Abstract
Human serum transferrin (hTF) binds two ferric iron ions which are delivered to cells in a transferrin receptor (TFR) mediated process. Critical to the delivery of iron to cells is the binding of hTF to the TFR and the efficient release of iron orchestrated by the interaction. Within the endosome, iron release from hTF is also aided by lower pH, the presence of anions, and a chelator yet to be identified. We have recently shown that three of the four residues comprising a loop in the N-lobe (Pro142, Lys144, and Pro145) are critical to the high-affinity interaction of hTF with the TFR. In contrast, Arg143 in this loop does not participate in the binding isotherm. In the current study, the kinetics of iron release from alanine mutants of each of these four residues (placed into both diferric and monoferric N-lobe backgrounds) have been determined ± the TFR. The R143A mutation greatly retards the rate of iron release from the N-lobe in the absence of the TFR but has considerably less of an effect in its presence. Our data definitively show that Arg143 serves as a kinetically significant anion binding (KISAB) site that is, by definition, sensitive to salt concentration and critical to the conformational change necessary to induce iron release from the N-lobe of hTF (in the absence of the TFR). This is the first identification of an authentic KISAB site in the N-lobe of hTF. The effect of the single R143A mutation on the kinetic profile of iron release provides a dramatic illustration of the dynamic nature of hTF.
The transferrins (TF)1 are a family of ~80 kDa bilobal iron binding glycoproteins. The two lobes (N- and C-lobe) are further divided into subdomains (NI and NII, CI and CII) that come together to form a deep cleft in which iron is bound (1). The iron is coordinated by identical ligands in each lobe: two tyrosine residues, a single histidine, and an aspartic acid residue as well as two oxygen atoms from the synergistic carbonate anion. Iron binds to TF with high affinity (10−22 M) at the neutral pH of the plasma (2). TF, with iron in each lobe, preferentially binds to specific TF receptors (TFR) on the cell surface (with nanomolar affinity), and the complex undergoes clathrin-dependent receptor-mediated endocytosis. The pH within the endosome is lowered to ~5.6, triggering a number of protonation events within TF and promoting the release of iron to a chelator that has yet to be identified (3–6). Apo TF (TF lacking iron in either lobe) remains bound to the TFR at low pH; the complex is recycled back to the cell surface, and upon fusion with the plasma membrane, apo TF dissociates from the TFR at the neutral pH of the plasma. The iron release process is influenced by several factors including (1) the low pH encountered within the endosome, (2) the presence of anions, (3) the presence of a chelator, (4) interaction with the TFR, and (5) lobe–lobe communication within TF itself. Since all of these elements are closely related, it has been difficult to accurately dissect and assess the impact of a single factor without perturbing the entire system (7).
The pronounced effects of anion binding on the thermodynamic, kinetic, and spectroscopic properties of human serum TF (hTF) have been commented on for many years. Minimally, there are two classes of anions, synergistic and nonsynergistic. The synergistic anion is an absolute requirement for high-affinity ferric iron binding. As previously elaborated (8, 9), virtually all synergistic anions adhere to an interlocking sites model in which an anion has a carboxylate group available to bind to an amino acid residue in hTF (now identified as Arg124 in the N-lobe and Arg456 in the C-lobe) and an electron donor group, one or two carbon atoms away, which binds to the ferric iron.
Nonsynergistic anions comprise a second distinct class, which by definition do not participate in or support high-affinity ferric iron binding. These anions influence iron release by binding to specific sites which are required for iron release (10, 11). Kretchmar and Raymond (12) noted the absolute necessity of the binding of such anions for iron to be released; at pH 7.4, when the ionic strength is extrapolated to zero, no iron is released. A major challenge in the study of hTF has been to identify the site(s) where nonsynergistic anions bind. Obviously, this requires a means to detect such a binding event. Spectroscopic techniques including EPR (10, 13–16), NMR (17), and UV-difference spectra (18–22) have been utilized. The EPR data suggest that a conformational change takes place in one or both lobes of hTF upon anion binding, resulting in a change in the relationship of iron to residues within the cleft (10, 16, 23). The UV-difference technique, championed by the laboratory of Dr. Wesley Harris, involves titration of apo hTF with various anions, the binding of which gives rise to negative electronic absorbance bands. Using this technique, the binding strength of many different anions (both synergistic and nonsynergistic) has been measured. The strength of binding to apo hTF follows the order PPi ~ diphosphonates>Pi>SO42−>HCO3−>Cl− > ClO4− (19, 24, 25). An important observation is that although divalent anions bind to hTF with much higher affinity (~100-fold greater), monovalent anions also bind (21). A review from the Sadler group (26) provides equilibrium constants for binding of 13 anions to apo hTF, ranging from log K ~ 0.82 to log K 7.45.
