Abstract
A number of bioactive dietary components are of particular interest in the field of epigenetics. Many of these compounds display anticancer properties and may play a role in cancer prevention. Numerous studies suggest that a number of nutritional compounds have epigenetic targets in cancer cells. Importantly, emerging evidence strongly suggests that consumption of dietary agents can alter normal epigenetic states as well as reverse abnormal gene activation or silencing. Epigenetic modifications induced by bioactive dietary compounds are thought to be beneficial. Substantial evidence is mounting proclaiming that commonly consumed bioactive dietary factors act to modify the epigenome and may be incorporated into an ‘epigenetic diet’. Bioactive nutritional components of an epigenetic diet may be incorporated into one’s regular dietary regimen and used therapeutically for medicinal or chemopreventive purposes. This article will primarily focus on dietary factors that have been demonstrated to influence the epigenome and that may be used in conjunction with other cancer prevention and chemotherapeutic therapies.
Keywords: acetylation, cancer, dietary, epigenetic, methylation, nutrigenomics
Dietary factors have become agents of strong interest in the field of epigenetics. A number of bioactive dietary components that appear to have potential to prevent disease and promote overall health have been identified [1–4]. In fact, several naturally occurring dietary phytochemicals have been demonstrated to have anticarcinogenic properties and may play a role in regulating biological processes [5,6]. Many studies have shown that natural products have epigenetic targets in cancer cells and can act as cancer preventive agents. Compounds found in dietary phytochemical preparations such as teas, garlic, soy products, herbs, grapes and cruciferous vegetables are now generally accepted to defend against the development of many different types of tumors as well as acting as epigenetic modulators that impact not only the initiation, but also the progression of oncogenesis [6–9].
Epigenetic mechanisms
The term epigenetics was coined in 1942 by developmental biologist Conrad H Waddington [10]. Epigenetics generally refers to heritable changes in gene expression and chromatin organization that are not due to alterations in the DNA sequence [11]. Epigenetic modifications typically occur by changes in DNA methylation, histone covalent modifications or by RNAi [12]. DNA methylation occurs when a methyl group is added to the 5-carbon (C5) position of a cytosine. The methylation of cytosine involves the transfer of a methyl group from S-adenosyl-L-methionine (SAM), a methyl precursor, to the cytosines in CpG dinucleotides [13]. DNA methylation has many roles in various cellular processes and may impact the transcription of genes by preventing the binding of key transcriptional factors [14–17]. Transcriptional silencing due to DNA methylation is thought to occur by the recruitment of methyl CpG-binding transcriptional repressors and by interfering with the DNA binding of transcriptional activators, which in turn results in a condensed chromatin state. In addition, methylated DNA may be bound by methyl-CpG-binding domain (MBD) proteins, which are essential for binding to 5-methylcytosine [16]. DNA methylation is catalyzed by enzymes known as DNA methyltransferases (DNMTs) and hypermethylation of CpG dinucleotides or CpG islands by DNMTs usually results in transcriptional gene silencing and gene inactivation [12]. The human genome contains four DNMT genes, DNMT1, DNMT2, DNMT3A and DNMT3B [18]. Altered DNMT expression and activity is seen in numerous diseases including autism, cardiovascular diseases, obesity, Type-2 diabetes and cancer [19–23]. In addition, global hypomethylation is associated with nearly all human cancers [24,25].
Histone modifications typically occur as post-translational modifications at the N-terminal of histones. These modifications include acetylation, methylation, phosphorylation, biotinylation and ubiquitination and are essential during development [26–28]. Histone modifications are catalyzed by enzymes such as histone methyltransferases (HMTs), histone demethylases (HDMs) histone acetyltransferases (HATs), and histone deacetylases (HDACs). HMTs act to add methyl groups to lysine and/or arginine residues in histones, while HDMs remove the methyl moieties. In turn, HATs catalyze the addition of acetyl groups to the lysine residues of histones, whereas HDACs are responsible for the removal of these groups [29,30]. Lysine methylation can cause either activation or repression of transcription, while arginine methylation typically activates transcription. Likewise, histone hyperacetylation results in the activation of normally repressed genes while hypoacetylation results in gene silencing. This is apparent in carcinogenesis where aberrant activity of HATs and HDACs are thought to trigger carcinogenic processes [31].
RNAi is the process by which dsRNA inhibits the accumulation of homologous transcripts from like genes [32]. RNAi or ncRNAs, in the form of antisense transcripts, can lead to transcriptional silencing by the formation of heterochromatin. The involvement of RNA in different silencing mechanisms has been described in detail in several organisms [33]. For example, in the yeast Schizosaccharomyces pombe, the deletion of different components of the RNAi machinery results in the loss of H3K9 methylation, as well as impairment of centromere function [33,34]. Similarly, DNA methylation and H3K9 methylation have been demonstrated in Arabidopsis thaliana and in α-thalassaemia [35,36]. RNAi has also been demonstrated to be involved in silencing genes associated with HIV-1, along with several types of cancers [37–41]. In addition, noncoding miRNAs can control the expression of DNMTs and other enzymes associated with epigenetic modifications, which affect mRNA translation and stability [42–44]. Exciting developments have indicated that RNAi-directed silencing of heterochromatic regions might trigger direct histone modifications and DNA methylation to specific loci, causing gene silencing [35,36,45,46].
Epigenetic modifications are of particular interest in the field of cancer research since their impact on the epigenome is involved in cell proliferation, differentiation and survival [27,47,48]. Furthermore, epigenetic modifications are often involved in transcriptional regulation and have been implicated both in tumor development and progression [40,49,50]. Epigenetic modifications causing transcriptional deregulation may result in the inappropriate expression or activation of transcription factors associated with oncogenes and/or the failure to express genes responsible for tumor suppression [51]. In fact, cancer cells have genome-wide aberrations in a number of epigenetic markers, including global hypomethylation, global downregulation of miRNAs, promoter-specific hypermethylation, histone deacetylation and upregulation of epigenetic machinery [52]. In addition, the impact of epigenomic processes in cancer is apparent by the finding that at least half of all tumor suppressor genes are inactivated through epigenetic mechanisms in tumorigenesis [16,53–55]. Bioactive dietary components consumed by ingesting natural products including fruits and vegetables can act as sources of vitamins and minerals. While this is an invaluable role, these agents have high potential for application to oncogenesis owing to in part to their anticarcinogenic properties [9,56]. A growing body of evidence suggests that dietary agents as well as non-nutrient components of fruits and vegetables can affect epigenetic processes and are involved in processes, including the reactivation of tumor suppressor genes, the initiation of apoptosis, the repression of cancer-related genes and the activation of cell survival proteins in different cancers [57–60]. Dietary phytochemicals such as tea polyphenols, genistein, sulforaphane (SFN), resveratrol, curcumin and others have been demonstrated to be effective agents against cancer and to act through epigenetic mechanisms that affect the epigenome [56,61].
Epigenetic diet phytochemicals affecting the epigenome
Dietary factors play a role in many normal biological processes and are also involved in the regulation of pathological progressions. Diseases linked to genetic and epigenetic modifications can be influenced by environmental and dietary factors. In particular, nutritional factors, drugs, chemicals used in pesticides, environmental compounds and inorganic contaminants (i.e., arsenic) can alter the epigenome, and may contribute to the development of abnormalities [62–66]. It has become increasingly clear that environmentally induced epigenetic changes can be mediated, in part, by diet [67–69]. This is especially illustrated by an early investigation conducted in an animal model, which demonstrated that dietary methyl deficiency of folate, choline and methionine altered DNA methylation patterns in the liver and induced hepatocarcinoma [70]. Evidence has also emerged indicating that giving mice that have developed methyl-deficiency-induced hepatocarcinoma a methyl-sufficient diet can lessen the occurrence of abnormal DNA methylation [71]. Because epigenetic variation can be influenced by dietary factors, it is reasonable to believe that investigating strategies that utilize dietary compounds to target epigenetic modifications may be worthwhile in preventing and treating diseases including cancer.
Dietary polyphenols
Polyphenols are present in fruits and vegetables and are a vital part of the human diet [56,72]. Plant polyphenols can be divided into at least ten different classes based on their chemical structure. These classes include: flavonoids, stilbenes, phenolic acids, benzoquinones, acetophenones, lignins and xanthones [72,73]. Polyphenols can range from simple molecules to highly complex compounds and are derived from either phenylalanine (a phenol intermediate) or its precursor shikimic acid. They typically contain one or more sugar residues linked to a hydroxyl group and occur in a conjugated form [72]. Some estimate that more than 8000 distinct dietary polyphenols exist [72]. Examples of common polyphenols include (−)-epigallocatechin-3-gallate (EGCG; found in green tea), curcumin (found in curry) and resveratrol (found in grapes). These dietary polyphenols have a protective role against diseases and also have a significant impact on cancer prevention [74,75]. In fact, various mechanisms have been identified that help to explain the preventive nature of polyphenols, including their ability to alter the epigenome in cancer cells by chromatin remodeling or by reactivating silenced genes [76–78]. The chemo-preventative potential of dietary polyphenols can be traced to their ability to inhibit DNMTs as well as their ability to act as histone modifiers. Both of these properties of dietary polyphenols can significantly change the epigenome of cancer cells and are viewed as attractive possibilities for anticancer therapeutics.
Tea polyphenols
Next to water, tea is the most consumed beverage worldwide with approximately 20 billion cups consumed daily [79]. The three most popular types of tea (green, black and oolong) are differentiated based on the degree of fermentation they undergo. Green tea leaves are dried and roasted but not fermented, whereas black tea leaves are well fermented and oolong tea leaves are only partially fermented [79,80]. Studies have indicated that compounds present in tea may reduce the risk of diseases including cancer. Teas contain polyphenolic compounds that serve to protect plants from photosynthetic stressors, reactive oxygen species and consumption by herbivores [56]. As previously stated, polyphenolic compounds in tea may actively reduce the risk of diseases such as cancer. One subcategory of polyphenols, catechins, is the most abundant of the bioactive compounds in green tea. These include (−)-epicatechin (EC), (−)-epicatechin-3-gallate (ECG), (−)-epigallocatechin (EGC), and EGCG [81]. While all of the aforementioned catechins have been found to share similar properties, the most efficient of these compounds in targeting factors like DNMTs is EGCG [77,78]. EGCG accounts for more than 50% of the active compounds in green tea and has been extensively studied for its anticarcinogenic properties [82]. While studies conducted by Chuang et al. report disagreeing results regarding the effects of EGCG on DNA methylation [83], an increasing number of investigations have suggested a positive correlation between the consumption of EGCG and the inhibition of oral, breast, prostate, gastric, ovarian, esophageal, skin, colorectal, pancreatic, and head and neck cancers [83–88].
