Abstract
SIRT6 is a histone deacetylase that has been proposed as a potential therapeutic target for metabolic disorders and the prevention of age-associated diseases. Thus the identification of compounds that modulate SIRT6 activity could be of great therapeutic importance. The aim of this study was to develop a screening method for the identification of novel modulators of SIRT6 from a natural plant extract. We immobilized SIRT6 onto the surface of magnetic beads, and assessed SIRT6 enzymatic activity on synthetic acetylated histone tails (H3K9Ac) by measuring products of the deacetylation process. The SIRT6 coated magnetic beads were then suspended in fenugreek seed extract (Trigonella foenumgraecum) as a bait to identify active ligands suppressing SIRT6 activity. While the whole extract also inhibited SIRT6 activity in a cell-based assay, the inhibitory effect of two flavonoids from this extract, quercetin and vitexin, was only detected in vitro. This is the first report for the use of protein-coated magnetic beads for the identification of an active ligand from a botanical matrix, and sets the basis for the de novo identification of SIRT6 modulators from complex biological mixtures.
1. Introduction
Epigenetics is the study of heritable alterations in gene expression caused by mechanisms other than changes in DNA sequence. Epigenetic regulators or chromatin-modifying enzymes, such as methyl transferases and histone acetyltransferases (HAT), control gene expression via histone modifications and (or) ATP-dependent nucleosome remodeling [1,2]. The nucleosome is the repeating minimal unit of chromatin that consists of 146 bp of DNA wrapped around the core histone octamer, consisting of two copies of each of histones H2A, H2B, H3 and H4. All of these core histones carry a flexible lysine-rich amino-terminal tail that extends from the nucleosome and is the subject to multiple posttranslational modifications including acetylation. HAT-mediated acetylation of histone tails correlates largely with active gene transcription, while deacetylation by histone deacetylases (HDAC) family is involved in the downregulation of gene expression [3]. Respective changes in acetylation are thought to control gene expression by altering chromatin structure [3].
HDACs are subdivided into four classes based on functional similarity: class I (HDAC1, 2, 3, and 8), class II (HDAC4, 5, 6, 7, 9, and 10), class III (NAD+- dependent SIRTuins; SIRT1-7) and class IV (HDAC11) [4–6]. Class III differs from other deacetylases in that lysine deacetylation is NAD+-dependent and leads to the generation of nicotinamide, which, in turn, inhibits their enzymatic activity [3]. It is important to note that the SIRTuin activities are controlled by the energy status of the cell, which is reflected by the cellular NAD+/NADH ratio. SIRT1 is the best-studied member of the SIRTuin family and has been proposed to be a target to reduce the inflammatory components of type 2 diabetes and metabolic diseases [7]. Interestingly, SIRT6 knockout mice show a premature aging phenotype and lifespan shortening, developing several acute degenerative processes by three weeks of age and dying prematurely at one month of age [8]. A t the molecular level, SIRT6 plays a role in DNA repair and chromosome stability, and deacetylates telomeric histone H3 at lysine 9 (H3K9) [9]. Lastly, SIRT6 has been proposed as a potential therapeutic target for metabolic disorders and the prevention of ageassociated diseases [10, 11].
The identification of new compounds that could modulate SIRT6 activity could be of great therapeutic importance. There is no known SIRT6 ligand to date other than nicotinamide. A well-established and robust approach for identification of novel drug candidates consists in the screening of constituents of medicinal plant and marine extracts. In fact, of the 1184 new drugs approved from 1981 and 2006, more than 70% were either derived from or structurally similar to organic material or nature based [12]. Compared to current approaches to assess SIRT6 activity, such as the radiolabeled ADP ribosylation assay [13] and the fluorescence deacetylase assay using an acetylated peptide substrate [14], the newly developed CycLex SIRT Deacetylase Fluoremetric Assay offers a great opportunity to screen for novel biologically active molecules acting on SIRT6. However, natural extracts with their diverse chemical compositions can potentially interfere in this screening assay, which relies on the use of a fluorescence-quenched acetylated peptide substrate that produces a fluorescent signal after deacetylation-coupled proteolytic cleavage of the peptide. This approach could be quite sensitive to autofluorescence, fluorescence-quenching or anti-proteolytic properties of natural extracts.
The aim of this study was to develop a screening method for the identification of novel modulators from natural plant extract that target SIRT6 deacetylase. In this approach, the targeted protein is immobilized onto the surface of magnetic beads and the active ligands are subsequently fished out by suspending the protein-coated beads directly in a crude plant extract. We have previously reported the development and use of human serum albumin- and HSP90-coated magnetic beads for the selective fractionation of interacting molecules present in a mixture of compounds [15,16]. Magnetic nano-particles have also been used as a novel high throughput screening method to determine whether a compound has an affinity for an immobilized target in a ‘yes’ or ‘no’ method [17]. Here, catalytically active SIRT6 was immobilized on magnetic beads and submitted to an H3K9Ac deacetylation-guided process in the presence of the fenugreek seed extract (Trigonella foenum-graecum). This extract was chosen because of its ability to lower blood glucose and cholesterol in type 1 and type 2 diabetic patients and experimental diabetic animals, as well as reducing high fat diet-induced body weight gain in obese mice [18,19]. The developed screening method identified the flavonoids, quercetin and vitexin, and showed that these compounds inhibited SIRT6 activity in vitro.
