Abstract
Alternative pre–messenger RNA splicing impacts development, physiology, and disease, but its regulation in humans is not well understood, partially due to the limited scale to which the expression of specific splicing events has been measured. We generated the first genome-scale expression compendium of human alternative splicing events using custom whole-transcript microarrays monitoring expression of 24,426 alternative splicing events in 48 diverse human samples. Over 11,700 genes and 9,500 splicing events were differentially expressed, providing a rich resource for studying splicing regulation. An unbiased, systematic screen of 21,760 4-mer to 7-mer words for cis-regulatory motifs identified 143 RNA 'words' enriched near regulated cassette exons, including six clusters of motifs represented by UCUCU, UGCAUG, UGCU, UGUGU, UUUU, and AGGG, which map to trans-acting regulators PTB, Fox, Muscleblind, CELF/CUG-BP, TIA-1, and hnRNP F/H, respectively. Each cluster showed a distinct pattern of genomic location and tissue specificity. For example, UCUCU occurs 110 to 35 nucleotides preceding cassette exons upregulated in brain and striated muscle but depleted in other tissues. UCUCU and UGCAUG appear to have similar function but independent action, occurring 5' and 3', respectively, of 33% of the cassette exons upregulated in skeletal muscle but co-occurring for only 2%.
Keywords: alternative splicing, whole-transcript microarrays, splicing regulatory motifs
Alternative splicing is a major mechanism for generating proteomic diversity, and as many as 74% of human multi-exon genes are alternatively spliced1. Many recent studies point to the importance of detection and measurement of alternative splicing. For example, more genetic variations in the CEU HapMap population manifest themselves through changes in transcript structure, including splicing, than gene transcription2. However, our knowledge of the differential expression of specific splicing events and characterization of corresponding cis-regulatory elements is limited. Likewise, while sequence targets of several individual RNA-binding splicing factors have been characterized, much remains to be learned about their regulatory mechanism within different human tissues.
PTB proteins are known to interact with pyrimidine rich elements, such as UCUCU3–5; the FOX proteins bind UGCAUG6,7; hnRNP A/B and hnRNP F/H bind GGGG and AGGGG, respectively, and GGG8,9; muscleblind (MBNL) proteins bind UGCU10; TIA-1 binds U-rich sequences11, and the CELF proteins12 and SUP-12 (RBM38)12,13 interact with UGUGU. Using microarrays14 and immunoprecipitation of cross-linked RNA15, Darnell and co-workers elucidated the binding element and regulatory influence of neuronal Nova RNA-binding proteins, showing that Nova binds either YCAY or YCATY (Y=pyrimidine), and causes exon inclusion or exclusion depending on the location of the motif relative to the exon.
Assessing the ratios of expression for two mutually exclusive splice forms ("splicing-event profiling") requires probes targeting both exons and junctions, e.g. probes monitoring both the cassette exon and the junction across the excluded exon. Using such arrays, mouse tissues have been profiled: Pan et al.16 (3,100 cassette exons, 10 tissues), Fagnani et al.17 (3,700 cassette exons, 27 tissues), and Sugnet et al.18 (6,700 events, 22 tissues). To further our understanding of alternative splicing expression and regulation, we generated the first human genome-wide alternative splicing-event compendium, monitoring 24,426 events in 48 tissues and cell lines, and undertook an unbiased, systematic search to identify and describe putative regulatory motifs.
Results
Microarrays for whole-transcript profiling
We designed microarrays monitoring 203,672 exons and 178,351 exon-exon junctions in 17,939 human genes. These microarrays report expression of both splicing event isoforms from 8,000 cassette exons, 3,950 alternative 5' splice sites, 3,672 alternative 3' splice sites, 3,770 multiple cassette exons, 3,123 mutually exclusive exons, and 1,890 inserted introns. The 'whole transcript' design used here is different from exon arrays19, junction arrays1, and cassette exon splicing arrays (e.g.16) in that it includes a constellation of probes targeting every exon and every junction, similar to the approach of Griffith et al.20 (Figure 1A). Although exon arrays provide an unbiased survey of transcript structure, they do not monitor connections between individual exons or many alternative 5' and 3' splice sites. Because they lack junction probes, they also generally monitor only one form of an alternative splicing event. The inability to monitor both of the two mutually exclusive forms prevents accurate measurement of expression ratios of the two forms. Cassette exon splicing arrays, with probes designed to monitor the inclusion and exclusion of known cassette exons are an economical option for profiling cassette exon splicing events but lack probes to profile other types of alternative splicing (e.g. alternative 5' and 3' splice sites), do not monitor alternative 5' and 3' exons, and do not permit discovery of novel alternative splice forms.
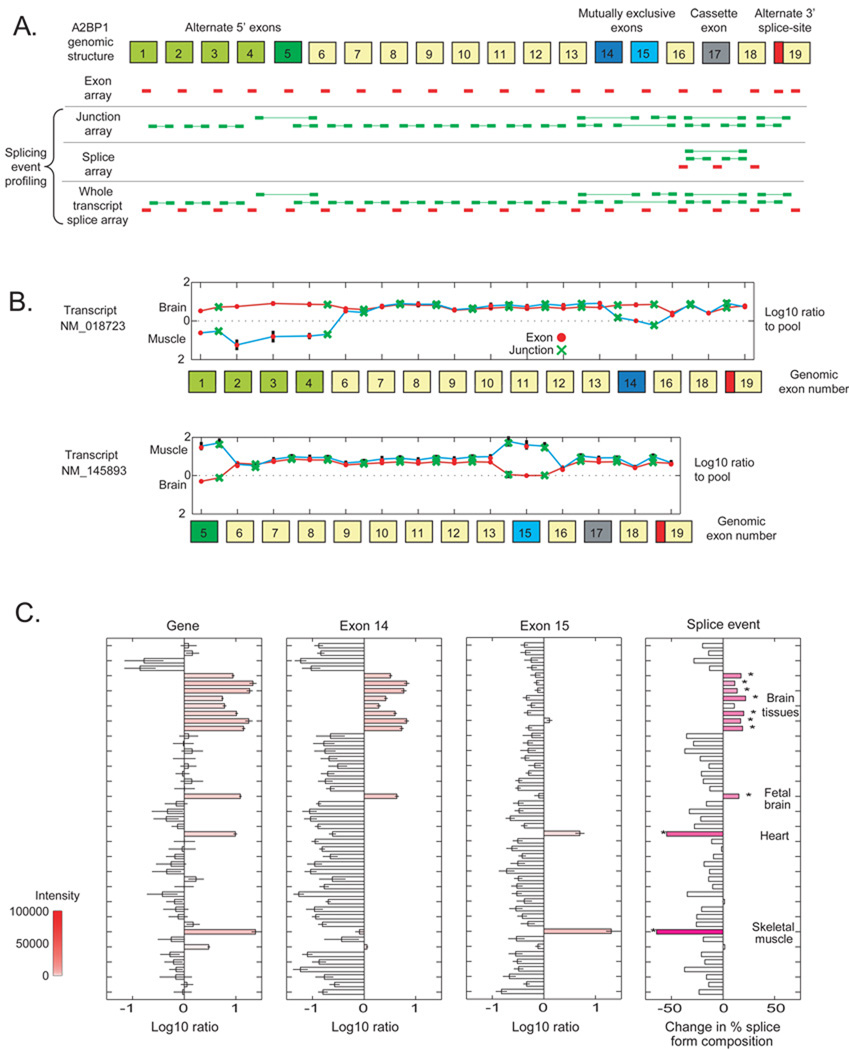
Figure 1.
Interrogation of A2BP1 (Fox-1) isoforms on the alternative splicing microarrays. A) The nineteen exons in the locus, including alternative 5' exons (green), mutually exclusive exons (blue), a cassette exon (gray), and an alternative 3' splice site (red). Exon (red) and exon-exon junction (green) probes are displayed for several splice-related microarrays. B) Expression (log10 fold-change to reference pool) of exons (red dots) and junctions (green crosses) in transcripts NM_018723 and NM_145893 in skeletal muscle (cyan line) and brain (red line). C) Expression of the gene, exon 15, exon 14, and the combined splice event expression level. Red shading represents microarray intensity values; dark pink shading (right) represents more significant p-values.