Epigallocatechin-3-gallate is thought to exert its anticancer effects through several different mechanisms much of which can be altered by epigenetic mechanisms; these include the induction of apoptosis and cell cycle arrest, inhibition of oxidative stress and angiogenesis, regulation of signal transduction and reduction of cancer cell proliferation [89–92]. EGCG can inhibit DNMT activity through direct enzyme interaction leading to demethylation and reactivation of genes previously silenced by methylation (see Figure 1 & Table 1) [77,93,94]. Early studies conducted by Fang et al. demonstrated that treatment of esophageal cancer cells with EGCG in fact lowered DNMT activity and caused a time- and dose-dependent reversal of hypermethylation in tumor suppressor genes including p16, RARβ, hMLH1 and MGMT [95]. A recent investigation involving A431 skin cancer cells revealed that EGCG treatment not only decreased global DNA methylation levels, but also decreased the levels of 5-methyl-cytosine, DNMT activity, mRNA and protein levels of DNMT1, DNMT3a and DNMT3b resulting in the re-expression of p16(INK4a) and p21/Cip1 mRNA and proteins [96]. In addition, in studies conducted by Li et al. it was found that EGCG is capable of reactivating estrogen receptor-α (ER-α) expression in ERα-negative MDA-MB-231 breast cancer cells [97]. This is of particular importance in breast cancer therapeutics where many treatment options utilize the ER pathway. Moreover, studies conducted by Berletch et al. indicated that EGCG treatment of MCF-7 breast cancer cells resulted in downregulation of the hTERT gene and a time dependent decrease in hTERT promoter methylation, which paradoxically leads to a decrease in telomerase activity in these cells [93]. In addition, EGCG has been demonstrated to demethylate the Wnt oncogene promoter in lung cells [98]. EGCG can also partially reverse the hypermethylation status of the RECK tumor suppressor gene in oral carcinoma cells, significantly enhancing the expression of RECK mRNA [99]. Moreover, treatment of LNCaP human prostate cancer cells with green tea polyphenol caused a time- and dose-dependent re-expression of GSTP1, whose overexpression has been associated with the development of several types of cancer [100]. In addition, Choi et al. have identified EGCG as a HAT inhibitor with global specificity for the majority of HAT enzymes [101]. Recently, EGCG has also been found to modulate miRNA expression in hepatocellular carcinoma cells (Figure 2 & Table 1) [102]. Furthermore, either EGCG or green tea polyphenols can inhibit carcinogenesis based on numerous in vivo studies; however, the effects on epigenetic mechanisms and the epigenome in vivo have not yet been clearly defined [103–105].
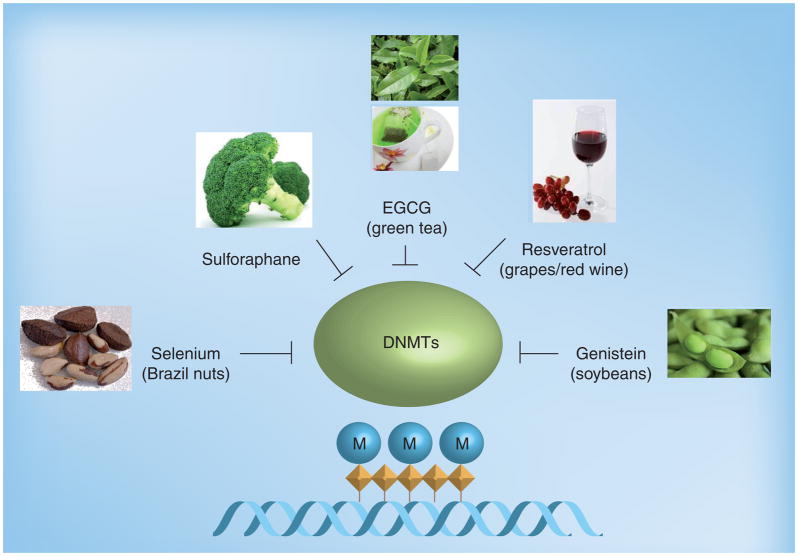
Figure 1. Dietary inhibitors of DNA methyltransferases.
DNMTs catalyze DNA methylation by adding a methyl group (CH3; indicated by M) to cytosines of CpG dinucleotides (diamond shapes). Hypermethylation of CpG dinucleotides or CpG islands by DNMTs usually results in transcriptional gene silencing and gene inactivation. Several bioactive compounds found in foods (e.g., EGCG in green tea) act as dietary inhibitors of DNA methyltransferases and also alter gene expression via epigenetic mechanisms.
DNMTs: DNA methyltransferases; EGCG: Epigallocatechin-3-gallate; M: Methylation.
Table 1.
Bioactive epigenetic diet compounds, food sources and epigenetic functions.
| Epigenetic diet compounds | Food sources | Epigenetic functions |
|---|---|---|
| EC, ECG, EGC and EGCG | Green tea | DNMT and HAT inhibitor, modulates miRNA |
| Resveratrol | Grapes, peanuts, mulberries, cranberries, blueberries | DNMT and HDAC inhibitor |
| Curcumin | Tumeric, curry | DNMT inhibitor and miRNA modulator |
| Genistein | Soybeans, fava beans | DNMT and HDAC inhibitor, enhances HATs, modulates miRNA |
| Isothiocyanates, sulforaphane | Broccoli, cabbage, kale, watercress | DNMT and HDAC inhibitor |
| Selenium | Brazilian nuts, chicken, game meat, beef | DNMT and HDAC inhibitor |
| Allyl mercaptan, organosulfur compounds | Garlic | HDAC inhibitor |
| Folate | Beans, grains, fortified breakfast cereals, pastas, green vegetables | Deficiencies alter DNA methylation patterns |
| Alcohol | Alcoholic beverages | High consumption increases promoter hypermethylation |
DNMT: DNA methyltransferase; EC: Epicatechin; ECG: Epicatechin-3-gallate; EGC: Epigallocatechin; EGCG: Epigallocatechin-3-gallate; HAT: Histone acetyltransferase; HDAC: Histone deacetylase.
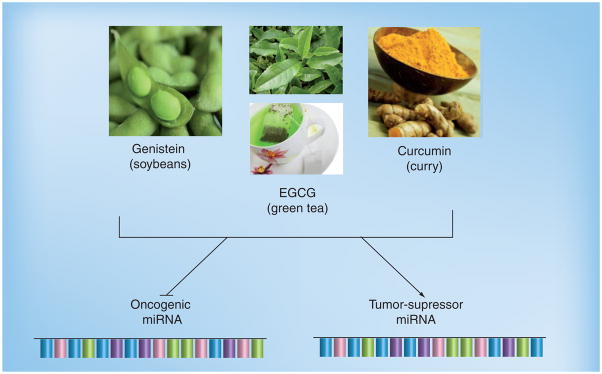
Figure 2. Dietary effectors of miRNAs.
miRNAs serve as regulators of gene expression. Evidence has been reported supporting the fact that dietary agents target the miRNA of oncogenes and/or tumor suppressors and impact the expression level of miRNAs.
EGCG: Epigallocatechin-3-gallate.
While considerable evidence has been presented showing the anticarcinogenic properties of green tea consumption, the EGCG compound is unstable under normal physiological conditions. To this end, synthetic analogs of EGCG have been studied in physiological conditions and show strong anticancer activity with more stability and efficacy [106]. Interestingly, studies conducted using EGCG and a prodrug of EGCG (pEGCG and EGCG octa-acetate) to enhance the bioavailability and stability of EGCG display inhibition of hTERT, the catalytic subunit of telomerase, through epigenetic mechanisms affecting the hTERT gene regulatory region in breast cancer cells [107,108]. Studies provide evidence that EGCG alone or combined with other epigenetic-modifying compounds such as HDAC inhibitors may be effective as cancer therapeutic agents and these lines of investigation are currently of considerable interest in the field of epi-genetics and in the development of an epigenetic diet for the purpose of cancer prevention.
Resveratrol
The dietary polyphenol resveratrol is naturally found in several plants including peanuts, mulberries, cranberries and blueberries, but is most abundant in the skin of grapes [109]. Resveratrol is also consumed in the form of red wine. Antioxidant, anti-inflammatory and anti-cancer properties of resveratrol occur through various molecular and biochemical pathways [110,111]. For instance, resveratrol has an impact on signaling pathways that control cell division, cell growth, apoptosis, angiogenesis and tumor metastasis [112–114]. Antiproliferative properties of resveratrol have been reported in liver, skin, breast, prostate, lung and colon cancer cells [115–117]. It has also been demonstrated that colon carcinoma cells treated with resveratrol inhibits cell migration, adhesion and invasion [118]. The beneficial effects of resveratrol have also been demonstrated in vivo in that this bio-active dietary component has been reported to reduce adenocarcinoma cell metastases in BALB/c mice thereby increasing the percentage survival of the mice [119,120].
While resveratrol has potential as a dietary anticancer agent, it displays less DNMT inhibitory activity than some of its dietary counterparts including EGCG (Figure 1 & Table 1). However, resveratrol is capable of preventing the epigenetic silencing of the BRCA1 tumor suppressor protein [121] and Papoutsis et al. demonstrated that resveratrol-treated MCF-7 cells partially restored monomethylated-H3K9, DNMT1, and MBD2 at the BRCA1 promoter [121]. Inhibition of DNMT has been shown in nuclear extracts from MCF-7 breast cancer cells treated with resveratrol although resveratrol was unable to reverse the methylation of certain tumor suppressor genes [60]. In addition, resveratrol was unable to inhibit RARβ2 and MGMT promoter methylation in MCF-7 cells [122,123].
Importantly, resveratrol is associated with activating SIRT-1 and p300, which are known HDAC inhibitors [124]. As aforementioned, HDACs are responsible for removing acetyl groups from the lysine residues of histones. There are to date at least 18 HDAC isozymes that are divided into different classes. Several studies have reported a link between class I HDACs and the development of malignant tumors, while this link has been observed substantially less in class II HDACs [125]. Class III HDACs are homologous to the Sir2 protein in yeast and are collectively known as Sir proteins or sirtuins. Sirtuins have specific inhibitors and are not responsive to class I and II HDAC inhibitors [126]. The SIRT-1-encoded proteins are necessary for chemoprevention mediated by resveratrol [127]. It is believed that resveratrol activates SIRT-1 by mimicking physiological pathways that stimulate SIRT-1 [128,129]. The activation of SIRT-1 by resveratrol negatively regulates expression of the antiapoptotic protein, Survivin, by deacetylating H3K9 within the promoter of its gene [130,131]. In addition, studies conducted by Wang et al. found that SIRT-1 mediates BRCA1 signaling in human breast cancer cells by altering H3 acetylation [130]. Furthermore, treatment of prostate cancer cells with resveratrol demonstrates enhanced p53 acetylation and apoptosis by inhibition of the MTA–NuRD complex [132]. In terms of aging, resveratrol has also been reported to extend the lifespan and improve the health of mice on a high-calorie diet [133]. Sirturins typically exhibit their activity through deacetylation of nonhistone proteins but are also important in the maintenance of histone acetylation patterns [126,134]. This evidence suggests that sirturins can affect normal gene expression through chromatin regulation and may provide a link between epigenetic changes associated with aging and obesity as well as epigenetic modifications in tumors.