2. Materials and Methods
2. Materials
2-(N-morpholino)ethanesulfonic acid (MES) was purchased from EMD-Calbiochem (Gibbstown, NJ). Adenosine, adenosine 5′-diphosphoribose (ADP ribose), adenosine 5′-monophosphate (AMP), p-aminohippuric acid, 1-ethyl-3-(3-methylaminopropyl)carbodiimide (EDC), gluteraldehyde, hydroxylamine hydrochloride, nicotinamide, nicotinamide adenine dinucleotide (NAD+), nicotinamide mononucleotide (NMN), potassium phosphate dibasic, pyridine (99.8%), sodium azide, sodium cyanoborohydride, and sodium phosphate monobasic were obtained from Sigma-Aldrich Chemical Co. (Milwaukee, WI). N-Hydroxysulfosuccinimide (Sulfo-NHS) was from Pierce (Rockford, IL). Acetylated histone H3 (K9) peptide (H3K9Ac) and acetylated histone H3 (K14) peptide (H3K14Ac) were from Upstate/Millipore (Temecula, CA). Solutions were prepared using purified water from a Millipore MilliQ system (Millipore Corporation, Bedford, MA). BcMag amine-terminated magnetic beads (50 mg/mL, 1 µm) were purchased from Bioclone, Inc. (San Diego, CA). The manual magnetic separator Dynal MPC-S was from Invitrogen Corporation (Carlsbad, CA).
2. Methods
2.1. Cell Culture
DT 40 chicken B-cells were cultured in RPMI 1640 (Mediatech) supplemented with 10% FBS (Gibco), 25mM HEPES, 2mM L-glutamine, Pen/Strep 50U/50µg/mL, and 1% chicken serum (Invitrogen). All cultures were maintained at 41°C with 5% CO2. SIRT6−/− cells were generated by gene-targeting (20) and will be described in detail elsewhere.
2.2. Cloning of expression construct
The expression plasmid pMBP-SIRT6 encodes a maltose-binding protein (MBP)-SIRT6 fusion protein. It was created by amplifying the full-length chicken SIRT6 cDNA from DT40 cDNA using primers SIRT6F (5′-CGCGGATCCATGGCGGTGAATTACGC-3′) / SIRT6R (5′-CGCTCGAGTCAGGTGAGGAGAGGCTC-3′), digesting the PCR product with BamHI and XhoI, and ligating is into the BamHI/XhoI sites of pMH6 [21].
2.3. Protein purification
Maltose binding protein (MBP) fusion proteins were expressed in Rosetta E. coli cells. Bacterial cultures were grown in 1000 mL of Millers/Luria-Bertani (LB) broth (10g/l tryptone, 5g/l yeast extract, 10g/l sodium chloride, 2g/l glucose) at 37°C. Protein production was induced by addition of isopropyl-D-thiogalactopyranoside (IPTG) to a final concentration of 50µM (Figure 1). After three hours, cells were pelleted, resuspended in lysis buffer (20mM Tris-HCl, pH 8.0, 200mM NaCl, 1mM EDTA, 5mM β-mercaptoethanol, 0.2mM ZnCl2, 10% glycerol, 1 mM PMSF, 10µg/mL leupeptin, 100µg/mL aprotinin, and 10µg/mL pepstatin A), and lysed by sonication (Branson digital sonifier). MBP-fusion proteins were bound in batch to 300 µl of amylose resin (New England Biolabs) and washed three times with lysis buffer. To cleave off the MBP tag, the resin was washed twice with Factor Xa reaction buffer (20mM Tris-HCl, pH 6.5, 50mM NaCl, 1mM CaCl2), and subsequently incubated with Factor Xa (New England Biolabs) for three hours at room temperature. The released SIRT6 protein was dialyzed against phosphate-buffered saline (PBS) at 4°C overnight, and subjected to size-exclusion chromatography using Sephadex G-200 column (Amersham Biosciences, Inc.). Fractions containing monomeric SIRT6 protein were pooled, dialyzed against storage buffer (25mM Tris-HCl, pH 8.0, 150mM KCl, 1mM MgCl2, 5µM ZnCl2, 2mM DTT, and 20% glycerol) and stored at −80°C.
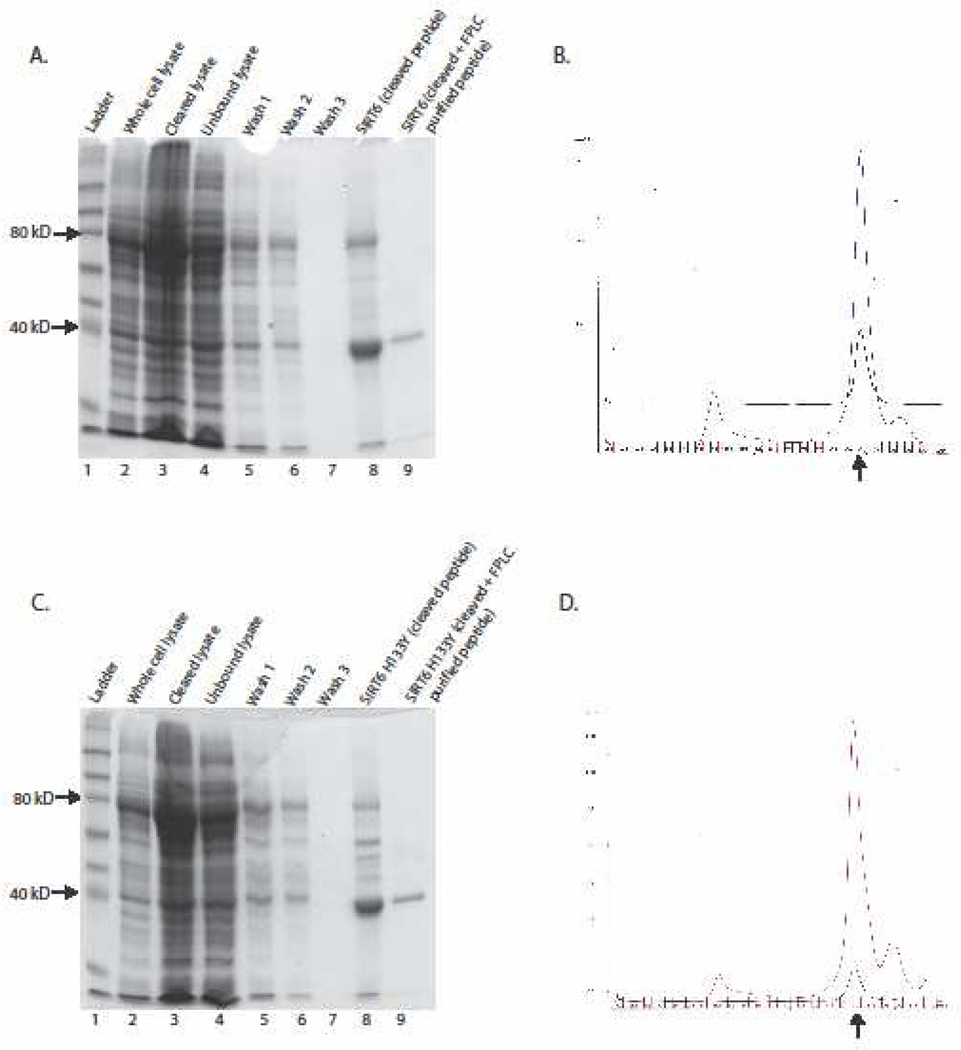
Figure 1.