Compendium of human alternative splicing events
A set of 48 human tissues and cell lines with dissimilar expression patterns were hybridized to the arrays, and 11,700 of 18,000 genes monitored by the array showed > 3-fold change in gene expression level relative to the pool (p < 0.01) in at least one tissue (Figure S1, Methods). Across the 48 tissue panel, 9,516 alternative splicing events were also significantly differentially expressed in at least one tissue (Figure S2; Methods), similar to the number of differentially expressed genes. Microarray data are available as GEO dataset GSE11863, and the entire compendium of alternative splicing expression, gene expression, probe and splicing event nucleotide sequences, genome browser tracks, and individual splicing event figures are available at http://rulai.cshl.edu/Rosetta_AS_supp/, with additional data at http://rulai.cshl.edu/cgi-bin/dbCASE/dbcase.cgi?process=home.
As an example of the data available for each gene, results for A2BP1 (Fox-1) are shown in Figure 1. A2BP1 regulates alternative splicing and is itself alternatively spliced, containing alternative 5’ exons, a pair of mutually exclusive exons, a cassette exon, and an alternative 3’ splice site6,21 (Figure 1A). A2BP1 is upregulated in muscle and brain (Figure 1B). The four alternative 5’ exons of transcript NM_018723 were detected in brain while the 5’ exon found in transcript NM_145893 was detected in muscle. Within the coding region, brain cells preferentially include the mutually exclusive exon found in NM_018723 whereas skeletal muscle cells include the NM_145893 form. Across all tissues, A2BP1 gene expression is highest in heart, skeletal muscle, and the nervous-system (Figure 1C). The first (5’) mutually exclusive exon is expressed in nervous-system tissues while the second (3’) mutually exclusive exon is found in heart and skeletal muscle. Expression of the gene, the first mutually exclusive exon, and the second mutually exclusive exon can be combined to estimate the relative abundance of each form across the tissue panel (Figure 1C, right; Methods). Of the two forms represented by the mutually exclusive exons, brain has a higher proportion of form NM_018723.
9,516 splice events are differentially expressed
Across the 48 tissue compendium, 9,516 alternative splicing events were significantly differentially expressed in at least one tissue (Figure S2; Methods), similar to the total number of differentially expressed genes (11,700). Of the monitored cassette exons, 42% were differentially expressed in at least one tissue (Figure 2A), while a much lower fraction (26%) of monitored alternative 3' splice sites were differentially expressed and a higher proportion (55%) of monitored inserted introns were differentially expressed. The samples with the highest number of differentially expressed alternative splicing events (Figure 2B) relative to the pool include the cell lines HeLa and HCT116, peripheral leukocytes, fetal brain, testis, mammary gland, and skeletal muscle, while the samples with the fewest include adipose, adrenal gland, and fetal lung. These rankings parallel those for gene expression; i.e. the same samples types were among those with the most and fewest differentially expressed genes.
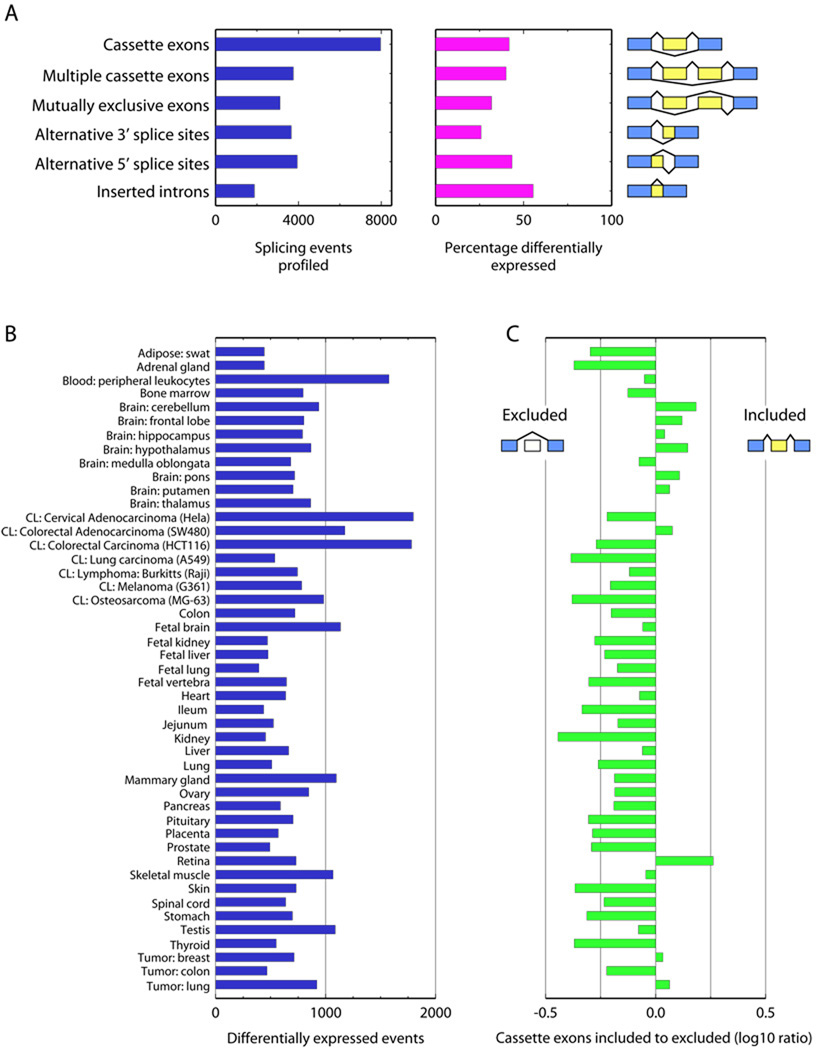
Figure 2.
A) The number of events monitored on the arrays and the percentage of each type differentially expressed in at least one tissue. B) The number of events differentially expressed in each tissue. C) The log10 ratio of the number of cassette exons differentially included to those differentially excluded.
Cassette exon inclusion/exclusion rates varied among the samples (Figure 2C). The ratio between the number of cassette exons differentially included to the number excluded was highest for SW480, breast and lung tumor, retina, and brain samples excluding medulla oblongata. To test whether this result might be due to the composition of the reference pool (normal adult tissues), we re-ratioed to each individual sample and to the average of all 48 samples, generating 49 in silico reference pools (Methods), and computed inclusion rates, exclusion rates, and ratios. While the number of cassette exons included or excluded depended on the reference sample, the inclusion-to-exclusion rankings of samples were similar regardless of the pool, with retina, and certain brain, tumor, and cell lines consistently ranking highest. These results agree with previous findings that brain tissues express the greatest number of tissue-specific exons19.
To provide an estimate of the accuracy of the quantitative predictions made by the splicing arrays and analysis tools, as part of a larger study22, we tested a sample of 23 predictions by semi-quantitative RT-PCR and found that 74% of the splice events called differentially expressed by the microarray at p-value < 0.1 show changes by PCR of at least 15% in differential expression, and correlations between the microarray and RT-PCR splice event proportionalities of r=0.88 (Methods, Table S1).