Curcumin
Curcumin, a diferuloylmethane, is a polyphenol that originates from the plant, Curcuma longa. Curcumin is the main component of the spice turmeric and is responsible for the yellow pigmentation of curry. This bioactive dietary component appears to have anti-inflammatory, antioxidant, antiangiogenic and anti-cancer properties and is used as a therapeutic agent in Indian and Chinese medicine [135,136]. Investigations indicate that curcumin inhibits DNMT activity (Figure 1 & Table 1) by covalently blocking the catalytic thiolate of C1226 of DNMT1 [137,138]. Moreover, there is evidence that curcumin may be an effective DNA hypomethylating agent that could facilitate the expression of inactive prometastatic and proto-oncogenes [16,77,137,139]. Curcumin also has epigenomic effects in that genomic DNA from leukemia cells show global hypomethylation after curcumin treatments [140]. In addition, studies conducted by Valinluck and Sowers indicated that curcumin induced anti-inflammatory effects. These effects stemmed from halogenated cytosine products that mimic 5-methylcytosine in DNA methylation. These data provide evidence of a link between inflammation and epigenetic alterations that are also seen in cancer [141].
Curcumin also functions as a histone modifying compound and as a HDAC and HAT inhibitor (Figure 3 & Table 1) [142]. While this inhibition is less than that of some other dietary epigenetic modifiers, Kang et al. found that the inhibition of curcumin-mediated HAT activity results in a decrease in global histone H3 and H4 acetylation in brain cells [142]. Furthermore, independent studies conducted by Cui and Pollack demonstrated that promoter hypoacetylation of several histones was curcumin-mediated and correlated to gene silencing [143,144]. In addition, numerous animal studies have built upon in vitro investigations and support the ability of curcumin to inhibit HATs and HDACs in several disease models including tumorigenesis [77,139,142,145–148]. While the inhibition of both HATs and HDACs may seem contradictory, recent investigations provide evidence that HAT inhibitors have a potential role in cancer therapies and that inhibition of both HATs and HDACs together may provide a potent strategy for cancer treatment [149]. Chemoprevention mediated by curcumin is mainly facilitated by the NF-κB and PI3K/AKT signaling pathway and typically induces cell cycle arrest and apoptosis [150]. Several groups have shown that curcumin is a potent inhibitor of p300/CBP activity in leukemia, hepatoma and cervical cancer cellular extracts [151,152]. There are also indications that curcumin prevents histone hyperacetylation induced by the MS-275 HDAC inhibitor in peripheral blood lymphocytes and cancer cells [151,153]. In addition, curcumin has been found to alter the miRNA expression profile in pancreatic cancer cell lines (Figure 2 & Table 1) [154,155].
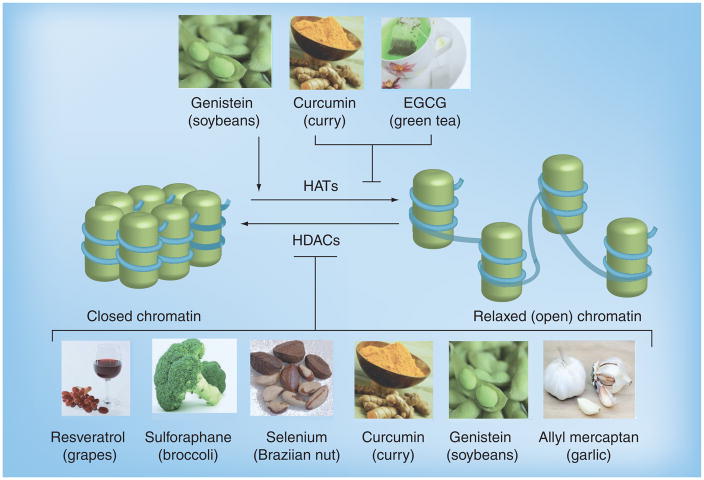
Figure 3. Dietary modifiers of histones.
Bioactive compounds including resveratrol, sulforaphane and curcumin have the ability to alter (indicated by cylinders) HATs as well as HDACs. These histone modifications cause conformational changes in chromatin structure that lead to changes in DNA accessibility. HATs induce a relaxed chromatin state allowing transcriptional factors access to DNA (blue cords) and activate gene expression whereas chromatin in its closed state is indicative of gene silencing and repression. Dietary compounds can inhibit and/or enhance (arrow) HATs and HDACs thereby altering gene expression.
EGCG: Epigallocatechin-3-gallate; HAT: Histone acetyltransferase; HDAC: Histone deacetylase.
The data showing the strong inhibitory activity of curcumin in carcinogenesis suggests its therapeutic abilities in cancer or its use in chemoprevention. An issue with using curcumin as a bioactive agent is that its insolubility and instability in water leads to low bioavailability. However, the bioavailability of curcumin can be enhanced by utilizing properties of dietary factors such as rubusoside (found in Chinese blackberry extract) and molecular compounds such as phosphatidylcholine (found in soy and egg yolks) thereby increasing its potential in cancer chemoprevention or therapy [156,157].
Isoflavones (genistein)
Isoflavones belong to the flavonoid group of compounds, the largest class of polyphenolic compounds [158]. Isoflavones are found in a number of plants including soybeans, fava beans and kudzu. Several isoflavones have been investigated and indications are that they have anti-angiogenic and anticancer properties. Genistein, a phytoestrogen primarily found in soybeans, is perhaps the most studied of these bioactive compounds. This estrogen-like compounds acts as a chemopreventative agent in several types of cancers [159]. In fact, moderate doses of genistein appear to induce inhibitory effects on cervical, prostate, colon and esophageal cancers [160–162]. Several mechanisms have been found to contribute to the anticarcinogenic properties of genistein including its ability to regulate gene transcription by affecting histone acetylation and/or DNA methylation [163].
Genistein treatment of esophageal squamous cell carcinoma partially reversed DNA hypermethylation and reactivated p16, RARβ and MGMT and a similar reversal was also seen in prostate cancer cells [162]. Further studies conducted in prostate cells indicate that genistein induces the expression of the tumor suppressor genes p16 and p21 by altering histone and promoter methylation [164,165]. Likewise, studies using breast cancer cells treated with a low (3.125 μM) concentration of genistein demethylated the promoter of the GSTP1 gene [166]. In addition, genistein-treated prostate and renal cells showed a reversal of hypermethylation of the BTG3 gene, a known tumor suppressor [167,168]. Moreover, genistein combined with DNA methylation inhibitors or other DNMTs can enhance the reactivation of genes silenced by methylation [163,169]. As evidence of this, Li et al. found that genistein inhibits DNMT1, 3a and 3b (Figure 1) and inhibits the expression of hTERT. Genistein also increases acetylation by enhancing HAT activity (Figure 3) [168]. Furthermore, investigations have demonstrated that genistein-mediated hypomethylation and hyperacetylation reactivate the expression of tumor suppressor genes in prostate cancer cells [164,165]. Genistein and other isoflavones have also been found to regulate miRNA expression in several cancer cell lines (Figure 2 & Table 1) [170,171].
Importantly, the effects of genistein have recently been tested in humans. For instance, in studies conducted by Qin et al., 34 healthy premenopausal women received either 40 mg or 140 mg of isoflavones, including genistein, daily through one menstrual cycle. Methylation assessment of five genes known to be methylated in breast cancer (p16, RASSIFA, RARβ2, ER and CCND2) was conducted on intraductal samples. The findings revealed hypermethylation (which typically leads to gene silencing) of cancer-related genes RARβ2 and CCND2 was increased after genistein treatment and correlated with serum genistein levels [172].
Isothiocyanates
Isothiocyanates are a category of dietary compounds present in cruciferous vegetables including broccoli, cabbage and kale. Isothiocyanates are characterized by a sulfur containing functional group (N=C=S). Commonly used isothiocyanates include: allyl isothiocyanate (AITC), benzyl isothiocyanate (BITC), phenethyl isothiocyanate (PEITC) and SFN [173]. Reports have indicated that isothiocyanates have proapoptotic and antiproliferative properties [174]. Several investigations have demonstrated evidence that isothiocyanates, including iberin, SFN and a SFN analog erucin, inhibit cancer cell growth and exhibit proapoptotic capabilities [174]. Treatment with isothiocyanates has also been reported to prevent esophageal tumorigenesis in rats [175]. Isothiocyanates are known to affect the epigenome and have anti-cancer properties. In fact, allyl-isothiocyanate, found in broccoli, has been reported to increase histone acetylation in mouse erythroleukemia cells. Phenylhexyl isothiocyanate (PHI), a synthetic isothiocyanate, acts as a HDAC inhibitor and has been demonstrated to hypomethylate p16 and to induce histone H3 hyperacetylation in myeloma cells [176]. PHI has also been reported to inhibit HDAC activity and plays a role in remodeling chromatin to activate p21 and induce cell cycle arrest in prostate cancer and leukemia cells [177,178]. In addition, prostate cancer cells treated with PEITC, found in watercress, demonstrated demethylation and re-expression of the GSTP1 gene [179].
One of the main isothiocyanate compounds is SFN. SFN is an isothiocyanate found in cruciferous vegetables such as broccoli. Investigations into the effects of dietary SFN have shown its anticarcinogenic activity in several cancers [173,180–182]. SFN has many effects that include inducing apoptosis, affecting the cell cycle and acting as a HDAC inhibitor (Figure 3 & Table 1). SFN-initiated HDAC inhibition has been found to have epigenomic effects in that it can increase global and local histone acetylation of a number of genes and is thought to be involved in the regulation of cancer-related genes [183–185]. Studies conducted in colorectal and prostate cancer cells show inhibition of HDAC activity due to SFN treatments. In addition, studies conducted by Myzak et al. using human subjects demonstrated that a dose of 68 g of broccoli sprouts was effective in inhibiting HDAC activity in peripheral blood mononucleocytes [186]. In addition, studies conducted by Meeran et al. indicated that SFN can inhibit DNMTs in breast cancer cells and that SFN inhibits hTERT in a dose and time-dependent manner [59]. This finding is significant since hTERT is overexpressed in approximately 90% of cancers.
Other dietary compounds affecting the epigenome
Selenium (Se) is a nutrient found in Brazil nuts, chicken, game meat and beef [187]. Other chemical forms of Se include selemethionine, selenocysteine, selenate and selenite. Se is an essential element with antioxidant, proapoptotic, DNA repair and anticancer properties [188–190]. Se is vital for human health and Se deficiencies have been linked to various human diseases including cancer [191]. In addition, several other selenoproteins (i.e., selenium binding protein-1) have been indicated as important in the development of cancers; however, their epigenetic effects have not been clearly defined [192].