(A) Purification of recombinant SIRT6. SIRT6 MBP fusion peptides (80kD) were bound onto amylose resin, washed and equilibrated in Factor Xa reaction buffer. Overnight incubation with Factor Xa enzyme released SIRT6 peptides (40kD) from MBP/amylose complex (lane 8). Recovered SIRT6 was further purified by size exclusion chromatogram (lane 9), dialyzed against storage buffer and stored at −80°C. (B.) For the initial FPLC analysis, cleaved single step purified SIRT6 was dialyzed against PBS. Blue line on size exclusion chromatograph shows single step purified fractionation of SIRT6 peptides. Monomeric fractions (indicated by arrow) of SIRT6 were collected and dialyzed against our storage buffer and reanalyzed by FPLC. Further purification of monomeric SIRT6 peptides by size exclusion chromatography is represented by black line. The same purification scheme was used for the using SIRT6 catalytic mutant (H133Y) (panels C and D).
2.4. Immobilization of SIRT6 onto the Surface of Magnetic Beads (MB)
2.4a. SIRT6 (NT)-MB
SIRT6 protein was immobilized onto the magnetic beads using a previously published method [16]. Briefly, 0.5 mL (25 mg) of BcMag amine-terminated magnetic beads was placed in a 2 mL microcentrifuge tube. After separation using a magnetic separator (Dynal MPC-S), the supernatant was discarded and the beads were washed three times with 1 mL of coupling buffer (pyridine [10mM, pH6.0]). BcMag beads were suspended in 1 mL of 5% gluteraldehyde solution in coupling buffer and rotated for 3h. After separation, BcMag beads were washed three times with 1 mL of coupling buffer. A suspension of 260 µg of SIRT-6 in 500 µL of coupling buffer containing 1% sodium cyanoborohydride was added to the activated BcMag beads. The reaction was left for 7 days at 4°C with gentle rotation. The supernatant was discarded and 200 µL of coupling buffer containing 100 mM hydroxylamine and 2.5% sodium cyanoborohydride was added. The mixture was shaken overnight at room temperature with gentle rotation. The supernatant was discarded and SIRT6 (NT)-MB was washed three times with 1 mL of storage buffer (phosphate buffer [10mM, pH7.4] containing 150 mM NaCl and 0.02% sodium azide). For comparison, the catalytically inactive SIRT6-H133Y mutant was also coupled with (NT)-MB according to the same protocol.
2.4b. SIRT6 (CT)-MB
The amine groups on the BcMag beads and the SIRT-6 protein were linked by a previously described method. Briefly, 0.5 mL (25 mg) of BcMag beads was rinsed with 1 mL of MES [100 mM, pH 5.5] in a 2 mL microcentrifuge tube. After magnetic separation, the supernatant was discarded, and the BcMag beads were suspended in 300 µL of MES [100 mM, pH 5.5] and 260 µg of SIRT6 protein. Fifty µL of a mixture of 10 mg of EDC and 15 mg of sulfo-NHS in 1 mL of water were added and the mixture was vortex-mixed for 5 min and left for 3 h at 4°C with gentle rotation. This was followed by the addition of 20 µL of 1 M hydroxylamine, and the mixture was left for 30 min at 4 °C with gentle rotation. The supernatant was discarded and the SIRT6 (CT)-MB was rinsed three times with 1 mL of storage buffer (phosphate buffer [10mM, pH7.4] containing 150 mM NaCl and 0.02% sodium azide). The SIRT6-H133Y (CT)-MB was made according to the same protocol.
2.5. Heat denaturation of SIRT6
SIRT6 solution (1.3 mg/mL) was heat-denatured in a boiling water bath for 10 min and then immobilized as reported above, resulting in the heat-inactivated SIRT6 MB.
2.6. Histone deacetylation assay by mass spectrometry
SIRT6 (NT) and SIRT6 (CT)-MB were incubated with 50 µL of HDAC assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT and 0.2 mM NAD+) containing 5 µg of acetylated histone H3(K9) peptide (amino acid residues 1–21; H3K9Ac) for 2 to 18 h at 37°C. After incubation, 10 µL of 120 µM p-aminohippuric acid was added as an internal standard. After magnetic separation, the supernatant was collected and analyzed by mass spectrometry using a system composed of an Agilent Technologies 1100 LC/MSD equipped with a G1322A degasser, G1312A binary pump, G1367A autosampler, G1316A column thermostat, G1315A diode array detector and G1946D mass spectrometer equipped with an electrospray ionization (ESI) interface. Selected ion monitoring (SIM) chromatograms were acquired using Chemstation software, Rev. A.10.02. For the separation of compounds, a reversed-phase Zorbax XDBC18 analytical column (50×4.6 mm i.d., 1.8 µm particles) was used. The column was operated at 25°C. The mobile phase was a mixture of 0.05% trifluoroacetic acid (TFA) in water (A) and 0.02% TFA in acetonitrile (B), in linear gradient mode, as followed: 2-min hold at 100% A, followed by a linear gradient to 20% B from 2–22 min and then up to 80% B within 0.1 min, held for 2 min, and then re-equilibrated at initial conditions for 4 min. The flow rate was 1.2 mL/min and the injection volume was 50 µL.