Transcription and alternative splicing regulation act on different genes
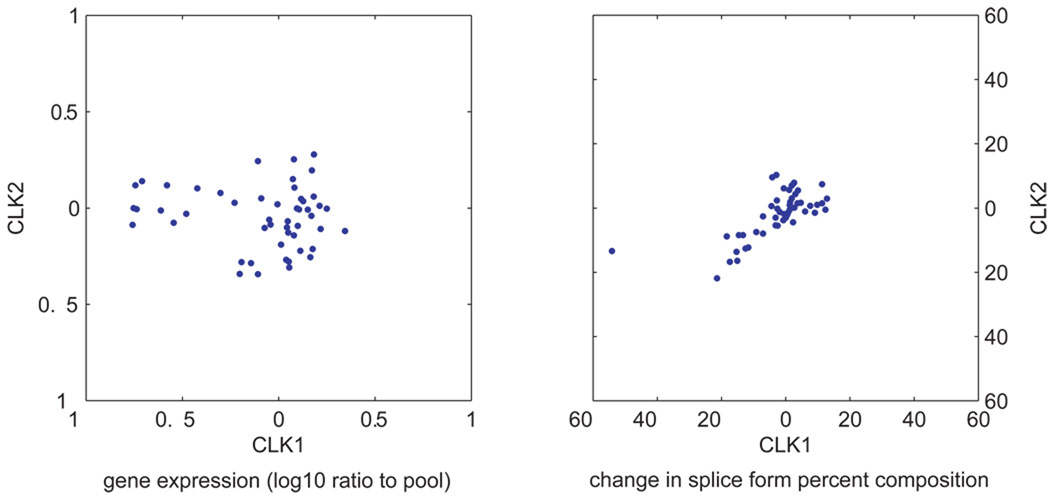
Similar to results from Pan et al.16, we did not detect significant enrichment of the differentially expressed genes in a given tissue and the genes with differentially expressed splicing events (p = 0.4). As one specific example, CLK1 (NM_004071) and CLK2 (NM_003993) contain cassette exons whose exclusion generates a protein that dimerizes but inhibits kinase activity23. Here, CLK1 and CLK2 showed uncorrelated gene expression (r=0.13) across the 48 samples but significantly correlated splicing expression (r=0.69) (Figure 3).
Figure 3.
CLK1 and CLK2 gene (left) and splice event expression (right).
Gene expression correlations are often used to define 'edges' of co-expression networks and infer functional associations. Here, gene-gene edges determined from correlated gene expression (r > 0.75) and those determined from correlated splice event expression show only 2% overlap (Supplementary Note). Although this overlap is statistically significant, the small overlap demonstrates these are largely different regulatory networks. Finally, we observed that splicing-event expression and gene expression clustered samples similarly but with a few exceptions, such as lung and breast tumor samples. These cluster with parental tissues (lung and breast) using gene expression but with cell lines using splicing expression, demonstrating that gene expression in these tumor samples is more similar to their normal parental tissues while splicing expression is more similar to cell lines (Supplementary Note).
Expression of specific splicing events
Similar to A2BP1 and CLK1/2, all splicing event tissue profiles are available at http://rulai.cshl.edu/Rosetta_AS_supp/. Ten alternative splicing profiles are highlighted in the Supplementary Note, including several associated with nervous system tissues (GSK3B, MAPT/Tau, APP, and CACNA1B), muscle tissues (MEF2C, CAMK2D, TPM1, and TPM2), splicing (NOVA1), and cancer (FGFR2).
The human gene CD44 encodes ten variable exons, one of which, CD44v6, is the target of bivatuzumab mertansine, an antibody for patients with advanced carcinoma24 that was discontinued due to skin toxicity in Phase I trials25. Although in our data CD44v6 is upregulated in tumors and cancer cell lines, it is expressed highest in skin, highlighting the potential value of this compendium as a public resource. Most microarray experiments use a probe or probe-set near the 3' end of each gene and consequently would not detect the isoform-specific variation of CD44.
De novo identification of 143 splicing regulatory elements
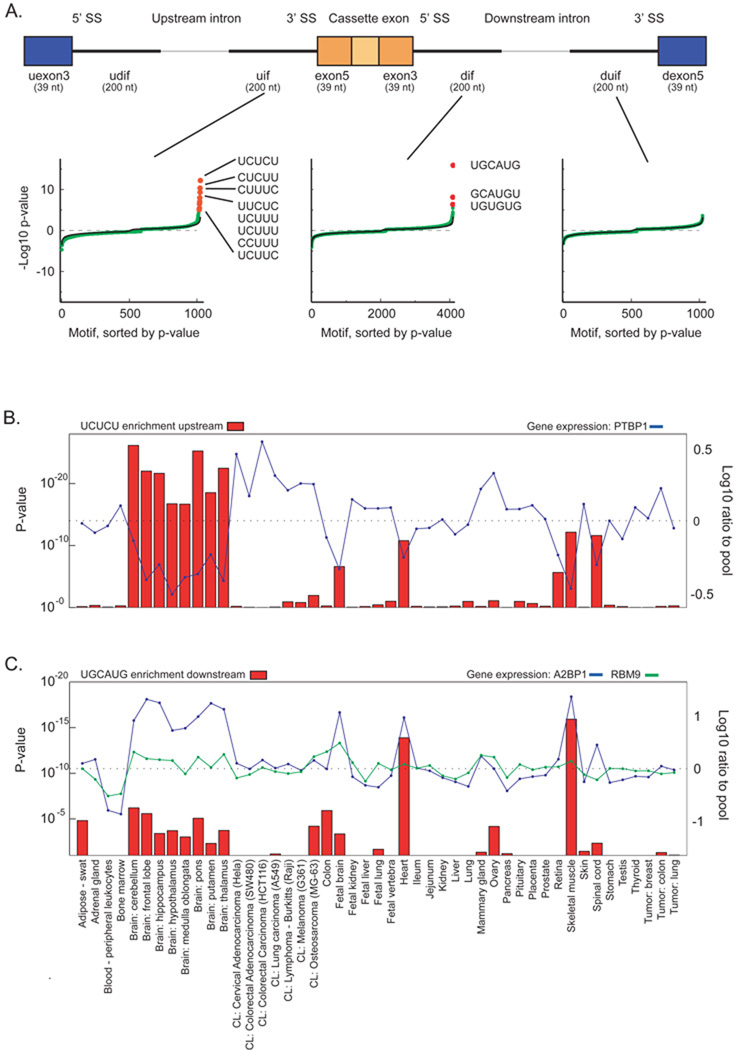
We next sought to discover regulatory elements in sequences in and adjacent to the tissue-regulated cassette exons. We extracted nucleotide sequence in eight regions ("neighborhoods") around regulated exons, and searched for over and under represented nucleotide "words" of size 4–7 nt, using neighborhood-specific sequences adjacent to all monitored cassette exons as a background set (Figure 4A, Methods). Examined neighborhoods were 200 nt for intronic regions and 39 nt exonic regions, and the hypergeometric distribution was used to calculate word enrichment p-values. In total, 33.5 million enrichment p-values were calculated.
Figure 4.
A) Identification of alternative splicing motifs enriched in eight regions adjacent cassette exons upregulated in skeletal muscle. Abbreviations: exon5 (exon3), 5' (3') portion of cassette exons; uif (dif), intronic fraction upstream (downstream); uexon3 (dexon5) and udif (duif) are the corresponding regions adjacent the upstream (downstream) exons. Sorted pentamer and hexamer enrichment in three exemplary regions are shown. Green points are −log10 p-values; magenta points are those with a Bonferroni-corrected p-value less than 0.01. Positive (negative) p-values indicate motif enrichment (depletion). The black line represents the average of 200 randomized runs. B) Enrichment of UCUCU in human tissues and cell lines. Left axis: enrichment of UCUCU in the 200 nt upstream of upregulated cassette exons. Right axis: gene expression of PTBP1. C) Enrichment of UGCAUG. Left axis: enrichment of UGCAUG in the 200 nt downstream of upregulated cassette exons. Right axis: gene expressions of Fox proteins A2BP1 and RBM9. Displayed p-values are not Bonferroni corrected.
Using a single tissue (skeletal muscle) to highlight the results, we observed that eight of 1,024 pentamers have a Bonferroni-corrected p-value < 0.01 for enrichment in the 200 nt intronic region upstream of cassette exons that are upregulated in skeletal muscle (Figure 4A). UCUCU is the most enriched, followed by other pyrimidine motifs. In the 200 nt intronic region following cassette exons, three of the 4,096 hexamers are significant. Strikingly, UGCAUG is most enriched, followed by GCAUGU and UGUGUG. No words were significantly enriched or depleted in the intronic region preceding the downstream 3' splice site.