In trials designed to test Se in nonmelanoma skin cancers, 1312 individuals at a high risk of developing this disease were given either 200 μg of Se or placebo orally per day for an average of 4.5 years [193,194]. This trial did not prevent skin cancer but did produce a significant 44% secondary decrease in lung cancer incidence [193]. Se has been linked to DNA methylation in cellular and animal models and Xiang et al. found that Se treatments caused partial promoter DNA demethylation and re-expression of GSTP1 in prostate cancer cells. This study also demonstrated that Se decreased histone deacetylase activity (Figure 3 & Table 1) and increased levels of acetylated H3K9, and decreased levels of methylated H3K9 [191]. In animal studies rats fed with selenium-rich diets induced significant DNA hypomethylation in the liver and colon [195,196]. Furthermore, Se deficiency has been demonstrated to cause global hypomethylation and promoter methylation of the p16 and p53 tumor suppressor genes [197]. Se can also decrease DNMT1 protein expression and inhibit DNMT1 (Figure 1 & Table 1) by direct interaction and indirectly by influencing homocysteine concentrations [198]. In addition, investigations have demonstrated that treatment of prostate cancer cells with selenite, an inorganic form of Se, can restore the expression of anticancer genes silenced by hypermethylation suggesting that epigenetic regulation by Se may play a role in cancer prevention [191]. Although these studies are intriguing, further studies involving the epigenetic influence of selenium are needed to fully appreciate the impact of selenium on the epigenome.
Garlic (Allium sativum) has been used for the prevention of disease for many years, and is thought to have antibacterial, antiviral and anti-inflammatory activities [199]. Garlic cloves contain several compounds including: vitamins A, B-complex, C, E, fiber, free amino acids, sulfur/organosulfur compounds and proteins. In addition, garlic also contains small amounts of selenium that may contribute to its chemopreventive properties. Garlic extracts and compounds have been used in experiments involving cancer treatment and prevention in isolated cell systems and in in vivo models [199]. These studies have shown that garlic acts to inhibit cell cycle progression, induce apoptosis, inhibit angiogenesis and modifies histones [200]. Studies conducted by Nian et al. revealed that garlic organosulfur compound, allyl mercaptan, inhibits histone deacetylase (Figure 3 & Table 1) and enhances Sp3 binding on the P21/WAF1 promoter, which results in elevated p21 protein expression and cell cycle arrest [200,201]. In addition, investigations conducted by Lea et al. demonstrate the induction of histone acetylation in cancer cells treated with garlic compounds [202,203].
Folic acid, or folate, is a B vitamin found in many beans, grains, fortified breakfast cereals, pastas and green vegetables. Folate is a key element in the methyl-metabolism pathway. Dietary methyl deficiency can alter hepatic DNA methylation patterns and induce liver cancer (Table 1) [70,71]. While folate has been studied extensively for its developmental effects, folate deficiencies lead to hypomethylated genomic DNA, which is associated with tumorigenesis [204,205]. Folate deficiencies are reported to contribute to the development of several different cancers including: breast, cervix, ovary, brain, lung and colorectal [206–208]. Folate regulates the biosynthesis, repair and methylation of DNA, whereas deficiencies in folate can induce carcinogenesis by augmenting these processes [207]. In addition, studies involving colorectal cancer indicated that folate deficiency can alter cytosine methylation in DNA leading to the activation of c-Myc, a known oncogene [209,210].
While most natural dietary products have shown beneficial effects on the epigenome, not all dietary components share this characteristic. In fact, alcohol consumption is associated with harmful epigenetic modifications as well as the development/progression of several human cancers. For example, colorectal cancer patients with high alcohol consumption had aprevalence of promoter hypermethylation of numerous genes when compared with patients with low alcohol consumption (Table 1) [211,212]. In addition, studies involving head and neck cancers demonstrated that promoter hypermethylation of MGMT and WNT pathway regulators occurred more frequently in heavy and light drinkers than in nondrinkers [213].
Other dietary factors including those found in coffee, cashews, tomatoes, parsley, milk thistle and rosemary, have also been reported to have epigenetic targets all of which could not be mentioned in this article. However, bioactive components of these nutrients may be of considerable interest and may have epigenetic targets in cancer [123,214–217].
Future perspective
Different mechanisms are involved in the maintenance of epigenetic states. Studies discussed herein have shown that dietary factors are likely to contribute to epigenetic alterations and in some cases may be able to reverse abnormal epigenetic states. In addition, while many of the aforementioned studies were conducted using a particular dietary factor, it is reasonable to believe that most may be consumed in combination and over a period of a lifetime. This may provide a rationale for studying nutrient epigenetic modifiers more in combination studies or the proposal of an ‘epigenetic diet’ focused on consuming products that show the ability to stimulate beneficial epigenetic modifications, including increased consumption of fruit, vegetables and those dietary components that are mentioned herein. This may be used from a chemopreventive standpoint to incorporate anticancer nutrients into one’s daily routine to impede disease mechanisms. From a therapeutic perspective many nutrients have been and are being studied for their ability to prevent and reduce the risk or severity of certain diseases and for their anticarcinogenic properties. The field of nutrigenomics involves studying how genes and dietary components interact to influence phenotype and can reveal how one responds to bioactive components based on genetics, nutrient-induced changes in DNA methylation and chromatin alterations, and nutrient-induced changes in gene expression, whereas the field of nutriepigenomics involves the lifelong remodeling of our epigenomes by nutritional factors [218–220]. Nutriepigenomic studies focusing on individual responses to bioactive components and individualized epigenetic diets consisting of bioactive dietary factors mentioned herein will be of particular interest in the future. Furthermore, future studies focusing on the clinical relevance and mechanism of epigenetic modification of bio-active dietary factors are needed to further assess the applicability of dietary factors as cancer preventive and chemopreventive agents.
Executive summary.
Epigenetic mechanisms
Epigenetic modifications typically occur by changes in DNA methylation, histone modifications, or by RNAi and can be influenced by dietary factors.
At least a half of all tumor suppressor genes are inactivated through epigenetic mechanisms in tumorigenesis.
Evidence suggests that dietary agents can affect epigenetic processes.
Epigenetic diet compounds
Dietary polyphenols such as tea polyphenols (i.e., epicatechin, epicatechin-3-gallate, epigallocatechin and epigallocatechin-3-gallate), resveratrol and curcumin can inhibit DNA methyltransferases and act as histone modifiers and demonstrate potential as anticancer therapeutic as well as chemopreventive agents.
Isoflavones such as genistein are found in soybeans, fava beans and kudzu and have been demonstrated to have anticancer properties that, in part, involve DNA methylation.
Isothiocyanates including sulforaphane are known to affect the epigenome and to have anticancer properties and act as a histone deacetylase inhibitor.
Other dietary factors including those found in Brazilian nuts, chicken, cereals, coffee, cashews, garlic, parsley, milk thistle and rosemary, have also been reported to have epigenetic targets in cancer. While most natural dietary products have shown beneficial effects on the epigenome, some dietary components (i.e., alcohol) are associated with harmful epigenetic modifications.
Future perspective
Many nutrients have been and are being studied for their chemopreventive and/or chemotherapeutic properties.
Numerous investigations provide a rationale for studying nutrient epigenetic modifiers further in combination studies. Furthermore, the proposal of an ‘epigenetic diet’ focused on consuming products that show the ability to stimulate beneficial epigenetic modifications will be of particular interest in the future.
Acknowledgments
The authors thank Drs Yuanyuan Li and Syed M Meeran for helpful comments on this manuscript.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was supported in part by grants from the National Cancer Institute (RO1 CA129415), the American Institute for Cancer Research, and the Norma Livingston Foundation. Tabitha M Hardy was supported by NIH National Institute of General Medical Sciences (NIGMS) Institutional Research and Academic Career Development Awards (IRACDA) Program 5K12GM088010 (Dr Bryan Noe [principal investigator]). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Knekt P, Järvinen R, Seppänen R, et al. Dietary flavonoids and the risk of lung cancer and other malignant neoplasms. Am J Epidemiol. 1997;146(3):223–230. doi: 10.1093/oxfordjournals.aje.a009257. [DOI] [PubMed] [Google Scholar]
- 2.Howells LM, Moiseeva EP, Neal CP, et al. Predicting the physiological relevance of in vitro cancer preventive activities of phytochemicals. Acta Pharmacol Sin. 2007;28(9):1274–1304. doi: 10.1111/j.1745-7254.2007.00690.x. [DOI] [PubMed] [Google Scholar]
- 3.Fowke JH. Head and neck cancer. A case for inhibition by isothiocyanates and indoles from cruciferous vegetables. Eur J Cancer Prev. 2007;16(4):348–356. doi: 10.1097/01.cej.0000236258.80522.fb. [DOI] [PubMed] [Google Scholar]
- 4.De Kok T, Van Breda S, Manson M. Mechanisms of combined action of different chemopreventive dietary compounds. Eur J Nutr. 2008;47:51–59. doi: 10.1007/s00394-008-2006-y. [DOI] [PubMed] [Google Scholar]
- 5.Kunnumakkara AB, Anand P, Aggarwal BB. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008;269(2):199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Ravindran J, Prasad S, Aggarwal B. Curcumin and cancer cells: how many ways can curry kill tumor cells selectively? AAPS J. 2009;11(3):495–510. doi: 10.1208/s12248-009-9128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho E, Clarke JD, Dashwood RH. Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J Nutr. 2009;139(12):2393–2396. doi: 10.3945/jn.109.113332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung KL, Khor TO, Huang M-T, Kong A-N. Differential in vivo mechanism of chemoprevention of tumor formation in azoxymethane/dextran sodium sulfate mice by PEITC and DBM. Carcinogenesis. 2010;31(5):880–885. doi: 10.1093/carcin/bgp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shu L, Cheung K-L, Khor T, Chen C, Kong A-N. Phytochemicals: cancer chemoprevention and suppression of tumor onset and metastasis. Cancer Metastasis Rev. 2010;29(3):483–502. doi: 10.1007/s10555-010-9239-y. [DOI] [PubMed] [Google Scholar]