Although Folajtar and Chasteen first described the curious nature and some attributes of the nonsynergistic anion binding sites (10), Egan et al. (27–30) proposed naming the sites to which they bind “kinetically significant anion binding” or KISAB sites to emphasize their allosteric effect on iron release. The original suggestion from Chasteen was reiterated, namely, that one or more such sites exist in each lobe, possibly near the metal-binding center. Consistent with the finding of Kretchmar and Raymond (12), the site must be occupied by an anion before any iron is released. To further complicate the situation, a chelator such as PPi can serve as both an anion and a chelator. The existence of KISAB sites was offered to explain the puzzling and often conflicting effects of anions on iron release. At neutral pH the competition between anions and chelators for binding to KISAB site(s) can result in a negative effect on iron release (as observed in the N-lobe of hTF at pH 7.4) (31). When iron-containing hTF is subjected to pH 5.6 (where the anion binding strength to the KISAB sites increases), we suggest that binding of the anions induces one or more conformational changes which prime hTF for iron release. It is important to point out that most of this previous work was carried out in the absence of the TFR and at pH 7.4.
By mutating each residue of a surface-exposed loop (Pro142-Arg143-Lys144-Pro145) in the NII subdomain of the N-lobe to alanine in a diferric hTF (Fe2hTF) construct, we have recently shown that the loop appears to be involved in the interaction with the soluble portion of the TFR (sTFR) at neutral pH (32). Specifically, we established that three of the four hTF mutants in the Fe2hTF background (P142A, K144A, and P145A) bound to the sTFR at pH 7.4 with affinities that were 5–7-fold weaker than the Fe2hTF control, very similar to the two monoferric controls. This indicated that binding of the N-lobe of these three mutants was compromised and implied a direct involvement of Pro142, Lys144, and Pro145 in the interaction with the sTFR(32). In contrast, the binding affinity of the R143A mutant was identical to the Fe2hTF control. Previously, surface plasmon resonance (SPR) binding studies revealed that mutation of any of three residues in the soluble portion of the transferrin receptor (Y123S, W124A, andD125K) resulted in a 5–20-fold decrease in binding affinity for Fe2hTF at pH 7.5 (33). In the cryo-EM model of the complex (34), these particular residues lie in the protease-like domain of the sTFR opposite the hTF Pro142-Arg143-Lys144- Pro145 loop. Significantly, at pH 6.3 the binding affinity of these sTFR mutants (Y123S, W124A, and D125K) for apo hTF was unaltered, indicating a pH-dependent effect of this portion of the sTFR on hTF binding. It was therefore of interest to measure the rate constants for iron release at pH 5.6 for the four hTF N-lobe mutants in both the absence and presence of the sTFR.
In the current study, rate constants for iron release and conformational change(s) from the four N-lobe mutants, P142A, R143A, K144A, and P145A in both a Fe2hTF and a monoferric N-lobe (FeNhTF) background in the absence and presence of the sTFR have been determined under our “standard conditions” at the putative endosomal pH of 5.6 (100 mM MES, pH 5.6, containing 300 mM KCl and 4 mM EDTA). These constructs allow us to determine rate constants for both lobes (Fe2hTF) or simply from the N-lobe (FeNhTF) with the C-lobe still present but lacking (and incapable of acquiring) iron. Additionally, salt titrations of the R143A mutant in both the FeNhTF and the isolated N-lobe background were carried out. Determination of the effect of salt on the rate constants has allowed us to definitively identify Arg143 as one important KISAB site in the N-lobe of hTF. This work clarifies a number of issues surrounding the relevance of previous observations in regard to the complexities of anion binding.