The substrates and metabolites were monitored in the positive-ion mode for SIM at m/z 922.4 ([M+3H]3+ of H3K9Ac), m/z 908.4 ([M+3H]3+ of deacetylated peptide; H3K9), m/z 664.1 (NAD+), m/z 123.1 (nicotinamide), m/z 602.1 (acetyl ADP ribose), m/z 335.1 (NMN), m/z 255.1 (nicotinamide riboside; NR), m/z 560.1 (ADP ribose), m/z 348.1 (AMP), and m/z 268.1 (adenosine). The internal standard was monitored at m/z 195.1 (p-aminohippuric acid).
2.7 Chromatin immunoprecipitation (ChIP) Assay
ChIP experiments were performed according to [22]. Briefly, DT40 chicken B cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Cells were lysed with SDS lysis buffer (1% SDS, 1% Triton-X 100 and 150mM NaCl), the chromatin was sheared to fragments of 200–1000 bp in length by sonication, and insoluble debris was removed by centrifugation [23]. Polyclonal antibodies against H3K9Ac or H3K27Ac (Upstate Biologicals) were used to immunoprecipitate DNA-histone complexes. Recovered DNA was quantified by real-time PCR using gene-specific primers that amplify the promoter region of CMYC, BCL2L1, CD3E, LYN, TOX, TMED, AND HSP90B1 (Table 1).
Table 1.
PCR primer list for SIRT-6 specific targets
| Gene symbol |
Sequence | orientation |
|---|---|---|
| C-MYC | 5’-CCTCCCCAGCAAGAACTACG-3’ | Fwd |
| 5’- AAGTAGGGCTGCACCGAGTC-3’ | Rev | |
| BCL2L1 | 5’-CTCCCGGTTACTGCTGGACA-3’ | Fwd |
| 5’-GGACTCATTGAGGGCGTCTC-3’ | Rev | |
| CD3E | 5’-AAGCATTTCTGCATCTTCCCAA-3’ | Fwd |
| 5’- TTGTGGGAGGCTCCTTAGCA-3’ | Rev | |
| LYN | 5’-ACATGCAGTGCAGATGGTCC-3’ | Fwd |
| 5‘-TGAGTCATCACATTGCAGCTCC-3’ | Rev | |
| TOX | 5’-AACGGGTTAGGTGAGCCCTT-3’ | Fwd |
| 5’- ACACCACAAAGTCTGGATGGG-3’ | Rev | |
| HSP90B1 | 5’-TTATGCCTGTTAAGAAATAGTCCAGAAA-3’ | Fwd |
| 5’- GTGGGTATGTGTACTTCCTAAGGTTTT-3’ | Rev |
3. Results and Discussion
3.1. Purification of bacterially-expressed SIRT6
To purify recombinant SIRT6 for our inhibitor screen, MBP-SIRT6 fusion protein and the catalytically inactive H133Y mutant thereof were expressed in Rosetta E. coli cells. After initial affinity purification over amylose resin, the MBP-tag was cleaved from resin-bound SIRT6 protein, and the released monomeric SIRT6 was purified by size-exclusion chromatography (Figure 1 Both, wildtype SIRT6 and the H133Y mutant showed very similar size distributions during this second purification step (Figure 1, B+D) with the majority of the protein being apparently monomeric, and both proteins maintained their monomeric state even after dialysis of the respective fractions. Misfolded proteins frequently form higher order aggregates, and thus present aberrant peaks of high molecular weight during size exclusion chromatography. The similarity of the purification patterns and the absence of aggregated proteins after the size selection step, suggests that both recombinant proteins are likely folded equally well.
3.2. Immobilization of SIRT6 to magnetic beads
The immobilization of SIRT6 was carried out either at the N-terminus (using gluteraldehyde) (SIRT6 (NT)) or C-terminus via a condensation reaction using the water-soluble carbodiimide, EDAC, (SIRT6 (CT)) onto the surface of amine-coated magnetic beads. To determine whether the immobilized recombinant SIRT6 retained its activity, a histone deacetylase assay was carried out using 200 µM NAD+ and synthetic peptides corresponding to the acetylated tail of H3K9Ac as the substrate. A time course experiment showed that maximal deacetylation occurred after 4 h at 37°C (data not shown). Although SIRT6 immobilization via both the N- and C-terminus was carried out successfully, only SIRT6 (CT) maintained its deacetylase function while SIRT6 (NT) was catalytically inactive (data not shown). This is consistent with a previous report that the N-terminal domain contributes to SIRT6 catalytic activity [24]. Hence, only the SIRT6 (CT)-MB was used for the rest of the study. The stability of the SIRT6 (CT)-MB was assessed by the determination of histone deacetylase activity. It was demonstrated that the SIRT6 (CT)-MB could be reused for a period of two weeks (data not shown), after which a loss of activity was observed.