- 10.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 11.Holliday R. Mechanisms for the control of gene activity during development. Biol Rev Camb Philos Soc. 1990;65(4):431–471. doi: 10.1111/j.1469-185x.1990.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 12.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22(2):91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 13.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 15.Altaf M, Saksouk N, Côté J. Histone modifications in response to DNA damage. Mutat Res. 2007;618(1–2):81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 16▪.Meeran S, Ahmed A, Tollefsbol T. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin Epigenetics. 2010;1(3):101–116. doi: 10.1007/s13148-010-0011-5. Synoptic review of bioactive dietary compounds with epigenetic and chemopreventive properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3(2):226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 18.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66(5):2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 19.Grafodatskaya D, Chung B, Szatmari P, Weksberg R. Autism spectrum disorders and epigenetics. J Am Acad Child Adolesc Psychiatry. 2010;49(8):794–809. doi: 10.1016/j.jaac.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Milagro FI, Campión J, Cordero P, et al. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J. 2011;25(4):1378–1389. doi: 10.1096/fj.10-170365. [DOI] [PubMed] [Google Scholar]
- 21.Maier S, Olek A. Diabetes: a candidate disease for efficient DNA methylation profiling. J Nutr. 2002;132(8):2440S–2443S. doi: 10.1093/jn/132.8.2440S. [DOI] [PubMed] [Google Scholar]
- 22.Chowdhury S, Erickson SW, Macleod SL, et al. Maternal genome-wide DNA methylation patterns and congenital heart defects. PLoS ONE. 2011;6(1):e16506. doi: 10.1371/journal.pone.0016506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nephew KP, Huang TH-M. Epigenetic gene silencing in cancer initiation and progression. Cancer Lett. 2003;190(2):125–133. doi: 10.1016/s0304-3835(02)00511-6. [DOI] [PubMed] [Google Scholar]
- 24.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 25.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doi A, Park I-H, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41(12):1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 28.Hassan YI, Zempleni J. Epigenetic regulation of chromatin structure and gene function by biotin. J Nutr. 2006;136(7):1763–1765. doi: 10.1093/jn/136.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhuri S, Cui Y, Klaassen CD. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol. 2010;245(3):378–393. doi: 10.1016/j.taap.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mottet D, Castronovo V. Histone deacetylases: target enzymes for cancer therapy. Clin Exp Metastasis. 2008;25(2):183–189. doi: 10.1007/s10585-007-9131-5. [DOI] [PubMed] [Google Scholar]
- 31.Lafon-Hughes L, Di Tomaso MV, Méndez-Acuña L, Martínez-López W. Chromatin-remodelling mechanisms in cancer. Mutat Res. 2008;658(3):191–214. doi: 10.1016/j.mrrev.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95(26):15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 34.Panning B, Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93(3):305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- 35.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299(5607):716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 36.Tufarelli C, Stanley JA, Garrick D, et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34(2):157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y-H, Liu S, Zhang G, et al. Knockdown of c-Myc expression by RNAi inhibits MCF-7 breast tumor cells growth in vitro and in vivo. Breast Cancer Res. 2005;7(2):R220–R228. doi: 10.1186/bcr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lakka SS, Gondi CS, Dinh DH, et al. Specific interference of urokinase-type plasminogen activator receptor and matrix metalloproteinase-9 gene expression induced by double-stranded RNA results in decreased invasion, tumor growth, and angiogenesis in gliomas. J Biol Chem. 2005;280(23):21882–21892. doi: 10.1074/jbc.M408520200. [DOI] [PubMed] [Google Scholar]
- 39.Fleming JB, Shen G-L, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res. 2005;3(7):413–423. doi: 10.1158/1541-7786.MCR-04-0206. [DOI] [PubMed] [Google Scholar]
- 40.Liang X, Yang X, Tang Y, et al. RNAi-mediated downregulation of urokinase plasminogen activator receptor inhibits proliferation, adhesion, migration and invasion in oral cancer cells. Oral Oncol. 2008;44(12):1172–1180. doi: 10.1016/j.oraloncology.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Yavari K, Taghikhani M, Ghannadi Maragheh M, Mesbah-Namin S, Babaei M. Downregulation of IGF-IR expression by RNAi inhibits proliferation and enhances chemosensitization of human colon cancer cells. Int J Colorectal Dis. 2010;25(1):9–16. doi: 10.1007/s00384-009-0783-2. [DOI] [PubMed] [Google Scholar]
- 42.Lujambio A, Portela A, Liz J, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29(48):6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: Tying it all together. Int J Biochem Cell Biol. 2009;41(1):87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Ducasse M, Brown M. Epigenetic aberrations and cancer. Mol Cancer. 2006;5(1):60. doi: 10.1186/1476-4598-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 46.Martínez MA, Gutiérrez A, Armand-Ugón M, et al. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS. 2002;16(18):2385–2390. doi: 10.1097/00002030-200212060-00002. [DOI] [PubMed] [Google Scholar]
- 47.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 49.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3(9):e157. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cairns P. Gene methylation and early detection of genitourinary cancer: the road ahead. Nat Rev Cancer. 2007;7(7):531–543. doi: 10.1038/nrc2170. [DOI] [PubMed] [Google Scholar]
- 51.Cox PM, Goding CR. Transcription and cancer. Br J Cancer. 1991;63(5):651–662. doi: 10.1038/bjc.1991.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taby R, Issa J-PJ. Cancer epigenetics. CA Cancer J Clin. 2010;60(6):376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 53.Issa J-P. Cancer prevention: epigenetics steps up to the plate. Cancer Prev Res. 2008;1(4):219–222. doi: 10.1158/1940-6207.CAPR-08-0029. [DOI] [PubMed] [Google Scholar]
- 54.Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59(1):67–70. [PubMed] [Google Scholar]
- 55.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 56.Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem Pharmacol. 2010;80(12):1771–1792. doi: 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landis-Piwowar KR, Milacic V, Dou QP. Relationship between the methylation status of dietary flavonoids and their growth-inhibitory and apoptosis-inducing activities in human cancer cells. J Cell Biochem. 2008;105(2):514–523. doi: 10.1002/jcb.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Tollefsbol TO. p16INK4a suppression by glucose restriction contributes to human cellular lifespan extension through SIRT1-mediated epigenetic and genetic mechanisms. PLoS ONE. 2011;6(2):e17421. doi: 10.1371/journal.pone.0017421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59▪.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS ONE. 2010;5(7):e11457. doi: 10.1371/journal.pone.0011457. Sulforaphane represses the catalytic component of telomerase, hTERT, through epigenetic modification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paluszczak J, Krajka-Kuzniak V, Baer-Dubowska W. The effect of dietary polyphenols on the epigenetic regulation of gene expression in MCF7 breast cancer cells. Toxicol Lett. 2010;192(2):119–125. doi: 10.1016/j.toxlet.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 61.Lee KW, Lee HJ, Lee CY. Vitamins, phytochemicals, diets, and their implementation in cancer chemoprevention. Crit Rev Food Sci Nutr. 2004;44(6):437–452. doi: 10.1080/10408690490886674. [DOI] [PubMed] [Google Scholar]
- 62.Singh KP, Dumond JW. Genetic and epigenetic changes induced by chronic low dose exposure to arsenic of mouse testicular Leydig cells. Int J Oncol. 2007;30(1):253–260. [PubMed] [Google Scholar]
- 63.Xie Y, Liu J, Benbrahim-Tallaa L, et al. Aberrant DNA methylation and gene expression in livers of newborn mice transplacentally exposed to a hepatocarcinogenic dose of inorganic arsenic. Toxicology. 2007;236(1–2):7–15. doi: 10.1016/j.tox.2007.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertram C, Khan O, Ohri S, Phillips DI, Matthews SG, Hanson MA. Transgenerational effects of prenatal nutrient restriction on cardiovascular and hypothalamic-pituitary-adrenal function. J Physiol. 2008;586(8):2217–2229. doi: 10.1113/jphysiol.2007.147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novikova SI, He F, Bai J, Cutrufello NJ, Lidow MS, Undieh AS. Maternal cocaine administration in mice alters DNA methylation and gene expression in hippocampal neurons of neonatal and prepubertal offspring. PLoS ONE. 2008;3(4):e1919. doi: 10.1371/journal.pone.0001919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andersen HR, Schmidt IM, Grandjean P, et al. Impaired reproductive development in sons of women occupationally exposed to pesticides during pregnancy. Environ Health Perspect. 2008;116(4):566–572. doi: 10.1289/ehp.10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skinner MK, Guerrero-Bosagna C. Environmental signals and transgenerational epigenetics. Epigenomics. 2009;1(1):111–117. doi: 10.2217/epi.09.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 70.Poirier LA. Methyl group deficiency in hepatocarcinogenesis. Drug Metab Rev. 1994;26(1–2):185–199. doi: 10.3109/03602539409029790. [DOI] [PubMed] [Google Scholar]
- 71.Pogribny IP, Ross SA, Wise C, et al. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res. 2006;593(1–2):80–87. doi: 10.1016/j.mrfmmm.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 72.Bravo L. Polyphenols: chemistry, dietary sources, metabolism, and nutritional significance. Nutr Rev. 1998;56(11):317–333. doi: 10.1111/j.1753-4887.1998.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 73.Harborne J. Plant Phenolics. Academic Press; London, UK: 1989. Methods in plant biochemistry. [Google Scholar]
- 74.Shukla Y, Singh R. Resveratrol and cellular mechanisms of cancer prevention. Ann NY Acad Sci. 2011;1215(1):1–8. doi: 10.1111/j.1749-6632.2010.05870.x. [DOI] [PubMed] [Google Scholar]