MATERIALS AND METHODS
Materials
Dulbecco’s modified Eagle’s medium-Ham F-12 nutrient mixture, antibiotic-antimycotic solution (100×), and trypsin were from the GIBCO-BRL Life Technologies Division of Invitrogen. Fetal bovine serum was obtained from Atlanta Biologicals. Ultroser G is a serum replacement from Pall BioSepra (Cergy, France). The Quik Change mutagenesis kit and pBlue-scriptII came from Stratagene. Methotrexate from Bedford laboratories was used for selection of plasmid containing baby hamster kidney (BHK) cells. All Corning tissue culture dishes, flasks, and expanded surface roller bottles were obtained from local distributors as were Ultracel 30 kDa MWCO microconcentrators (Amicon). Ni-NTA resin came from Qiagen. Hi-prep 26/60 Sephacryl S-200HR and S-300HR columns were obtained from Amersham Pharmacia. EDTA was from Mann Research Laboratories Inc. NTA and ferrous ammonium sulfate were from Sigma. Novex 6%TBE–urea gels, 2× TBE–urea gel sample buffer, and 5× TBE–urea gel running buffer were from Invitrogen.
Production and Purification of hTF and the Four N-Lobe Mutants
As described previously, mutations (P142A, R143A, K144A, and P145A) were introduced into the pNUT construct containing the cDNA coding for N-His-tagged nonglycosylated hTF (Fe2hTF) using the QuikChange site-directed mutagenesis kit (32). Each of the mutations was also placed in the FeNhTF background which prevents iron binding in the C-lobe by strategic mutation of the two Tyr iron binding ligands (Y426F and Y517F) (35). The R143A mutation was also introduced into the pNUT construct coding for the N-lobe of hTF (residues 1–337) with an N-terminal hexahistidine tag. The forward mutagenic primer with the underlined nucleotides indicating substitutions resulting in the mutation is as follows: 5′ CCT GAG CCA GCT AAA CCT CTT GAG AAA GCA GTG GCC 3′. The presence of the mutation was confirmed by sequence analysis of the entire coding region of the plasmid prior to proceeding.
Transfection of the plasmid into BHK cells followed by selection with methotrexate resulted in cells which secrete recombinant hTF, mutants of hTF, and the sTFR into the tissue culture medium (35, 36). As described in detail (35), Fe3+ (NTA)2 is added to the tissue culture medium immediately upon collection to iron load and thereby stabilize the recombinant hTF throughout the purification process. The volume of the collected medium was reduced using a tangential flow device with a 30 kDa cutoff membrane. The concentrated medium was centrifuged (6000g) to remove particulates, and the supernatant was diluted by addition of 5× start buffer to the sample (1×, final concentration: 50 mM Tris, pH 7.5, 300 mM NaCl, 20 mM imidazole, 10% glycerol, and 0.05% sodium azide). All of the His-tagged hTFs as well as the sTFR were then captured by passage over a QiagenNi-NTA column; in each case the proteins were displaced from the column by addition of 250 mM imidazole to the start buffer. Passage over either a Sephacryl S-200HR (for hTF) or S-300HR(for sTFR) gel filtration column in 100 mM NH4HCO3 completed the purification. A competitive immunoassay was used to monitor protein production (37). SDS–PAGE on 10% gels was used to evaluate (and confirm) sample homogeneity.
Complexes of Fe2hTF/sTFR and FeNhTF/sTFR (and mutants thereof) were made by our standard protocol (36) in which a molar excess of hTF is added to 1.5 mg of sTFR, and the sample is loaded onto a Sephacryl S-300HR column in 100 mM NH4HCO3 to isolate the complex and remove excess hTF. Fractions containing the complex were concentrated to a nominal 15 mg/mL (with respect to hTF, ε280=1.2 mg mL−1 cm−1 (38)) using Ultracel microconcentrators (30 kDa MWCO).