3.3. Histone deacetylation assay on SIRT6 (CT)-MB
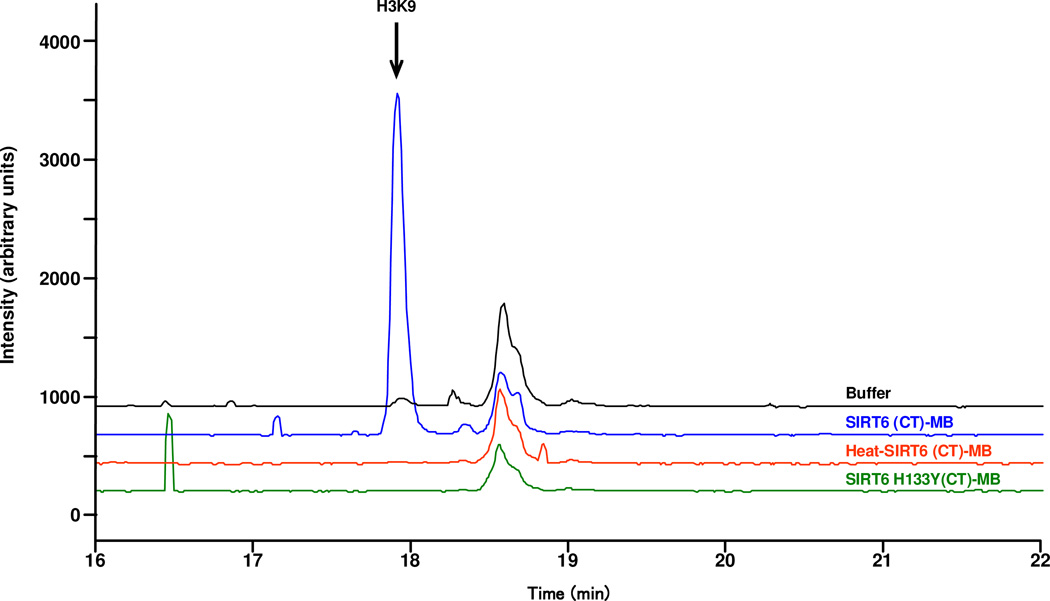
To evaluate the specificity of SIRT6 catalytic deacetylase activity in the deacetylation of the acetylated tail of H3K9, we monitored the production of H3K9 in the SIRT6 (CT)-MB, the catalytic mutant of SIRT6 (H133Y) and the heat-inactivated wildtype SIRT6. H3K9 was only observed with the SIRT6 (CT)-MBs, whereas immobilized H133Y mutant and heat-inactivated SIRT6 were found to be inactive (Figure 2). It is noteworthy that incubation of SIRT6 (CT)-MB with an H3K14Ac tail, containing an acetylated lysine that is not thought to be a SIRT6 substrate, failed to elicit a deacetylation signal (data not shown), further demonstrating the selectivity of the SIRT6 (CT)-MB.
Figure 2.
Incubation of 1 µg of H3K9Ac with buffer alone (Tris [50 mM, pH 8.0] containing 150 mM NaCl, 0.2 mM NAD and 1mM DTT at 37°C for 4 hours, SIRT-6 CT, H133Y SIRT-6 (CT)-MB and heat inactivated SIRT-6 (CT)-MB.
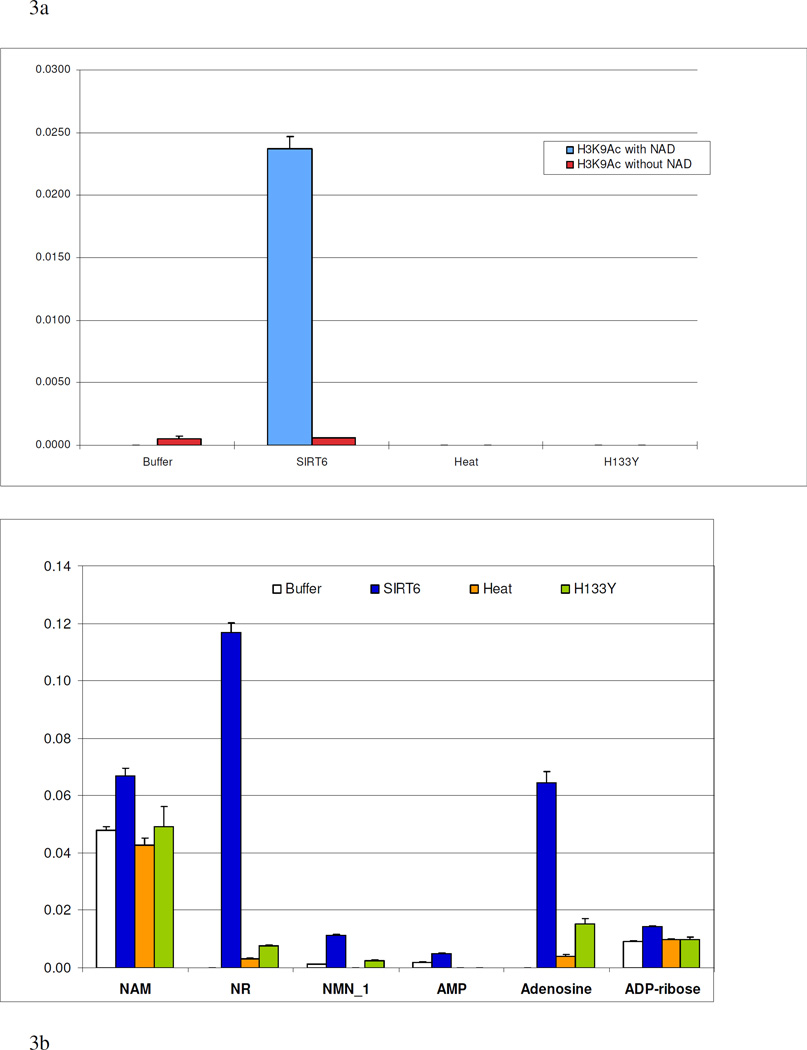
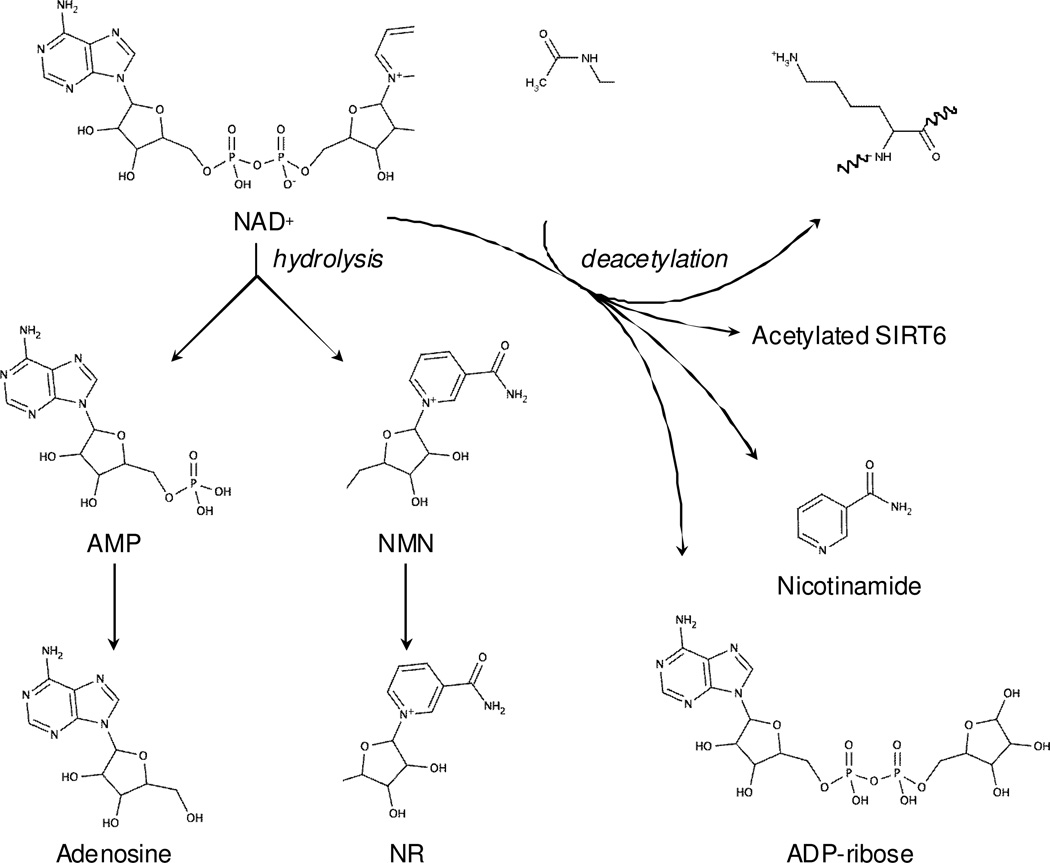
In a second set of experiments, wildtype SIRT6, the H133Y mutant, and heat-inactivated SIRT6 were immobilized on MBs and incubated with H3K9Ac in the presence or absence of 200 µM NAD+ for 4 h at 37°C (Figure 3a). After removal of the beads, the supernatants were resolved on a reversed-phase Zorbax XDB-C18 analytical column to detect and quantify NAD+, NAM, NR, NMN, AMP, adenosine, and ADP-ribose (Scheme I), as well as H3K9Ac and H3K9. Consistent with our earlier results, the deacetylation of H3K9Ac occurred only with the wildtype SIRT6 (CT)-MBs in the presence of NAD+. The levels of NAD+ and related breakdown products were subsequently determined by mass spectrometry (Figure 3b). Large amounts of NR and adenosine were detected in the supernatant of wildtype SIRT6 (CT)-MBs incubated with H3K9Ac and NAD+ but they were not