- 75.Cui X, Jin Y, Hofseth AB, et al. Resveratrol suppresses colitis and colon cancer associated with colitis. Cancer Prev Res. 2010;3(4):549–559. doi: 10.1158/1940-6207.CAPR-09-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aggarwal BB, Shishodia S. Molecular targets of dietary agents for prevention and therapy of cancer. Biochem Pharmacol. 2006;71(10):1397–1421. doi: 10.1016/j.bcp.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 77.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137(1):223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 78.Lee WJ, Shim J-Y, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005;68(4):1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 79.Shen FM, Chen HW. Element composition of tea leaves and tea infusions and its impact on health. Bull of Environ Contam Toxicol. 2008;80(3):300–304. doi: 10.1007/s00128-008-9367-z. [DOI] [PubMed] [Google Scholar]
- 80.Fujihara T, Nakagawa-Izumi A, Ozawa T, Numata O. High-molecular-weight polyphenols from oolong tea and black tea: purification, some properties, and role in increasing mitochondrial membrane potential. Biosci Biotechnol Biochem. 2007;71(3):711–719. doi: 10.1271/bbb.60562. [DOI] [PubMed] [Google Scholar]
- 81.Graham HN. Green tea composition, consumption, and polyphenol chemistry. Prev Med. 1992;21(3):334–350. doi: 10.1016/0091-7435(92)90041-f. [DOI] [PubMed] [Google Scholar]
- 82.Lin J-K, Liang Y-C, Lin-Shiau S-Y. Cancer chemoprevention by tea polyphenols through mitotic signal transduction blockade. Biochem Pharmacol. 1999;58(6):911–915. doi: 10.1016/s0006-2952(99)00112-4. [DOI] [PubMed] [Google Scholar]
- 83.Chuang JC, Yoo CB, Kwan JM, et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol Cancer Ther. 2005;4(10):1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- 84.Shanmugam MK, Kannaiyan R, Sethi G. Targeting cell signaling and apoptotic pathways by dietary agents: role in the prevention and treatment of cancer. Nutr Cancer. 2011;63(2):161–173. doi: 10.1080/01635581.2011.523502. [DOI] [PubMed] [Google Scholar]
- 85.Chen P-N, Chu S-C, Kuo W-H, Chou M-Y, Lin J-K, Hsieh Y-S. Epigallocatechin-3 gallate inhibits invasion, epithelial-mesenchymal transition, and tumor growth in oral cancer cells. J Agric Food Chem. 2011;59(8):3836–3844. doi: 10.1021/jf1049408. [DOI] [PubMed] [Google Scholar]
- 86.Tu S-H, Ku C-Y, Ho C-T, et al. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits nicotine- and estrogen-induced α9-nicotinic acetylcholine receptor upregulation in human breast cancer cells. Mol Nutr Food Res. 2011;55(3):455–466. doi: 10.1002/mnfr.201000254. [DOI] [PubMed] [Google Scholar]
- 87.Kürbitz C, Heise D, Redmer T, et al. Epicatechin gallate and catechin gallate are superior to epigallocatechin gallate in growth suppression and anti-inflammatory activities in pancreatic tumor cells. Cancer Sci. 2011;102(4):728–734. doi: 10.1111/j.1349-7006.2011.01870.x. [DOI] [PubMed] [Google Scholar]
- 88.Kim JW, Amin AR, Shin DM. Chemoprevention of head and neck cancer with green tea polyphenols. Cancer Prev Res. 2010;3(8):900–909. doi: 10.1158/1940-6207.CAPR-09-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang C, Lambert J, Sang S. Antioxidative and anti-carcinogenic activities of tea polyphenols. Arch Toxicol. 2009;83(1):11–21. doi: 10.1007/s00204-008-0372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90▪▪.Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010;17:2141–2151. doi: 10.2174/092986710791299966. (−)-epigallocatechin-3-gallate combined with histone deacetylase inhibition reactivates the estrogen receptor and may be useful in chemoprevention and therapeutics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Farabegoli F, Papi A, Bartolini G, Ostan R, Orlandi M. (−)-epigallocatechin-3-gallate downregulates P-gP and BCRP in a tamoxifen resistant MCF-7 cell line. Phytomedicine. 2010;17:356–362. doi: 10.1016/j.phymed.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 92.Balasubramanian S, Adhikary G, Eckert RL. The Bmi-1 polycomb protein antagonizes the (−)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival. Carcinogenesis. 2010;31(3):496–503. doi: 10.1093/carcin/bgp314. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 93.Berletch JB, Liu C, Love WK, Andrews LG, Katiyar SK, Tollefsbol TO. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103(2):509–519. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee AH, Fraser ML, Meng X, Binns CW. Protective effects of green tea against prostate cancer. Expert Rev Anticancer Ther. 2006;6(4):507–513. doi: 10.1586/14737140.6.4.507. [DOI] [PubMed] [Google Scholar]
- 95.Fang MZ, Wang Y, Ai N, et al. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63(22):7563–7570. [PubMed] [Google Scholar]
- 96.Nandakumar V, Vaid M, Katiyar SK. (−)-epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32(4):537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Y, Yuan Y-Y, Meeran S, Tollefsbol T. Synergistic epigenetic reactivation of estrogen receptor-α (ERα) by combined green tea polyphenol and histone deacetylase inhibitor in ERα-negative breast cancer cells. Mol Cancer. 2010;9(1):274. doi: 10.1186/1476-4598-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao Z, Xu Z, Hung M-S, et al. Promoter demethylation of WIF-1 by epigallocatechin-3-gallate in lung cancer cells. Anticancer Res. 2009;29(6):2025–2030. [PubMed] [Google Scholar]
- 99.Kato K, Long NK, Makita H, et al. Effects of green tea polyphenol on methylation status of RECK gene and cancer cell invasion in oral squamous cell carcinoma cells. Br J Cancer. 2008;99(4):647–654. doi: 10.1038/sj.bjc.6604521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pandey M, Shukla S, Gupta S. Promoter demethylation and chromatin remodeling by green tea polyphenols leads to re-expression of GSTP1 in human prostate cancer cells. Int J Cancer. 2010;126(11):2520–2533. doi: 10.1002/ijc.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Choi K-C, Jung MG, Lee Y-H, et al. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009;69(2):583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 102.Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem. 2010;21(2):140–146. doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 103.Tran P, Kim S-A, Choi H, Yoon J-H, Ahn S-G. Epigallocatechin-3-gallate suppresses the expression of HSP70 and HSP90 and exhibits anti-tumor activity in vitro and in vivo. BMC Cancer. 2010;10(1):276. doi: 10.1186/1471-2407-10-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sagara Y, Miyata Y, Nomata K, Hayashi T, Kanetake H. Green tea polyphenol suppresses tumor invasion and angiogenesis in N-butyl-(-4-hydroxybutyl) nitrosamine-induced bladder cancer. Cancer Epidemiol. 2010;34(3):350–354. doi: 10.1016/j.canep.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 105.Zhang D, Al-Hendy M, Richard-Davis G, et al. Green tea extract inhibits proliferation of uterine leiomyoma cells in vitro and in nude mice. Am J Obs Gynecol. 2010;202(3):289, e1–e9. doi: 10.1016/j.ajog.2009.10.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lam WH, Kazi A, Kuhn DJ, et al. A potential prodrug for a green tea polyphenol proteasome inhibitor: evaluation of the peracetate ester of (−)-epigallocatechin gallate [(−)-EGCG] Bioorganic Med Chem. 2004;12(21):5587–5593. doi: 10.1016/j.bmc.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 107.Meeran SM, Patel SN, Chan T-H, Tollefsbol TO. A novel prodrug of epigallocatechin-3-gallate: Differential epigenetic hTERT repression in human breast cancer cells. Cancer Prev Res. 2011 doi: 10.1158/1940-6207.CAPR-11-0009. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Landis-Piwowar KR, Huo C, Chen D, et al. A novel prodrug of the green tea polyphenol (−)-epigallocatechin-3-gallate as a potential anticancer agent. Cancer Res. 2007;67(9):4303–4310. doi: 10.1158/0008-5472.CAN-06-4699. [DOI] [PubMed] [Google Scholar]
- 109.Das D, Mukherjee S, Ray D. Resveratrol and red wine, healthy heart and longevity. Heart Fail Rev. 2010;15(5):467–477. doi: 10.1007/s10741-010-9163-9. [DOI] [PubMed] [Google Scholar]
- 110.Athar M, Back JH, Kopelovich L, Bickers DR, Kim AL. Multiple molecular targets of resveratrol: anti-carcinogenic mechanisms. Arch Biochem Biophys. 2009;486(2):95–102. doi: 10.1016/j.abb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5(6):493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 112.Bishayee A, Waghray A, Barnes K, et al. Suppression of the inflammatory cascade is implicated in resveratrol chemoprevention of experimental hepatocarcinogenesis. Pharmaceut Res. 2010;27(6):1080–1091. doi: 10.1007/s11095-010-0144-4. [DOI] [PubMed] [Google Scholar]
- 113.Bishayee A, Politis T, Darvesh AS. Resveratrol in the chemoprevention and treatment of hepatocellular carcinoma. Cancer Treat Rev. 2010;36(1):43–53. doi: 10.1016/j.ctrv.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 114.Kraft TE, Parisotto D, Schempp C, Efferth T. Fighting cancer with red wine? Molecular mechanisms of resveratrol. Crit Rev Food Sci Nutr. 2009;49(9):782–799. doi: 10.1080/10408390802248627. [DOI] [PubMed] [Google Scholar]
- 115.Mao Q-Q, Bai Y, Lin Y-W, et al. Resveratrol confers resistance against taxol via induction of cell cycle arrest in human cancer cell lines. Mol Nutr Food Res. 2010;54(11):1574–1584. doi: 10.1002/mnfr.200900392. [DOI] [PubMed] [Google Scholar]
- 116.Vanamala J, Reddivari L, Radhakrishnan S, Tarver C. Resveratrol suppresses IGF-1 induced human colon cancer cell proliferation and elevates apoptosis via suppression of IGF-1R/Wnt and activation of p53 signaling pathways. BMC Cancer. 2010;10(1):238. doi: 10.1186/1471-2407-10-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu P-L, Tsai J-R, Charles AL, et al. Resveratrol inhibits human lung adenocarcinoma cell metastasis by suppressing heme oxygenase 1-mediated nuclear factor-κB pathway and subsequently downregulating expression of matrix metalloproteinases. Mol Nutr Food Res. 2010;54(S2):S196–S204. doi: 10.1002/mnfr.200900550. [DOI] [PubMed] [Google Scholar]
- 118.Wu H, Liang X, Fang Y, Qin X, Zhang Y, Liu J. Resveratrol inhibits hypoxia-induced metastasis potential enhancement by restricting hypoxia-induced factor-1[α] expression in colon carcinoma cells. Biomed Pharmacother. 2008;62(9):613–621. doi: 10.1016/j.biopha.2008.06.036. [DOI] [PubMed] [Google Scholar]
- 119.Weng YL, Liao HF, Li AF, Chang JC, Chiou RY. Oral administration of resveratrol in suppression of pulmonary metastasis of BALB/c mice challenged with CT26 colorectal adenocarcinoma cells. Mol Nutr Food Res. 2010;54(2):259–267. doi: 10.1002/mnfr.200900049. [DOI] [PubMed] [Google Scholar]