The compendium can also be used to find enrichment of specific motifs across all 48 tissues and cell lines. For example, the motif UCUCU, which has been associated with PTB3–5, is enriched in the intronic region preceding upregulated cassette exons occurs in brain (p-value < 10−26 in cerebellum), spinal cord, retina, heart, and skeletal muscle (Figure 4B & C). In this data set, PTBP1 gene expression shows a dramatic anti-correlation with UCUCU enrichment, corroborating the inhibitory role of PTBP1. The UGCAUG motif, associated with Fox proteins A2BP1 and RBM96, is enriched in the intronic region following upregulated cassette exons in skeletal muscle (p-value < 10−16) and heart, with limited enrichment in other tissues, including brain, adipose, and colon. A2BP1 expression correlates highly with UGCAUG enrichment, corroborating the splicing enhancer role of A2BP1. Finally, while we showed above that muscular and neuronal tissues express different A2BP1 isoforms, we observed UGCAUG enrichment in both tissues, although highest in muscle and heart.
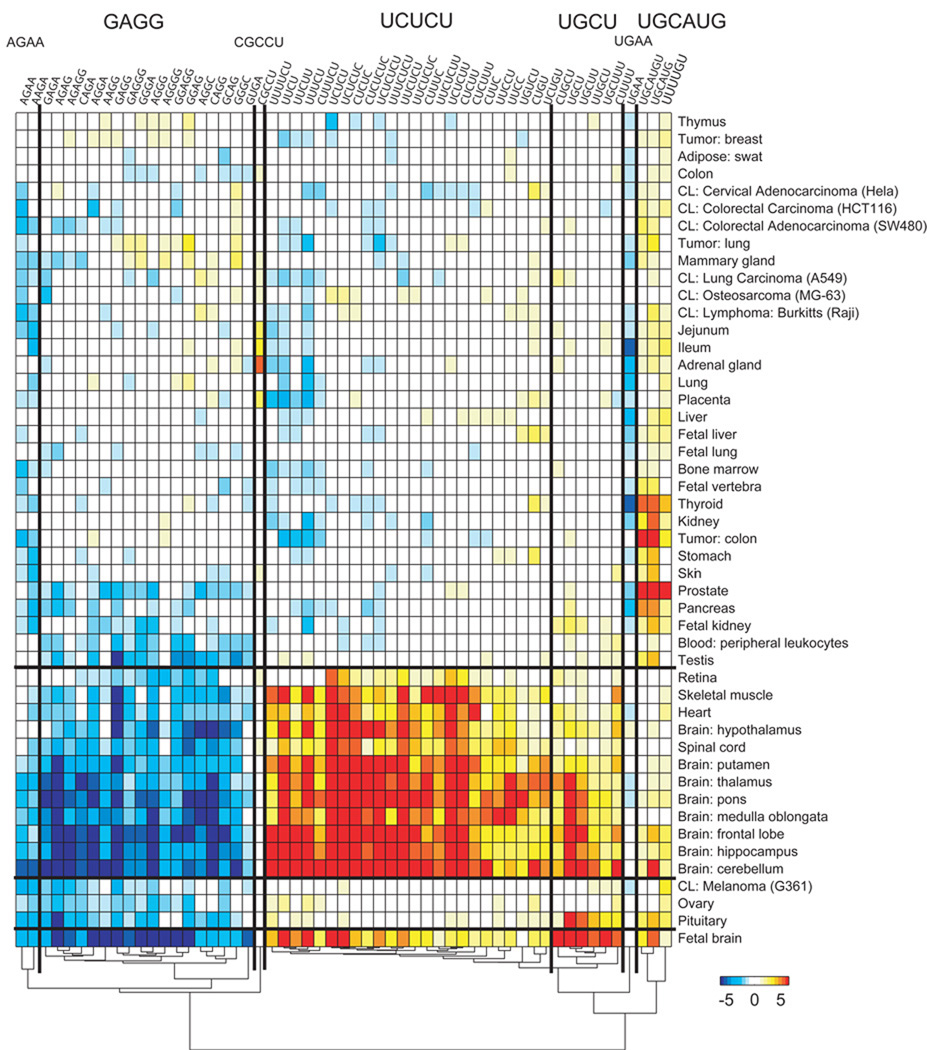
The systematic analysis of all words, in all samples, in all neighborhoods identified 143 significant motifs (p < 1e-3, Bonferroni-corrected; Methods). Two prominent motifs exist upstream of cassette exons (Figure 5). A UC-rich cluster, exemplified by UCUCU, is most enriched in brain and muscle tissues and the AG-rich cluster, including GAGG, AGAGG, and AGGG, is depleted in cassette exons upregulated in brain. Other clusters are enriched in brain (UGCU) and in several tissues (UGCAUG). Smaller clusters include motifs AGAA, CGCCU, and UGAA.
Figure 5.
Motif enrichment upstream of cassette exons upregulated in human tissues and cell lines. Values (−log10 e-value) are positive for enrichment and negative for depletion. Motifs and tissues are clustered using agglomerative clustering of the enrichment values.
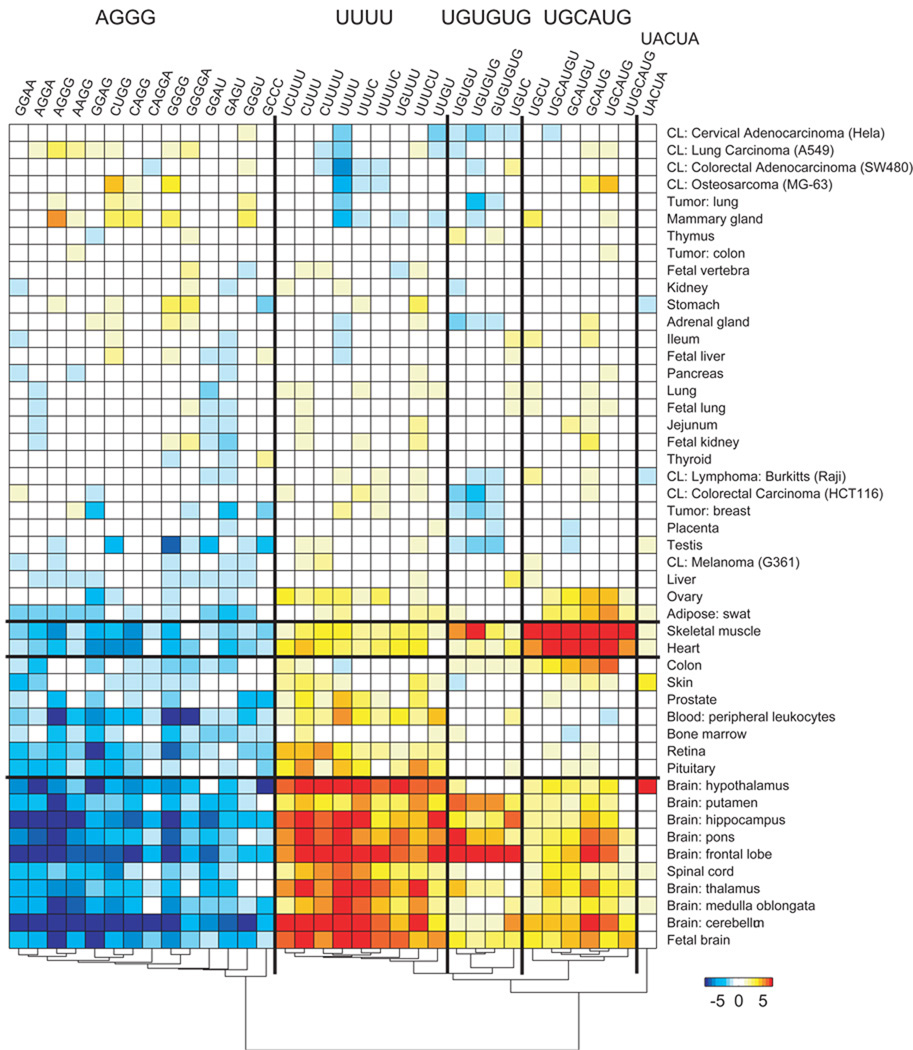
Motifs in the intronic neighborhood following upregulated cassette exons cluster into five groups (Figure 6). UGUGUG is enriched in muscle and brain subsections, UGCAUG is enriched primarily in heart and skeletal muscle, and UUUU is enriched in brain tissues. AG-rich motifs are depleted downstream of cassette exons upregulated in several tissues, including brain, peripheral leukocytes, bone marrow, and striated muscle. UACUA is enriched in hypothalamus.