detectable in the absence of NAD+. In contrast, supernatants from the matrices containing SIRT6 H133Y or heat-inactivated SIRT6 were found to be largely devoid of any of NR and Adenosine even in the presence of NAD+. NAM and ADP-ribose were also detected in larger amounts in wildtype SIRT6 (CT)-MBs incubated with H3K9Ac and NAD+ when compared to control matrices, however, the difference between wildtype and control was not as large as what was observed for Adenosine or NR. Therefore, the determination of adenosine levels was correlated with SIRT6 catalytic activity in our system.
Figure 3.
Incubation of 1 µg of H3K9Ac in (Tris [50 mM, pH 8.0] containing 150 mM NaCl, 1mM DTT) at 37°C for 4 hours with and without NAD. A. the measurement of H3K9 in the incubates on the SIRT6 (CT)-MB, the H133Y SIRT6 (CT)-MB, the heat-inactivated SIRT6 (CT)-MB and buffer alone (area of H3K9 over the internal standard). B. the determination of the levels of NAD, NAM, NR, NMN, ADP-ribose, AMP and Adenosine in the incubates (area of the shown substrate (compound) over the internal standard).
Scheme 1.
To confirm that nicotinamide is an inhibitor of SIRT6, we evaluated its ability to block the deacetylation of H3K9Ac. SIRT-6 (CT)-MBs were incubated with NAD, the acetylated tail of H3K9, and 100 µM nicotinamide and supernatants were analyzed by HPLC. As expected, a statistically significant decrease (40%) in the amount of deacetylated H3K9 was observed (data not shown). SIRT6 is known to have mono-ADP-ribose polymerase activity [25] and, therefore, the effect of benzamide, a commonly inhibitor for ADP-ribose polymerases of the PARP family, on SIRT-6 deacetylase activity was tested. Importantly, the level of H3K9Ac deacetylation remained unaltered when benzamide (100 µM) was added to the reaction, indicating that the deacetylase activity of SIRT-6 does not require ADP-ribose polymerase function.