- 120.Jang M, Cai L, Udeani GO, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275(5297):218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 121.Papoutsis AJ, Lamore SD, Wondrak GT, Selmin OI, Romagnolo DF. Resveratrol prevents epigenetic silencing of BRCA-1 by the aromatic hydrocarbon receptor in human breast cancer cells. J Nutr. 2010;140(9):1607–1614. doi: 10.3945/jn.110.123422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stefanska B, Rudnicka K, Bednarek A, Fabianowska-Majewska K. Hypomethylation and induction of retinoic acid receptor β 2 by concurrent action of adenosine analogues and natural compounds in breast cancer cells. Eur J Pharmacol. 2010;638(1–3):47–53. doi: 10.1016/j.ejphar.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 123.Paluszczak J, Krajka-Kuzniak V, Malecka Z, et al. Frequent gene hypermethylation in laryngeal cancer cell lines and the resistance to demethylation induction by plant polyphenols. Toxicol In Vitro. 2011;25(1):213–221. doi: 10.1016/j.tiv.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 124.Howitz KT, Bitterman KJ, Cohen HY, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 125.Hoshino I, Matsubara H. Recent advances in histone deacetylase targeted cancer therapy. Surgery Today. 2010;40(9):809–815. doi: 10.1007/s00595-010-4300-6. [DOI] [PubMed] [Google Scholar]
- 126.Pruitt K, Zinn RL, Ohm JE, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2(3):e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boily G, He XH, Pearce B, Jardine K, Mcburney MW. SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene. 2009;28(32):2882–2893. doi: 10.1038/onc.2009.147. [DOI] [PubMed] [Google Scholar]
- 128.Feige JN, Lagouge M, Canto C, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8(5):347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 129.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280(17):17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 130.Wang R-H, Zheng Y, Kim H-S, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32(1):11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stünkel W, Peh BK, Tan YC, et al. Function of the SIRT1 protein deacetylase in cancer. Biotechnol J. 2007;2(11):1360–1368. doi: 10.1002/biot.200700087. [DOI] [PubMed] [Google Scholar]
- 132.Kai L, Samuel SK, Levenson AS. Resveratrol enhances p53 acetylation and apoptosis in prostate cancer by inhibiting MTA1/NuRD complex. Int J Cancer. 2010;126(7):1538–1548. doi: 10.1002/ijc.24928. [DOI] [PubMed] [Google Scholar]
- 133.Baur JA, Pearson KJ, Price NL, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444(7117):337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Haigis MC, Guarente LP. Mammalian sirtuins – emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 135.Maheshwari RK, Singh AK, Gaddipati J, Srimal RC. Multiple biological activities of curcumin: a short review. Life Sci. 2006;78(18):2081–2087. doi: 10.1016/j.lfs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 136.Goel A, Aggarwal BB. Curcumin, the golden spice from Indian saffron, is a chemosensitizer and radiosensitizer for tumors and chemoprotector and radioprotector for normal organs. Nutr Cancer. 2010;62(7):919–930. doi: 10.1080/01635581.2010.509835. [DOI] [PubMed] [Google Scholar]
- 137.Liu Z, Xie Z, Jones W, et al. Curcumin is a potent DNA hypomethylation agent. Bioorganic Med Chem Lett. 2009;19(3):706–709. doi: 10.1016/j.bmcl.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 138.Kuck D, Singh N, Lyko F, Medina-Franco JL. Novel and selective DNA methyltransferase inhibitors: Docking-based virtual screening and experimental evaluation. Bioorganic Med Chem. 2010;18(2):822–829. doi: 10.1016/j.bmc.2009.11.050. [DOI] [PubMed] [Google Scholar]
- 139.Fu S, Kurzrock R. Development of curcumin as an epigenetic agent. Cancer. 2010;116(20):4670–4676. doi: 10.1002/cncr.25414. [DOI] [PubMed] [Google Scholar]
- 140.Liu H-L, Chen Y, Cui G-H, Zhou J-F. Curcumin, a potent anti-tumor reagent, is a novel histone deacetylase inhibitor regulating B-NHL cell line Raji proliferation. Acta Pharmacol Sin. 2005;26(5):603–609. doi: 10.1111/j.1745-7254.2005.00081.x. [DOI] [PubMed] [Google Scholar]
- 141.Valinluck V, Sowers LC. Inflammation-mediated cytosine damage: a mechanistic link between inflammation and the epigenetic alterations in human cancers. Cancer Res. 2007;67(12):5583–5586. doi: 10.1158/0008-5472.CAN-07-0846. [DOI] [PubMed] [Google Scholar]
- 142.Kang S-K, Cha S-H, Jeon H-G. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. Stem Cells Dev. 2006;15(2):165–174. doi: 10.1089/scd.2006.15.165. [DOI] [PubMed] [Google Scholar]
- 143.Cui L, Miao J, Furuya T, Li X, Su X-Z, Cui L. PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryotic Cell. 2007;6(7):1219–1227. doi: 10.1128/EC.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pollack BP, Sapkota B, Boss JM. Ultraviolet radiation-induced transcription is associated with gene-specific histone acetylation. Photochem Photobiol. 2009;85(3):652–662. doi: 10.1111/j.1751-1097.2008.00485.x. [DOI] [PubMed] [Google Scholar]
- 145.Sng JCG, Taniura H, Yoneda Y. Histone modifications in kainate-induced status epilepticus. Eur J Neurosci. 2006;23(5):1269–1282. doi: 10.1111/j.1460-9568.2006.04641.x. [DOI] [PubMed] [Google Scholar]
- 146.Chiu J, Khan ZA, Farhangkhoee H, Chakrabarti S. Curcumin prevents diabetes-associated abnormalities in the kidneys by inhibiting p300 and nuclear factor-κB. Nutrition. 2009;25(9):964–972. doi: 10.1016/j.nut.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 147.Tikoo K, Meena RL, Kabra DG, Gaikwad AB. Change in post-translational modifications of histone H3, heat-shock protein-27 and MAP kinase p38 expression by curcumin in streptozotocin-induced type I diabetic nephropathy. Br J Pharmacol. 2008;153(6):1225–1231. doi: 10.1038/sj.bjp.0707666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Morimoto T, Sunagawa Y, Kawamura T, et al. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J Clin Invest. 2008;118(3):868–878. doi: 10.1172/JCI33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee Y-H, Hong SW, Jun W, et al. Anti-histone acetyltransferase activity from allspice extracts inhibits androgen receptor-dependent prostate cancer cell growth. Biosci Biotechnol Biochem. 2007;71(11):2712–2719. doi: 10.1271/bbb.70306. [DOI] [PubMed] [Google Scholar]
- 150.Reuter S, Eifes S, Dicato M, Aggarwal BB, Diederich M. Modulation of anti-apoptotic and survival pathways by curcumin as a strategy to induce apoptosis in cancer cells. Biochem Pharmacol. 2008;76(11):1340–1351. doi: 10.1016/j.bcp.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 151▪.Marcu MG, Jung YJ, Lee S, et al. Curcumin is an inhibitor of p300 histone acetyltransferase. Med Chem. 2006;2(2):169–174. doi: 10.2174/157340606776056133. Shows that curcumin acts as a histone demethylase inhibitor. [DOI] [PubMed] [Google Scholar]
- 152.Balasubramanyam K, Varier RA, Altaf M, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279(49):51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 153.Kutluay SB, Doroghazi J, Roemer ME, Triezenberg SJ. Curcumin inhibits herpes simplex virus immediate-early gene expression by a mechanism independent of p300/CBP histone acetyltransferase activity. Virology. 2008;373(2):239–247. doi: 10.1016/j.virol.2007.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7(3):464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 155.Ali S, Ahmad A, Banerjee S, et al. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70(9):3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 156.Zhang F, Koh GY, Jeansonne DP, et al. A novel solubility-enhanced curcumin formulation showing stability and maintenance of anticancer activity. J Pharm Sci. 2011;100(7):2778–2789. doi: 10.1002/jps.22512. [DOI] [PubMed] [Google Scholar]
- 157.Gupta NK, Dixit VK. Bioavailability enhancement of curcumin by complexation with phosphatidyl choline. J Pharm Sci. 2011;100(5):1987–1995. doi: 10.1002/jps.22393. [DOI] [PubMed] [Google Scholar]
- 158.Valls J, Millán S, Martí MP, Borràs E, Arola L. Advanced separation methods of food anthocyanins, isoflavones and flavanols. J Chromatog A. 2009;1216(43):7143–7172. doi: 10.1016/j.chroma.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 159.Barnes S. Effect of genistein on in vitro and in vivo models of cancer. J Nutr. 1995;125(Suppl 3):777S–783S. doi: 10.1093/jn/125.3_Suppl.777S. [DOI] [PubMed] [Google Scholar]
- 160.Vardi A, Bosviel R, Rabiau N, et al. Soy phytoestrogens modify DNA methylation of GSTP1, RASSF1A, EPH2 and BRCA1 promoter in prostate cancer cells. In vivo. 2010;24(4):393–400. [PubMed] [Google Scholar]
- 161.Lattrich C, Lubig J, Springwald A, Goerse R, Ortmann O, Treeck O. Additive effects of trastuzumab and genistein on human breast cancer cells. Anti-Cancer Drugs. 2011;22(3):253–261. doi: 10.1097/CAD.0b013e3283427bb5. [DOI] [PubMed] [Google Scholar]
- 162▪.Fang MZ, Chen D, Sun Y, Jin Z, Christman K, Yang CS. Reversal of hypermethylation and reactivation of p16INK4a, RARβ, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005;11(19):7033–7041. doi: 10.1158/1078-0432.CCR-05-0406. Genistein acts as a DNA methyltransferase inhibitor and regulator of cancer-related genes. [DOI] [PubMed] [Google Scholar]
- 163.Li Y, Liu L, Andrews LG, Tollefsbol TO. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int J Cancer. 2009;125(2):286–296. doi: 10.1002/ijc.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Majid S, Kikuno N, Nelles J, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68(8):2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 165.Kikuno N, Shiina H, Urakami S, et al. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123(3):552–560. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- 166.King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008;49(1):36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- 167.Majid S, Dar AA, Shahryari V, et al. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-cell translocation gene 3 in prostate cancer. Cancer. 2010;116(1):66–76. doi: 10.1002/cncr.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Majid S, Dar AA, Ahmad AE, et al. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30(4):662–670. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Raynal NJ, Charbonneau M, Momparler LF, Momparler RL. Synergistic effect of 5-aza-2-deoxycytidine and genistein in combination against leukemia. Oncol Res. 2008;17:223–230. doi: 10.3727/096504008786111356. [DOI] [PubMed] [Google Scholar]
- 170.Li Y, Vandenboom TG, Kong D, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69(16):6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Parker LP, Taylor DD, Kesterson J, Metzinger DS, Gercel-Taylor C. Modulation of microRNA associated with ovarian cancer cells by genistein. Eur J Gynaecol Oncol. 2009;30(6):616–621. [PubMed] [Google Scholar]
- 172.Qin W, Zhu W, Shi H, et al. Soy isoflavones have an antiestrogenic effect and alter mammary promoter hypermethylation in healthy premenopausal women. Nutri Cancer. 2009;61:238–244. doi: 10.1080/01635580802404196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cheung K, Kong A-N. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 2010;12(1):87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fimognari C, Lenzi M, Hrelia P. Chemoprevention of cancer by isothiocyanates and anthocyanins: mechanisms of action and structure-activity relationship. Curr Med Chem. 2008;15(5):440–447. doi: 10.2174/092986708783503168. [DOI] [PubMed] [Google Scholar]