Figure 6.
Motif enrichment downstream of cassette exons upregulated in human tissues and cell lines. Values (−log10 e-value) are positive for enrichment and negative for depletion. Motifs and tissues are clustered using agglomerative clustering of the enrichment values.
Enrichment occurs in only two other neighborhood and regulation combinations. Immediately upstream of downregulated cassette exons, UCUCU enrichment occurs in all samples except brain, spinal cord, muscle, heart, and leukocytes (online material). Coupled with PTBP1 gene expression, these data corroborate the role of PTB in silencing downstream exons. In the intron downstream of the exon preceding upregulated cassette exons (the 5' splice site), AU-rich motif enrichment occurs in brain and G-rich motif enrichment occurs in HeLa and HCT116, while G-rich motif depletion occurs in brain. G-rich motifs are highly enriched at the 5' splice site preceding cassette exons relative to constitutive exons. G-rich motifs may act to silence cassette exons in brain when positioned in the 5' splice site following a cassette exon and may be a generic 5' splice-site defining factor8,9. This suggests a similar role for G-rich motifs near the 5' splice-site upstream of cassette exons.
This set of predicted alternative splicing motifs is in excellent agreement with existing studies, including tissue-specific roles for UGUGU, UCUUUC, UGCAUG, UGUGUC, UGCAUG and UUUUU 18,26–30. In other cases, motifs identified in the literature, such as ACUAAC 30,31 lie just below our threshold of statistical significance. Fagnani et al.17 identified pyrimidine-rich intronic motifs adjacent brain-enriched cassette exons, but with enrichment spread over several genomic regions and with other motifs, such as UGCAUG, scoring lower.
High resolution map of RNA alternative splicing regulation
We expected motif enrichment to occur non-uniformly across the neighborhoods, especially within the 200 nt intronic regions. To more precisely predict where individual motifs exert a regulatory impact, we examined each motif's frequency at each nucleotide position in each of the eight neighborhoods, using a Gaussian wavelet for smoothing (Methods).
UGCUAG enrichment occurs 10–80 nt downstream of cassette exons upregulated in muscle
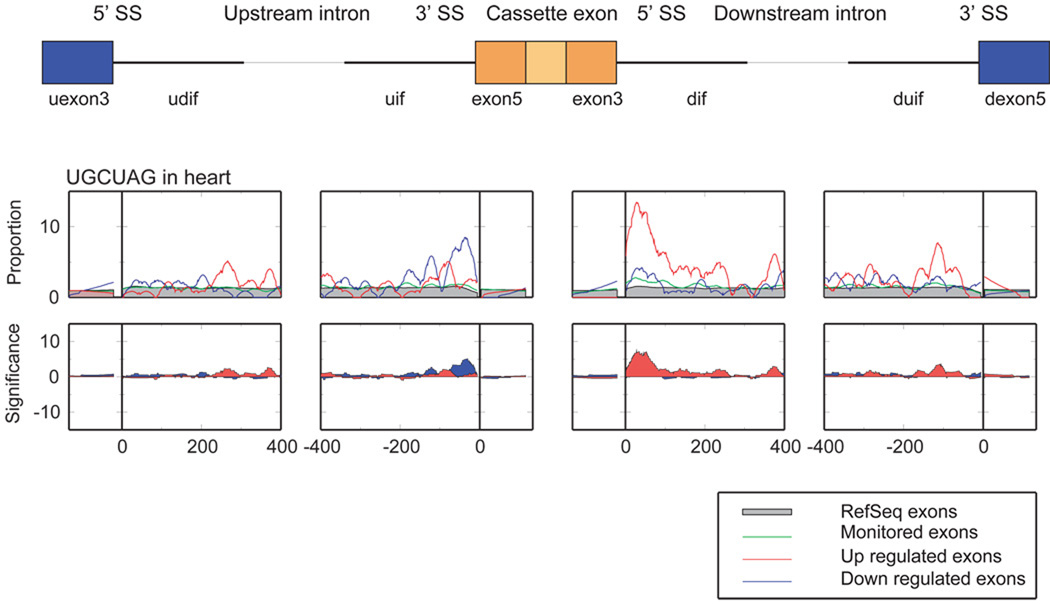
The motif UGCAUG has been associated with Fox proteins (below,6). We found UGCAUG enrichment highest in the intronic neighborhood following cassette exons upregulated in striated muscle (Figure 4C). When examined at higher resolution, the regulatory influence of UGCAUG in muscle varies across and within the eight neighborhoods (Figure 7). A broad, highly significant enrichment of UGCAUG occurs from 10 to 80 nt downstream of cassette exons upregulated in heart and skeletal muscle. A second enrichment peak occurs from 65 to 15 nt preceding cassette exons downregulated in heart, while less enrichment occurs in skeletal muscle in this region (online material). Using RT-PCR, RNAi and cDNA over-expression, we validated the position-specific influence of UCGAUG on cassette exon inclusion, identified Fox alternative splicing targets, and found that the targets are enriched for genes involved in neuromuscular function32.
Figure 7.
Higher resolution enrichment of UGCAUG adjacent heart regulated cassette exons. Middle: smoothed fraction of the cassette exons with UGCAUG. Red, heart upregulated cassette exons; blue, heart downregulated; green, all monitored exons; filled gray regions, RefSeq exons. Bottom: hypergeometric probability (−log10 p-value) for up- and downregulated exons relative to all monitored exons.