Urea Gel Analysis
The extent of iron removal from the hTF constructs using our standard iron removal conditions was examined by urea gel electrophoresis (39). Briefly, hTF samples (1 μg/μL) were incubated in 100 mM MES buffer, pH 5.6, containing 300mM KCl and 4 mM EDTA, and the reaction was stopped by addition of 2× TBE–urea gel sample buffer (final concentration of sample 0.5 μg/μL). Approximately 2.5 μg of sample was loaded per lane, and the gels were electrophoresed for 2.25 h at 125 V. Protein bands were visualized by staining with Coomassie blue.
Iron Release Kinetics
The kinetics of iron release from each of the hTF constructs was monitored at pH 5.6 on an Applied Photophysics SX.18MV stopped-flow spectrofluorometer under our standard conditions (36). As described in detail previously (36, 40), one syringe contained iron-loaded hTF (375 nM) in 300 mM KCl, and the other syringe contained 200 mM MES buffer, pH5.6, 300 mM KCl, and 8 mM EDTA. The mixture has a final pH of 5.6. To determine the effect of salt concentration, the hTF constructs were taken up in KCl of different concentrations between 50 and 600 mM and rapidly mixed with 200 mM MES buffer, pH 5.6, and 8 mM EDTA containing an equivalent concentration of KCl. All samples were excited at 280 nm, and the intrinsic tryptophan fluorescence emission was monitored using a high-pass 320 nm cut-on filter. The data were typically normalized to a fluorescence intensity of 1.0 prior to fitting; rate constants were determined by fitting the change in fluorescence intensity versus time using Origin software (version 7.5) to iron release models which have been described in detail (7).
RESULTS
Kinetic Analysis of the Fe2hTF Mutants (in the Absence of the sTFR)
The rate constants for iron release at pH 5.6 from the Fe2hTF control and the four N-lobe mutants (P142A, R143A, K144A, and P145A) in the Fe2hTF background (in the presence of 300 mM KCl and 4 mM EDTA) are given in Table 1. Under the stated conditions, we have established that k1 reports iron release from the N-lobe (k1N) and k2 is the rate constant for iron release from the C-lobe (k2C) (7). Additionally, we have shown that iron release from this construct goes through this pathway 96%of the time, precluding the need to consider the alternative pathway (k1C and k2N) in the fit. Thus the fits describe the pathway Fe2hTF → FeChTF → apo hTF for the curve shown in Figure 1A. In the absence of the sTFR, mutation of either Pro142 or Pro145 to alanine had only a small effect on the rate constant for iron release from either lobe of Fe2hTF (Table 1). Similar to the control, the curve for the K144A mutant in the Fe2hTF background fit to the A → B → C model; however, k1N was accelerated (2.2-fold) in comparison to k1N for Fe2hTF although, again, the rate constant for iron release from the C-lobe (k2C) was minimally affected. Significantly, the R143A mutant in the Fe2hTF background yielded a progress curve that fit well to a simple A → B model (Figure 1B), providing only a single rate constant which was indistinguishable from the rate constant for iron release from the C-lobe (Table 1). As shown in Figure 1C, the iron release curve for the R143A Fe2hTF mutant (red curve) compared to the Fe2hTF control (black curve) is clearly missing the first fast event (corresponding to iron release from the N-lobe).
Table 1.
Rate Constants for Iron Release from the Control, Fe2hTF, and the Four N-Lobe Mutants in the Fe2hTF Backgrounda
| protein | iron release
|
FCd | Fapod | |
|---|---|---|---|---|
| k1N (min−1)b | k2C (min−1) | |||
| Fe2hTF,c N = 14 | 17.7 ± 2.2 | 0.65 ± 0.06 | 0.30 ± 0.07 | 1.0 |
| P142A Fe2hTF, N = 2 | 13.0 ± 0.8 | 0.64 ± 0.04 | 0.20 ± 0.00 | 1.0 |
| R143A Fe2hTF, N = 2 | 0.65 ± 0.01 | 1.0 | ||
| K144A Fe2hTF, N = 2 | 39.5 ± 0.7 | 0.57 ± 0.04 | 0.13 ± 0.01 | 1.0 |
| P145A Fe2hTF, N = 2 | 15.5 ± 0.03 | 0.61 ± 0.03 | 0.22 ± 0.01 | 1.0 |
The control and three of the four mutants fit best to an A → B → C model yielding two rate constants assigned to iron release from the N-lobe and the C-lobe, respectively.