- 175.Wilkinson JT, Morse MA, Kresty LA, Stoner GD. Effect of alkyl chain length on inhibition of N-nitrosomethylbenzylamine-induced esophageal tumorigenesis and DNA methylation by isothiocyanates. Carcinogenesis. 1995;16(5):1011–1015. doi: 10.1093/carcin/16.5.1011. [DOI] [PubMed] [Google Scholar]
- 176.Lu Q, Lin X, Feng J, et al. Phenylhexyl isothiocyanate has dual function as histone deacetylase inhibitor and hypomethylating agent and can inhibit myeloma cell growth by targeting critical pathways. J Hematol Oncol. 2008;1:6. doi: 10.1186/1756-8722-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Beklemisheva AA, Fang Y, Feng J, Ma X, Dai W, Chiao JW. Epigenetic mechanism of growth inhibition induced by phenylhexyl isothiocyanate in prostate cancer cells. Anticancer Res. 2006;26(2A):1225–1230. [PubMed] [Google Scholar]
- 178.Ma X, Fang Y, Beklemisheva A, et al. Phenylhexyl isothiocyanate inhibits histone deacetylases and remodels chromatins to induce growth arrest in human leukemia cells. Int J Oncol. 2006;28:1287–1293. [PubMed] [Google Scholar]
- 179.Wang LG, Beklemisheva A, Liu XM, Ferrari AC, Feng J, Chiao JW. Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Mol Carcinogenesis. 2007;46(1):24–31. doi: 10.1002/mc.20258. [DOI] [PubMed] [Google Scholar]
- 180.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharm Res. 2007;55(3):224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pledgie-Tracy A, Sobolewski MD, Davidson NE. Sulforaphane induces cell type-specific apoptosis in human breast cancer cell lines. Mol Cancer Ther. 2007;6(3):1013–1021. doi: 10.1158/1535-7163.MCT-06-0494. [DOI] [PubMed] [Google Scholar]
- 182.Keum Y-S, Khor TO, Lin W, et al. Pharmacokinetics and pharmacodynamics of broccoli sprouts on the suppression of prostate cancer in transgenic adenocarcinoma of mouse prostate (TRAMP) mice: implication of induction of Nrf2, HO-1 and apoptosis and the suppression of Akt-dependent kinase pathway. Pharm Res. 2009;26(10):2324–2331. doi: 10.1007/s11095-009-9948-5. [DOI] [PubMed] [Google Scholar]
- 183.Bhamre S, Sahoo D, Tibshirani R, Dill DL, Brooks JD. Temporal changes in gene expression induced by sulforaphane in human prostate cancer cells. Prostate. 2009;69(2):181–190. doi: 10.1002/pros.20869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Telang U, Brazeau DA, Morris ME. Comparison of the effects of phenethyl isothiocyanate and sulforaphane on gene expression in breast cancer and normal mammary epithelial cells. Exp Biol Med. 2009;234(3):287–295. doi: 10.3181/0808-RM-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dashwood RH, Ho E. Dietary agents as histone deacetylase inhibitors: sulforaphane and structurally related isothiocyanates. Nutr Rev. 2008;66:S36–S38. doi: 10.1111/j.1753-4887.2008.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Myzak MC, Tong P, Dashwood W-M, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med. 2007;232(2):227–234. [PMC free article] [PubMed] [Google Scholar]
- 187.Lemire M, Fillion M, Barbosa F, Jr, Guimarães JR, Mergler D. Elevated levels of selenium in the typical diet of Amazonian riverside populations. Sci Total Environ. 2010;408(19):4076–4084. doi: 10.1016/j.scitotenv.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 188.Fischer JL, Lancia JK, Mathur A, Smith ML. Selenium protection from DNA damage involves a Ref1/p53/Brca1 protein complex. Anticancer Res. 2006;26(2A):899–904. [PubMed] [Google Scholar]
- 189.Combs GF, Jr, Clark LC, Turnbull BW. An analysis of cancer prevention by selenium. Biofactors. 2001;14(1):153–159. doi: 10.1002/biof.5520140120. [DOI] [PubMed] [Google Scholar]
- 190.Redman C, Scott JA, Baines AT, et al. Inhibitory effect of selenomethionine on the growth of three selected human tumor cell lines. Cancer Lett. 1998;125(1–2):103–110. doi: 10.1016/s0304-3835(97)00497-7. [DOI] [PubMed] [Google Scholar]
- 191.Xiang N, Zhao R, Song G, Zhong W. Selenite reactivates silenced genes by modifying DNA methylation and histones in prostate cancer cells. Carcinogenesis. 2008;29(11):2175–2181. doi: 10.1093/carcin/bgn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhuo P, Diamond AM. Molecular mechanisms by which selenoproteins affect cancer risk and progression. Biochim Biophys Acta (BBA) 2009;1790(11):1546–1554. doi: 10.1016/j.bbagen.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Clark LC, Combs GF, Turnbull BW, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. JAMA. 1996;276(24):1957–1963. [PubMed] [Google Scholar]
- 194.Hirsch FR, Lippman SM. Advances in the biology of lung cancer chemoprevention. J Clin Oncol. 2005;23(14):3186–3197. doi: 10.1200/JCO.2005.14.209. [DOI] [PubMed] [Google Scholar]
- 195.Uthus E, Ross S, Davis C. Differential effects of dietary selenium (Se) and folate on methyl metabolism in liver and colon of rats. Biol Trace Element Res. 2006;109(3):201–214. doi: 10.1385/BTER:109:3:201. [DOI] [PubMed] [Google Scholar]
- 196.Davis CD, Uthus EO. Dietary folate and selenium affect dimethylhydrazine-induced aberrant crypt formation, global DNA methylation and one-carbon metabolism in rats. J Nutr. 2003;133(9):2907–2914. doi: 10.1093/jn/133.9.2907. [DOI] [PubMed] [Google Scholar]
- 197.Davis CD, Uthus EO, Finley JW. Dietary selenium and arsenic affect DNA methylation in vitro in Caco-2 cells and in vivo in rat liver and colon. J Nutr. 2000;130(12):2903–2909. doi: 10.1093/jn/130.12.2903. [DOI] [PubMed] [Google Scholar]
- 198.Uthus EO, Yokoi K, Davis CD. Selenium deficiency in Fisher-344 rats decreases plasma and tissue homocysteine concentrations and alters plasma homocysteine and cysteine redox status. J Nutr. 2002;132(6):1122–1128. doi: 10.1093/jn/132.6.1122. [DOI] [PubMed] [Google Scholar]
- 199.Alpers DH. Garlic and its potential for prevention of colorectal cancer and other conditions. Curr Opin Gastroenterol. 2009;25(2):116–121. doi: 10.1097/MOG.0b013e32831ef221. [DOI] [PubMed] [Google Scholar]
- 200.Nian H, Delage B, Ho E, Dashwood RH. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ Mol Mutagen. 2009;50(3):213–221. doi: 10.1002/em.20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Nian H, Delage B, Pinto JT, Dashwood RH. Allyl mercaptan, a garlic-derived organosulfur compound, inhibits histone deacetylase and enhances Sp3 binding on the P21WAF1 promoter. Carcinogenesis. 2008;29(9):1816–1824. doi: 10.1093/carcin/bgn165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lea MA, Randolph VM, Lee JE, Desbordes C. Induction of histone acetylation in mouse erythroleukemia cells by some organosulfur compounds including allyl isothiocyanate. Int J Cancer. 2001;92(6):784–789. doi: 10.1002/ijc.1277. [DOI] [PubMed] [Google Scholar]
- 203.Lea MA, Ibeh C, Han L, Desbordes C. Inhibition of growth and induction of differentiation markers by polyphenolic molecules and histone deacetylase inhibitors in colon cancer cells. Anticancer Res. 2010;30(2):311–318. [PubMed] [Google Scholar]
- 204.Cole BF, Baron JA, Sandler RS, et al. Folic acid for the prevention of colorectal adenomas. JAMA. 2007;297(21):2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 205.Mai A, Massa S, Rotili D, et al. Histone deacetylation in epigenetics: an attractive target for anticancer therapy. Med Res Rev. 2005;25(3):261–309. doi: 10.1002/med.20024. [DOI] [PubMed] [Google Scholar]
- 206.Kim Y-I. Folate and colorectal cancer: an evidence-based critical review. Mol Nutr Food Res. 2007;51(3):267–292. doi: 10.1002/mnfr.200600191. [DOI] [PubMed] [Google Scholar]
- 207.Duthie S. Folate and cancer: how DNA damage, repair and methylation impact on colon carcinogenesis. J Inherited Metab Dis. 2011;34(1):101–109. doi: 10.1007/s10545-010-9128-0. [DOI] [PubMed] [Google Scholar]
- 208.Yang Q, Bostick RM, Friedman JM, Flanders WD. Serum folate and cancer mortality among U.S. adults: findings from the third national health and nutritional examination survey linked mortality file. Cancer Epidemiol Biomark Prev. 2009;18(5):1439–1447. doi: 10.1158/1055-9965.EPI-08-0908. [DOI] [PubMed] [Google Scholar]
- 209.Lu R, Wang X, Sun D-F, et al. Folic acid and sodium butyrate prevent tumorigenesis in a mouse model of colorectal cancer. Epigenetics. 2008;3(6):330–335. doi: 10.4161/epi.3.6.7125. [DOI] [PubMed] [Google Scholar]
- 210.Marsit CJ, Mcclean MD, Furniss CS, Kelsey KT. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int J Cancer. 2006;119(8):1761–1766. doi: 10.1002/ijc.22051. [DOI] [PubMed] [Google Scholar]
- 211.Van Engeland M, Weijenberg MP, Roemen GM, et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: The Netherlands cohort study on diet and cancer. Cancer Res. 2003;63(12):3133–3137. [PubMed] [Google Scholar]
- 212.Giovannucci E, Rimm EB, Ascherio A, Stampfer MJ, Colditz GA, Willett WC. Alcohol, low-methionine-low-folate diets, and risk of colon cancer in men. J Natl Cancer Inst. 1995;87(4):265–273. doi: 10.1093/jnci/87.4.265. [DOI] [PubMed] [Google Scholar]
- 213.Puri SK, Si L, Fan C-Y, Hanna E. Aberrant promoter hypermethylation of multiple genes in head and neck squamous cell carcinoma. Am J Otolaryngol. 2005;26(1):12–17. doi: 10.1016/j.amjoto.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 214.Lee WJ, Zhu BT. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006;27(2):269–277. doi: 10.1093/carcin/bgi206. [DOI] [PubMed] [Google Scholar]
- 215.Davis CD, Milner JA. Biomarkers for diet and cancer prevention research: potentials and challenges. Acta Pharmacol Sin. 2007;28(9):1262–1273. doi: 10.1111/j.1745-7254.2007.00678.x. [DOI] [PubMed] [Google Scholar]
- 216.Sun Y, Jiang X, Chen S, Price BD. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 2006;580(18):4353–4356. doi: 10.1016/j.febslet.2006.06.092. [DOI] [PubMed] [Google Scholar]
- 217.Eliseeva ED, Valkov V, Jung M, Jung MO. Characterization of novel inhibitors of histone acetyltransferases. Mol Cancer Ther. 2007;6(9):2391–2398. doi: 10.1158/1535-7163.MCT-07-0159. [DOI] [PubMed] [Google Scholar]
- 218.Davis CD, Milner J. Frontiers in nutrigenomics, proteomics, metabolomics and cancer prevention. Mutat Res. 2004;551(1–2):51–64. doi: 10.1016/j.mrfmmm.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 219.Davis CD, Hord NG. Nutritional ‘omics’ technologies for elucidating the role(s) of bioactive food components in colon cancer prevention. J Nutr. 2005;135(11):2694–2697. doi: 10.1093/jn/135.11.2694. [DOI] [PubMed] [Google Scholar]
- 220.Gallou-Kabani C, Vigé A, Gross M-S, Junien C. Nutri-epigenomics: lifelong remodelling of our epigenomes by nutritional and metabolic factors and beyond. Clin Chem Lab Med. 2007;45(3):321–327. doi: 10.1515/CCLM.2007.081. [DOI] [PubMed] [Google Scholar]