UCUCU enrichment occurs from 110 to 35 nt preceding regulated cassette exons
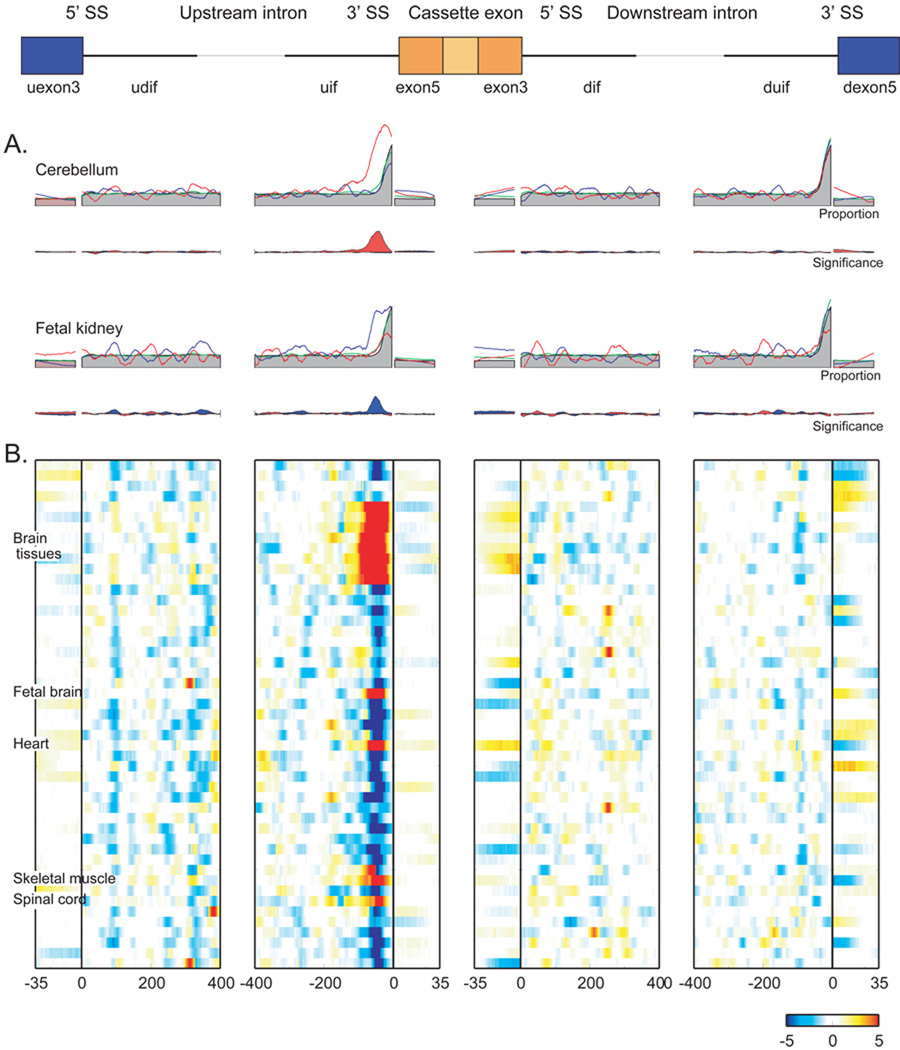
UCUCU has been associated with PTB proteins (below,3–5). The motif UCUCU frequently occurs directly upstream of 3' splice sites as part of the polypyrimidine tract. Indeed, when we examined constitutive and cassette exons, both showed high UCUCU occurrence at the 3' splice site relative to the average intronic rate. However, enrichment upstream of constitutive exons extends only 35 nt from the 3' splice site into the upstream intron, whereas tissue-varying cassette exons show extended enrichment (Figure 8A). Exons upregulated in cerebellum show marked UCUCU enrichment from −95 to −15 nt (Figure 8A). Fetal kidney shows UCUCU enrichment in a similar location, −75 to −20 nt, but intriguingly, this enrichment surrounds downregulated cassette exons. Almost every tissue shows UCUCU enrichment in the similar location from −110 to −35 nt preceding cassette exons (Figure 8B). To our knowledge this is the first high-resolution map of UCUCU location and splice regulatory influence. Brain tissues, spinal cord, retina, and striated muscle show enrichment preceding upregulated cassette exons while other samples show enrichment preceding downregulated cassette exons. Thus, we find that UCUCU enrichment occurs in the polypyrimidine tract immediately preceding the 3' splice site upstream of all exons but enrichment from −110 to −35 nt occurs only upstream of tissue-regulated cassette exons.
Figure 8.
Higher resolution enrichment of UCUCU adjacent regulated cassette exons. A) Smoothed fraction of the cassette exons with UGCAUG in cerebellum (upper) and fetal kidney (lower). Red, upregulated cassette exons; blue, downregulated; green, all monitored exons; filled gray regions, RefSeq exons. Maximum significance in cerebellum/uif is p-value < 1e-22; maximum proportion is 52. B) Hypergeometric probability (−log10 p-value) for up- and down-regulated exons relative to all monitored exons. At each point, the most significant p-value associated with either up or down regulated exons is shown. Samples are ordered alphabetically as per Figure 5.
Exonic splicing elements act to define exons (e.g.,33–35). While we examined both introns and exons for motifs associated with tissue-varying cassette exons, only intronic motifs passed the p-value cutoff. While this might suggest intronic motifs play a greater role in tissue-specific cassette exon regulation, we searched only 39 nt of each exonic region compared to 200 nt for each intronic region, and the smaller window will lessen the significance of exonic motifs. Indeed, we see evidence for exonic UCUCU enrichment at the 3' edge of cassette exons expressed at higher levels in brain and heart but at less significant p-values (Figure 8B). We also did not build cross-species conservation into our statistical model for motif detection. To assess whether more complex motif-detection methods would significantly alter our results, we tested several other published methods, both pre-filtering sequences using cross-species conservation (e.g., 36), and filtering motifs based on frequency in neighborhoods adjacent to alternate and constitutive exons (e.g., 37). The results were largely similar in terms of the motifs identified, associated samples, and associated locations (Supplementary Note). For example, the p-value for UGCUAG enrichment downstream of skeletal muscle enriched exons changes from 1e-16 to 1e-12 when using conservation and is still the most significant word. The next enriched word in both cases is the related word GCAUGU at 1e-09 and 1e-07, respectively, while exonic motifs remain below significance.
UGCU, UGUGU, and AG-rich motifs show different localization
Other previously established relationships between RNA binding proteins and motifs include muscleblind proteins binding UGCU10, and CELF12 and SUP-12 (RBM38)12,13 proteins interacting with UGUGU. Here UGCU enrichment occurs upstream of cassette exons upregulated in brain, from −100 to −5 nt upstream of the 3' splice site (online material). Less significant enrichment is found 5 to 50 nt downstream of brain-upregulated cassette exons. Skeletal muscle and heart show enrichment 30 to 110 nt following upregulated cassette exons but not upstream. UGUGU shows consistent enrichment 10 to 100 nt following brain and spinal cord upregulated cassettes. hnRNP A/B and hnRNP F/H bind purine-rich motifs GGGG and AGGGG, respectively8,9. In our data, AGGG and similar purine motifs are depleted preceding brain upregulated cassette exons (Figure 5). When examined at higher resolution, depletion of AG-rich motifs occurs over a wide region. In cerebellum, the depletion ranges from 150 nt preceding the cassette exon to over 100-nt beyond the exon.
FOX and PTB act on non-overlapping sets of cassette exons
As both the UCUCU and UGCAUG words appear to regulate alternative splicing in some of the same samples, but only when positioned in precise regions around exons, we explored whether they occur around the same cassette exons. In cassette exons upregulated in skeletal muscle, we counted occurrences of UCUCU in the region −110 to −35 nt upstream of the cassette exon and UGCAUG in the region from 10 to 80 nt downstream of the cassette exon (Figure S3). At least one of the motifs occurs adjacent to 33% of cassette exons upregulated in skeletal muscle. However, we were surprised that they co-occur adjacent to only 2% of the exons, not significantly below the 3% expected by chance (p= 0.2), suggesting these two motifs, both enriched in muscle, represent independent regulatory mechanisms. In cerebellum, as in skeletal muscle, UGCAUG and UCUCU are both associated with upregulated cassette exons. 10% and 23% of the cassette exons upregulated in cerebellum contain the motif in the identified region, respectively, and 31% of the exons contain at least one. The motifs co-occur adjacent only 2% of the exons, the percentage expected by chance, suggesting independent action as in skeletal muscle. In fetal kidney, on the other hand, UGCUAG is not enriched adjacent to up- or down-regulated cassette exons, likely reflecting the lower levels of Fox-1 expression, while UCUCU enrichment occurs upstream of down-regulated cassette exons, likely reflecting higher levels of PTB expression.
