzyme: An Asymmetric Dimeric Replicative Complex with Leading and Lagging Strand Polymerases. Cell. 2001;105:925–934. doi: 10.1016/s0092-8674(01)00400-7. [DOI] [PubMed] [Google Scholar]
- 35.Hingorani MM, O'Donnell ME. ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J Biol Chem. 1998;273:24550–24563. doi: 10.1074/jbc.273.38.24550. [DOI] [PubMed] [Google Scholar]
- 36.Kumar R, Nashine VC, Mishra PP, Benkovic SJ, Lee TH. Stepwise loading of yeast clamp revealed by ensemble and single-molecule studies. Proc. Natl. Acad. Sci. U. S. A. 2010;107:19736–19741. doi: 10.1073/pnas.1014139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertram JG, Bloom LB, Turner J, O'Donnell ME, Beechem JM, Goodman MF. Pre-Steady State Analysis of the Assembly of Wild Type and Mutant Circular Clamps of Escherichia coli DNA Polymerase III onto DNA. J Biol Chem. 1998;273:24564–24574. doi: 10.1074/jbc.273.38.24564. [DOI] [PubMed] [Google Scholar]
- 38.McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 39.Sexton DJ, Kaboord BF, Berdis AJ, Carver TE, Benkovic SJ. Dissecting the Order of Bacteriophage T4 DNA Polymerase Holoenzyme Assembly. Biochemistry. 1998;37:7749–7756. doi: 10.1021/bi980088h. [DOI] [PubMed] [Google Scholar]
- 40.Zhuang Z, Berdis AJ, Benkovic SJ. An alternative clamp loading pathway via the T4 clamp loader gp44/62-DNA complex. Biochemistry. 2006;45:7976–7989. doi: 10.1021/bi0601205. [DOI] [PubMed] [Google Scholar]
- 41.Breier AM, Weier HU, Cozzarelli NR. Independence of replisomes in Escherichia coli chromosomal replication. Proc Natl Acad Sci U S A. 2005;102:3942–3947. doi: 10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McHenry CS, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli purfication and resolution into subunits. J Biol Chem. 1977;252:6478–6484. erratum1978. 253:645. [PubMed] [Google Scholar]
- 43.Ingraham J. Growth of the Bacterial Cell. Sinauer Assoc., Inc.; 1983. p. 269. [Google Scholar]
- 44.Johanson KO, McHenry CS. Adenosine 5'-O-(3-thiotriphosphate) can support the formation of an initiation complex between the DNA polymerase III holoenzyme and primed DNA. J Biol Chem. 1984;259:4589–4595. [PubMed] [Google Scholar]
- 45.Kim S, Dallmann HG, McHenry CS, Marians KJ. τ protects β in the leading-strand polymerase complex at the replication fork. J Biol Chem. 1996;271:4315–4318. doi: 10.1074/jbc.271.8.4315. [DOI] [PubMed] [Google Scholar]
- 46.Bruck I, O'Donnell ME. The DNA Replication Machine of a Gram-positive Organism. J Biol Chem. 2000;275:28971–28983. doi: 10.1074/jbc.M003565200. [DOI] [PubMed] [Google Scholar]
- 47.Trakselis MA, Berdis AJ, Benkovic SJ. Examination of the Role of the Clamp-loader and ATP Hydrolysis in the Formation of the Bacteriophage T4 Polymerase Holoenzyme. J Mol Biol. 2003;326:435–451. doi: 10.1016/s0022-2836(02)01330-x. [DOI] [PubMed] [Google Scholar]
- 48.Johanson KO, Haynes TE, McHenry CS. Chemical Characterization Purification of the β Subunit of the DNA Polymerase III Holoenzyme from an Overproducing Strain. J Biol Chem. 1986;261:11460–11465. [PubMed] [Google Scholar]
- 49.Griep MA, McHenry CS. Glutamate Overcomes the Salt Inhibition of DNA Polymerase III Holoenzyme. J Biol Chem. 1989;264:11294–11301. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.