3.4. Inhibition of SIRT6 activity by plant extracts
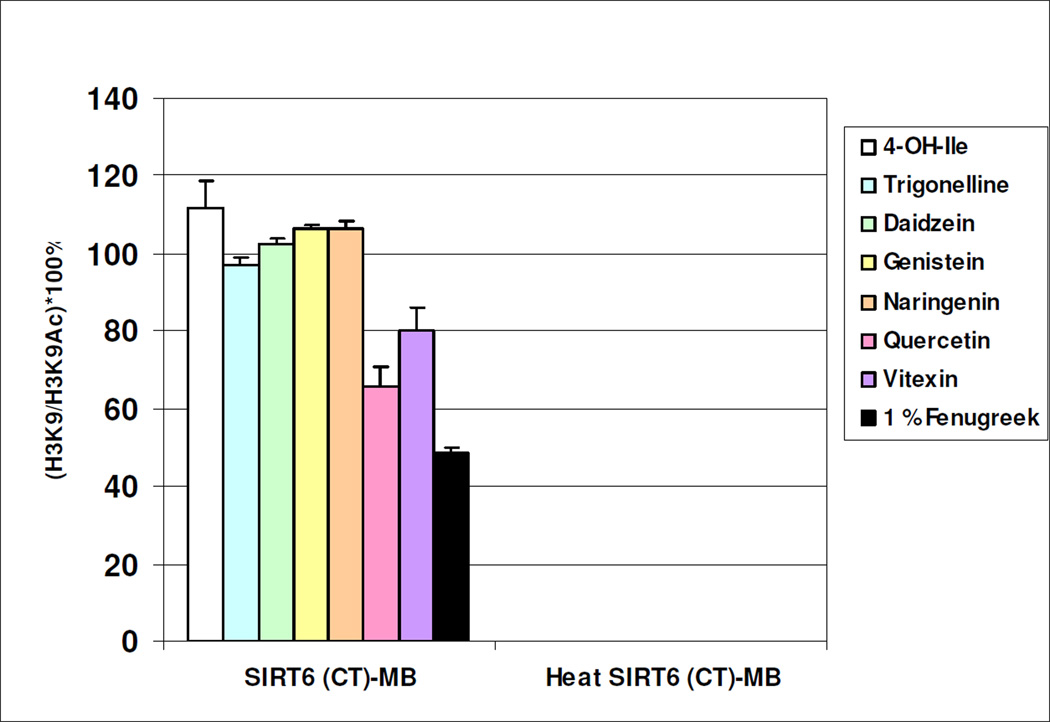
To determine whether our assay is suitable to identify inhibitors of SIRT6 activity from a complex biological mixture, fenugreek seed extract (Trigonella foenum-graecum) was added to the reaction mix. Interestingly, the deacetylation of H3K9Ac by SIRT6 (CT)-MB was significantly inhibited (>50%, P< 0.05) by 1% fenugreek seed extract (Fig. 4). To identify the SIRT6 inhibitor within this complex mixture, a candidate approach was used. The major components of this extract, including 4-hydroxyisoleucine (4-OH-Ile), trigonelline, naringenin, quercetin and vitexin were then individually tested along with genistein and diadezin, two flavonoids that are absent from fenugreek seed extract. Of all these compounds tested, only quercetin and vitexin were active and inhibited SIRT6-mediated H3K9Ac deacetylation, although to a lower degree than the complete fenugreek seed extract (Fig. 4). Furthermore, the combination of vitexin and quercetin together did not elicit a more potent inhibitory activity than quercetin alone (data not shown). Thus we conclude that the inhibition of SIRT6 by fenugreek extract involves additional yet to be identified components.
Figure 4.
SIRT6-MB was incubated with 50 µL of HDAC assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT and 0.2 mM NAD+) containing 5 µg of acetyl-histone H3(K9) peptide (amino acid residues 1–21; H3K9Ac) and 1% fenugreek seed extract for 4h at 37°C.
3.4. ChIP assay
To test whether fenugreek extract and individual active components thereof can inhibit SIRT6 activity in the cellular context, we treated DT40 cells with fenugreek extract or vehicle and performed ChIP assays to quantify H3K9Ac levels in the promoters of genes regulated by SIRT6. SIRT6−/− DT40 cells served as an important control, as any effects in these cells would be independent of SIRT6. Based on genome-wide SIRT6 chromatin immunoprecipitation experiments (D.R.W., S.F. unpublished data) we chose C-MYC, BCL2L1, CD3E, LYN, TOX, HSP90B1 for our analysis, as these genes likely represent direct targets of SIRT6. Primer pairs were designed to amplify fragments located ±1.5kb from the transcriptional start site (Table 1). The changes in the levels of H3K9Ac and H3K27Ac are represented as the ratios of amounts detected in fenugreek extracted treated cells to those treated with the vehicle alone (Fig. 5). Interestingly, treatment of SIRT6-deficient cells with 0.5% fenugreek seed extract for 24 h led to a strong decrease in the levels of H3K9Ac in the promoter regions of all genes tested as compared to untreated cells (Fig. 5A). This effect which could be caused by a histone acetyltransferase inhibitor in the plant extract, and sets the baseline upon which the changes in the SIRT6-proficient cells will need to be interpreted. Strikingly the relative acetylation levels are strongly increased upon treatment with fenugreek extract in the very same genes in wildtype cells. This strongly suggested that this extract had an inhibitory effect on SIRT6’s deacetylase activity. Importantly, the ratios of signal intensities for H3K27Ac remained comparable between wildtype and SIRT6 deficient cells, indicating that our observations are specific for H3K9Ac. In summary, we conclude that active compound(s) present in fenugreek seed extract are able to attenuate SIRT6 deacetylase activity, but do also alter the epigenetic landscape by SIRT6 independent mechanisms.
Figure 5.