De novo predicted relationships between RNA-binding proteins and binding elements
The large number of samples and splicing events in the compendium can be leveraged to predict associations between trans-acting splicing regulatory proteins and binding elements. For 135 of the motifs identified (Figure 5 & 6), we calculated a signed, normalized rank-order metric representing the similarity between the motif enrichment/depletion (p-value) tissue-profiles and the gene expression profiles of RNA-binding proteins (Figures S4–5, Methods). The results of this calculation represent de novo predictions of potential protein-motif partners and capture many known relationships. PTBP1 has the highest absolute score, with a negative value, for UCUCU upstream of cassette exons and is also negatively associated with pyrimidine-rich motifs downstream of upregulated cassette exons, as previously established4,38. Upstream of upregulated exons, UCUCU is enriched when PTBP1 expression is low but not when PTBP1 is high (online material). Upstream of downregulated exons, UCUCU is more often enriched when PTBP1 is expressed. Thus, cassette exons preceded by UCUCU are downregulated when PTBP1 is expressed and upregulated when PTBP1 is absent. Upstream of cassette exons, HNRPF has the highest positive score for the purine motif AGGG, and HNRPF and HNRPH1 are known to interact with AG-rich and G-motifs8,9. Downstream of cassette exons, the CELF protein CUGBP2 ranks highest for exon inclusion using UGUGUG, and Fox proteins A2BP1 and RBM9 rank first and second, respectively, for UGCAUG. SFRS6, SFRS7, and HNRPF score positively for AGGG. The fact that expression of Fox, CELF, and HNRPF are positively associated with their motifs surrounding up-regulated cassette exons implies that the exons are preferentially included when the protein is expressed, and suggests a mechanism where the protein acts to enhance inclusion of exons that are otherwise skipped.
The RNA-binding protein NOVA1 targets YCATY and YCAY motifs, with a position-dependent influence39,40. We examined our data for this degenerate motif, summing the values for all four words of YCATY (CCATC, CCATT, TCATC, TCATT) and calculating the hypergeometric probability against the count of the similar set of words in the background. Exons downregulated in brain are enriched in YCATY upstream whereas exons upregulated in brain are depleted of YCATY upstream and enriched downstream (Figure S6). We observe NOVA1 levels almost 10-fold higher than the pool in brain and spinal cord, corroborating a regulatory mechanism in which NOVA1 expression causes exon skipping when bound to upstream YCATY and causes exon inclusion when bound to YCATY downstream of the exon. These results are fully consistent with previous reports on the position-specific role of NOVA140.
Discussion
This is the first genome-scale compendium of human alternative splicing-events and the largest alternative splicing-event compendium of any species with each isoform of 24,426 splicing events measured in 48 tissues and cell lines. These data provide a resource for further studies of the expression and regulation of the transcriptome with detail not available previously. We also provide a catalog of annotated spicing events; the mapping of all probes to the hg18 human genome, viewable in the UCSC browser; and the expression of each probe, gene, exon, splicing event and all word enrichments in each tissue. While not investigated here, these data will be of substantial value in mapping and analyzing alternative 5' and 3' exon usage and tissue-specific alternative polyadenylation.
The splicing event expression compendium was then used to identify candidate regulatory motifs, identifying 143 motifs in five main groups that are associated with tissue-varying alternative splicing. For each motif, we measured the enrichment in each tissue and neighborhood combination and determined the regulatory impact (inclusion or exclusion) on the transcription of the associated cassette exon. In cerebellum, for example, over 30% of the preferentially included cassette exons are adjacent to either a PTB (−110 to −35 nt upstream) or Fox (10 to 80 nt downstream) motif.
Our results extend previous studies of motifs in brain and muscle to 48 diverse samples, and present tissue- and position-specific maps of each motif's likely regulatory influence, at a resolution that to our knowledge is only currently available for YCAY (Nova)40. With the more precise location of motif regulatory impact, we are able to show that co-occurrence rate of the UGCAUG (Fox) and UCUCU (PTB) motifs around tissue-regulated cassette exons is similar to that expected by chance, suggesting they act independently.
Finally, we used splicing array data to identify RNA regulatory motifs and predict binding elements for RNA-binding proteins de novo. While these predictions will require further validation, the fact that many established associations rank highly (e.g., UGCAUG with Fox-1, UCUCU with PTB1, and UGUGUG with CELF), suggests these results could be used as part of a prioritization scheme for further experimentation.
Methods
Microarray pattern generation
We downloaded genome and transcript sequences from the National Center for Biotechnology Information on January 8, 2003. We aligned RefSeq, mRNA, EST41 and patent transcripts to the genome using sim442 and identified all exons and exon-exon junctions. We also aligned mouse transcripts to the human genome to identify possible splice forms not yet present in human transcript databases43. An optimized 60 nt probe was designed to monitor each exon and a 36 nt probe was centered across each exon-exon junction44. Exons less than 60 nt were monitored using shorter probes attached to T-stilts. Probes predicted to perform poorly, such as those associated with repeat sequences or containing unacceptable GC-content, were discarded. In total, 383,014 oligonucleotide probes to monitor 203,672 exons and 178,351 exon-exon junctions in 17,939 genes were synthesized to a 17 array set, with each array containing 23,107 non-control probes. Arrays were ordered from Agilent (California). Probes have been mapped to the hg18 genome and can be viewed with the UCSC genome browser using the supplementary probe alignment file.
Tissue preparation and hybridization
48 diverse human tissues and cell lines were hybridized to this array set. Samples were purchased as pools from multiple donors, typically over 10 (Clontech, Mountainview, CA). We isolated polyA+ RNA from each sample, amplified and labeled the entire transcript structure with Cy3 and Cy544, and hybridized all samples against a common reference pool in a dye-swamp replicate experiment45. Pooled RNA from 20 diverse disease-free adult tissue pools comprised the reference pool. Single array intensities were normalized spatially and for overall intensity; fluor-reverse pairs were combined; and intensity, fold-change (log10-ratio), and uncertainty values were output for each probe and sample46. Tissue-specific probe intensities were further normalized by the relative pool intensity, for the specific probe, to the average intensity of the pool for the probe when hybridized against other samples.
Gene expression
We determined gene expression by averaging values from three selected probes per gene. Probes were prioritized by location in a constitutively transcribed exons or junctions and then by correlation with the average of all constitutive-element monitoring probes across the 48 samples. From the three probes, log10 expression ratios represent the error-weighted average ratio to the pool and intensities as the average probe intensity. P-values are assigned to fold-change values using the uncertainty and reflect the probability of no differential expression between the sample and the pool.
Alternative splicing event profiling
We identified putative mutually exclusive alternative splicing events found in our alignment of RefSeq, GenBank, dbEST, patent, and mouse transcripts to the human build 35 version 1 genome assembly47. As per the microarray design process, transcripts were aligned to the genome, exons and exon-exon junction coordinates were identified. For each gene, we examined the exon and junction coordinates for mutually exclusive splicing events, including cassette exons (single and multiple), mutually exclusive exons, alternative 5' and 3' splice sites, and retained introns. The exon and junction nucleotide sequence specific to each form was extracted; probes were associated with each form based on sequence identity.
For alternative splicing events for which both inclusion and exclusion were monitored, we calculated a fold change (log10-ratio) to the reference pool and intensity for each form. If multiple probes reported on a form, we combined log10 ratios using error-weighted averaging and calculated a median intensity. To estimate the proportion of given splice form relative to the pool, we used "method 1 + 2" of Ule et al.14, similar to a method described by Fehlbaum et al.48. For a given alternative splicing event, log-ratios monitoring event inclusion and exclusion are normalized by gene expression to isolate splicing changes from transcription changes. These normalized inclusion and exclusion log-ratios are compared. The output is a measure of the change in percentage of one splice form, such that a change from 80% of total to 20% of total is reported as −60. If no change in alternative splicing occurs, the method is unable to assign proportionality.
Error estimate and significance calls for differential splice expression
We added an uncertainty, p-value, and filtering to the alternative splicing event profiling. Events were flagged for which the gene intensities were below the noise level in either sample, for which the event intensities were below the noise level in both samples, or for which either the gene or event intensities were near saturation levels in either sample. We flagged events showing intensity more than 10-fold higher than the gene intensity as this represents possible cross-hybridization. In true cases of differential alternative splicing, the log10-ratios monitoring the splice event inclusion and exclusion should have opposite signals after removal of the gene expression changes; we therefore flagged those not consistent. We calculated an uncertainty with each log ratio measurement and using standard error propagation49 calculated a p-value for each splicing event monitored, representing the probability that the splicing event is not differentially expressed, after accounting for gene expression changes. Splice events with a change of at least magnitude 5, a p-value less than 0.3, and no flags are called significantly differentially expressed.
Validation of splicing event profiling
In conjunction with the Cooper Lab (Baylor), we established validation rates for this platform. We profiled mouse heart at different development stages22. We tested 23 microarray predictions (with p-value <0.1 and passing the intensity and cross-hybridization filters described above) and found 17 (74%) showed RT-PCR determined proportionality changes exceeding 15 percentage points (e.g. isoform A changes from 60% to 45% of the total) and the correlation between microarray and RT-PCR splice event profiles was r=0.88 (Table S1).
Re-ratio of microarray data
The microarray values were derived from two-color hybridizations of the sample and a common reference pool. For each sample, the resultant values included an intensity, log10 ratio-to-pool, and log10 ratio-to-pool uncertainty. To define a different pool in silico using one of the existing samples, the log10 ratio-to-"new pool" was calculated for each sample as the difference between the log10 ratio-to-pools from the sample and from the new pool sample. The log10-ratio-to-"new pool" uncertainty was calculated as the square root of the sum of the squares of each log10 ratio-to-pool uncertainty.
Motif identification
For each tissue, we identified up- and downregulated cassette exons and searched associated genomic regions for over and underrepresented motifs. Up- and downregulated cassette exons were identified that passed the described intensity and consistency filters and showed a minimum percent inclusion change of 5 and a p-value lower than 0.3. For each cassette exon, we extracted the repeat-masked nucleotide sequence in eight regions, including the 3' edge of the previous exon, the intronic region adjacent the upstream 5' splice site, the intronic region adjacent the upstream 3' splice site, the 5' region of the cassette exon, the 3' region of the cassette exon, the intronic region adjacent the downstream 5' splice site, the intronic region adjacent the downstream 3' splice site, and the 5' edge of the following exon. For intronic and exonic regions, we extracted 200 nt and 39 nt, respectively, based on previous studies18,40,50 and required introns to be at least 300 nt to limit the impact of neighboring splice sites.
The neighborhood-specific background set included sequences from the set of cassette exons monitored on the microarrays. For each neighborhood in each tissue, we counted the occurrence of every word, size 3 through 7. Then, for each word, tissue, and region, we used the cumulative hypergeometric distribution to assign a p-value representing the probability for observing that number of occurrences in the foreground versus the number in the background set. In cases where we examined enrichment (inverse cumulative hypergeometric) and depletion (cumulative hypergeometric), the most significant p-value was retained and a sign, positive or negative, was assigned to the p-value based on whether enrichment or depletion, respectively, was most significant. Counting the number of motifs, rather than the number of cassette exons with the motif, was more sensitive for motifs that commonly occur in a given region, such as UCUCU upstream of cassette exons.
To identify motifs, such as those in plotted Figure 5, we identified words with signed hypergeometric p-values more significant than 1e-3 (Bonferroni-corrected) in any tissue in the specific neighborhood, minus words found in longer words with more significant p-values. Using randomized exon annotations, we calculated a false discovery rate (FDR) for each motif in each neighborhood. UUUUU and AAAAA had significantly higher FDRs and were excluded from further analysis.
For the high resolution enrichment study, the number of motifs at each position was convolved with a Gaussian wavelet. The Gaussian wavelet was width 40, with maximum value of one at position zero, falling to 0.26 at +/− 20. The smoothed counts were calculated at each position for tissue-upregulated cassette exons, tissue-downregulated cassette exons, and the background sets. We again used the hypergeometric distribution to calculate p-values for enrichment and depletion from the smoothed motif counts, in both tissue-up and tissue-down sets, versus the corresponding motif counts from the set of all cassette exons monitored.
Co-regulation of motifs and RNA-binding protein transcript expression
We considered RNA-binding proteins found in the Panther mRNA splicing gene set and those classified by Interpro as having a RNP domain. Muscleblind proteins (e.g., MBNL2) are not included in these sets; they are found in 24 genes in the Panther double-stranded DNA binding set, so we included this set as well. For each significant motif in each genomic region, we calculated a non-parametric score by sorting tissue enrichment ranking based on the gene expression rankings for each protein, and identifying the greatest divergence from random. The maximum value for each motif was normalized to one. We preserved the sign to represent positive and negative divergence. Clustering in figures is agglomerative clustering.
Supplementary Material
Acknowledgements
We thank Chris Armour, Chris Raymond, David Haynor, Valur Emilsson, Phil Garrett-Engele, Auinash Kalsotra, Fritz Roth, Patrick Loerch, Ronghua Chen, Carol Rohl, Martin Tompa, Eric Schadt, and Lee Lim for input, Rosetta’s Gene Expression Laboratory microarray hybridization facility for microarray data, and Sonia Carlson for project management. Funding for TAC provided by R01GM076493.
References
- 1.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Kwan T, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 3.Chan RC, Black DL. Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro. Mol Cell Biol. 1995;15:6377–6385. doi: 10.1128/mcb.15.11.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 5.Gooding C, Roberts GC, Smith CW. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. Rna. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 6.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Contreras R, et al. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho TH, et al. Muscleblind proteins regulate alternative splicing. Embo J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forch P, et al. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 12.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroyanagi H, Ohno G, Mitani S, Hagiwara M. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol. 2007;27:8612–8621. doi: 10.1128/MCB.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ule J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 15.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 16.Pan Q, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Fagnani M, et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugnet CW, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark TA, et al. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffith M, et al. ALEXA: a microarray design platform for alternative expression analysis. Nat Methods. 2008;5:118. doi: 10.1038/nmeth0208-118. [DOI] [PubMed] [Google Scholar]
- 21.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalsotra A, et al. A post natal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Submitted to PNAS. 2008 doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan PI, Stojdl DF, Marius RM, Bell JC. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol Cell Biol. 1997;17:5996–6001. doi: 10.1128/mcb.17.10.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rupp U, et al. Safety and pharmacokinetics of bivatuzumab mertansine in patients with CD44v6-positive metastatic breast cancer: final results of a phase I study. Anticancer Drugs. 2007;18:477–485. doi: 10.1097/CAD.0b013e32801403f4. [DOI] [PubMed] [Google Scholar]
- 25.Riechelmann H, et al. Phase I trial with the CD44v6-targeting immunoconjugate bivatuzumab mertansine in head and neck squamous cell carcinoma. Oral Oncol. 2008 doi: 10.1016/j.oraloncology.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Brudno M, et al. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29:2338–2348. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci U S A. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeo GW, Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeo GW, et al. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das D, et al. A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing. Nucleic Acids Res. 2007;35:4845–4857. doi: 10.1093/nar/gkm485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voelker RB, Berglund JA. A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing. Genome Res. 2007;17:1023–1033. doi: 10.1101/gr.6017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, et al. The functional landscape of mouse gene expression. J Biol. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goren A, et al. Comparative analysis identifies exonic splicing regulatory sequences--The complex definition of enhancers and silencers. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Fairbrother WG, et al. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3' splice site selection in vivo. Rna. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc Natl Acad Sci U S A. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 41.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kan Z, Castle J, Johnson JM, Tsinoremas NF. Detection of novel splice forms in human and mouse using cross-species approach. Pac Symp Biocomput. 2004:42–53. doi: 10.1142/9789812704856_0005. [DOI] [PubMed] [Google Scholar]
- 44.Castle J, et al. Optimization of oligonucleotide arrays and RNA amplification protocols for analysis of transcript structure and alternative splicing. Genome Biol. 2003;4:R66. doi: 10.1186/gb-2003-4-10-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes TR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 46.Weng L, et al. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- 47.Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 48.Fehlbaum P, Guihal C, Bracco L, Cochet O. A microarray configuration to quantify expression levels and relative abundance of splice variants. Nucleic Acids Res. 2005;33:e47. doi: 10.1093/nar/gni047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor JR. An Introduction to Error Analysis. Vol. 270. Mill Valley, CA: University Science Books; 1982. [Google Scholar]
- 50.Fairbrother WG, Holste D, Burge CB, Sharp PA. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.