(A) Chromatin immunoprecipitation assays of 0.5% fenugreek treated SIRT6+/+ (solid black bar) and SIRT6−/− cells (grey bars) using anti-H3K9Ac and anti-H3K27Ac antisera. Promoter regions of the indicated genes were tested by quantitative real-time PCR for the presence in respective immunoprecipitates. Absolute values of DNA amounts amplified from fenugreek treated samples were divided by the absolute values of DNA amounts amplified from cell treated with vehicle. All assays were performed in triplicate. A student’s paired t-test was conducted to determine statistical significance of differences between SIRT6-proficient and –deficient cells at each gene promoter (* : p<0.05). (B) ChIP analysis from quercetin treated B cells.
As quercetin is a major component of fenugreek seed extract and was shown to inhibit deacetylase activity in silico (see above), we evaluated the effect of this flavonoid on H3K9Ac levels in the promoter regions of the same SIRT6 target genes described above. Wildtype and SIRT6−/− DT40 cell lines were treated with 100µM of quercetin for 24 h, and H3K9Ac and H3K27Ac levels were analyzed by ChIP. Interestingly, in contrast to the whole extract, this compound showed little effect on the SIRT6 dependent H3K9Ac levels, with the exception of the TMED promoter which showed a clear SIRT6-dependent increase (Fig. 5B). The basis of the striking specific increase in H3K9Ac and H3K27Ac in the LYN promoter of quercetin treated SIRT6-deficient cells remains unclear. Overall, these findings are in contrast to our in vitro observations, and it is possible that the quercetin inhibitory activity on immobilized SIRT6 (Fig. 4) is masked by the complex intracellular environment. Moreover, it is possible that in the cellular context additional compounds present in the fenugreek seed extract are key mediators of quercetin’s effects. Physical and biological characterization of these compounds is currently being carried out and will be reported elsewhere.
4. Conclusion
In this report, we describe the development of a novel SIRT6 deacetylase assay and showed that it can be used to screen a medicinal plant extract, and fractions thereof, for inhibitors with good sensitivity and accuracy. Quercetin and vitexin compounds were identified as SIRT6 inhibitors from an active extract from the fenugreek seeds. In principle, this mass spectrometry assay format is not limited to the identification of catalytic inhibitors of SIRT6 but could also allow detecting compounds acting as allosteric modifiers of this deacetylase.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging. We would like to thank Dr. Michel Bernier for critical review of the manuscript and Dr.Irving W.Wainer for his continued support.
Footnotes
There are no financial conflicts.
References
- 1.Lois S, Blanco N, Martinez-Balbas M, de la Cruz X. BMC Genomics. 2007;8:252. doi: 10.1186/1471-2164-8-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson CL, Laniel MA. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Nature Reviews Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 4.Blander G, Guarente L. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 5.Johnstone RW. Nat Rev Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 6.Verdin E, Dequiedt F, Kasler HG. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 7.Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh da Y, Lu M, Milne JC, Westphal C, Bandyopadhyay G, Olefsky JM. Am J Physiol Endocrinol Metab. 2010;298(3):E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 9.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodgers JT, Puigserver P. Cell Metabolism. 2006;3:77–82. doi: 10.1016/j.cmet.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack D, Guimaraes A, Marinelli B, Widstrom JD, Nir T, Clish CB, Vaitheesvaran B, Illiopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newmann DJ, Cragg GM. J.Nat.Prod. 2007;70(3):461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 13.Liszt G, Ford E, Kurtev M, Guarente L. J.Biol.Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 14.Merrick CJ, Duraising MT. Eukaryotic Cell. 2007;6:2081–2091. doi: 10.1128/EC.00114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moaddel R, Marszall M, Bigi F, Yang Q, Duan X, Wainer IW. Anal.Chem. 2007;79:5414–5417. doi: 10.1021/ac070268+. ASAP article. [DOI] [PubMed] [Google Scholar]
- 16.Marszall MP, Moaddel R, Kole S, Gandhari M, Bernier M, Wainer IW. Anal. Chem. 2008;80:7571–7575. doi: 10.1021/ac801153h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.N. Jonker A, Krestchmer J, Kool A, Fernandez D, Kloos JG, Krabbe H, Lingeman H. Irth. Anal Chem. 2009;81:4263. doi: 10.1021/ac9000755. [DOI] [PubMed] [Google Scholar]
- 18.Xue W, Li X, Zhang J, Liu Y, Wang Z, Zhang R. Asia Pac J Clin Nutr. 2007;16 Suppl 1:422–426. [PubMed] [Google Scholar]
- 19.Handa T, Yamaguchi K, Sono Y, Yazawa K. Biosci. Biotechnol. Biochem. 2004;69:1186–1188. doi: 10.1271/bbb.69.1186. [DOI] [PubMed] [Google Scholar]
- 20. S.D.F. unpublished data. [Google Scholar]
- 21.Rodgers KK, Villey IJ, Ptaszek L, Corbett E, Schatz DG, Coleman JE. Nucleic Acid Res. 1999;27:2938–2946. doi: 10.1093/nar/27.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wurster AL, Pazin MJ. Mol.Cell Biol. 2008;28:7274–7285. doi: 10.1128/MCB.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Pazin MJ, Ravid K. Mol.Cell Biol. 2004;24:428–441. doi: 10.1128/MCB.24.1.428-441.2004. (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tennen R, Berber E, Chua KF. Mechanisms of Ageing and Development. 2010;131:185–192. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosi C, Guerin K, Marien M, Koek W, Rollet K. Brain Research. 2004;996:1–8. doi: 10.1016/j.brainres.2003.09.072. [DOI] [PubMed] [Google Scholar]
- 26.Lee WJ, Chen YR, Tseng TH. Oncol. Rep. 2011;25:583–591. doi: 10.3892/or.2010.1097. [DOI] [PubMed] [Google Scholar]