Averages and 95%confidence intervals for kinetic runs performed on N = 2–14 different days. On each day at least three kinetic traces were averaged prior to fitting.
From ref 7.
Fluorescence intensity constants (7).
Figure 1.

Iron release from Fe2hTF and the R143A Fe2hTF mutant. (A) Iron release progress curve from Fe2hTF (black curve) fit to the A → B → C model (red line). Residuals shown in green. (B) Iron release progress curve from theR143A Fe2hTF mutant (black curve) fit to the A → B model (red line). Residuals shown in green. (C) Overlay of the iron release progress curves of Fe2hTF and the R143A Fe2hTF mutant. Note the loss of the initial fast event from the R143A mutant. All samples (375 nM in 300 mM KCl) were rapidly mixed with 200 mM MES, pH 5.6, 300 mM KCl, and 8 mM EDTA and excited with 280 nm light, and the emission was monitored using a 320 nm cut-on filter.
Kinetic Analysis of the FeNhTF Mutants (in the Absence of the sTFR)
Rate constants for iron release (under our standard conditions) from each mutant in the FeNhTF background are presented in Table 2. The control, FeNhTF, fits to the A → B → C → D model as shown in Figure 2A. Previously, we assigned k1 to iron release from the N-lobe, k2 to an initial conformational change, and k3 to a second and final conformational change to achieve the fully apo conformation (7). The P142A and P145A mutants both fit to the A → B → C → D model. The rate of iron release from the N-lobe (k1) of these two mutants is ~2-fold slower than the rate of iron release from the N-lobe of FeNhTF, while the rates of the two conformational changes (k2 and k3) are essentially equivalent to the control (Table 2). Similarly, K144A fit to the A → B → C → D model; however, the rate of iron release from this mutant was slightly faster (~1.4 times) than the rate constant for iron release from the N-lobe of the control; again, k2 and k3 were similar to the control. Significantly, and in sharp contrast to P142A, K144A, and P145A, the R143A mutant fit to the A → B → C model (yielding two rate constants) with the iron release step, k1, appearing to be absent (Figure 2B). Instead, there are two rate constants which are similar to those assigned to the slow conformational changes of the FeNhTF control. We suggest that the rate constant for iron removal may occur at a rate that is contained within or obscured by one of these slower events. An overlay of the kinetic curves for the FeNhTF control and the R143A FeNhTF mutant is presented in Figure 2C and highlights the difference in the early part of the curve corresponding to iron release.
Table 2.
Rate Constants for the Kinetics of Iron Removal from the Control, FeNhTF, and the Four N-Lobe Mutants in the FeNhTF Backgrounda
| protein | iron release, k1 (min−1)b | conformational change
|
FBd | FCd | FD(apo)d | |
|---|---|---|---|---|---|---|
| k2 (min−1) | k3 (min−1) | |||||
| FeNhTF,c N = 11 | 24.8 ± 5.5 | 5.8 ± 2.1 | 1.1 ± 0.2 | 0.39 ± 0.07 | 0.63 ± 0.07 | 1.0 |
| P142A FeNhTF, N = 3 | 13.4 ± 1.2 | 4.6 ± 0.3 | 0.85 ± 0.03 | 0.38 ± 0.06 | 0.67 ± 0.05 | 1.0 |
| R143A FeNhTF, N = 2 | 5.6 ± 0.7 | 0.98 ± 0.1 | 0.29 ± 0.01 | 1.0 | ||
| K144A FeNhTF, N = 2 | 34.2 ± 8.9 | 6.6 ± 1.8 | 0.69 ± 0.03 | 0.32 ± 0.01 | 0.43 ± 0.01 | 1.0 |
| P145A FeNhTF, N = 2 | 15.1 ± 2.5 | 5.7 ± 1.1 | 0.77 ± 0.00 | 0.46 ± 0.03 | 0.65 ± 0.03 | 1.0 |
The control and three of the four mutants fit best to an A → B → C → D model yielding three rate constants.
Averages and 95% confidence intervals for kinetic runs performed on N=2–11 different days. Each day at least three kinetic traces were averaged prior to fitting.
From ref 7.
Fluorescence intensity constants (7).
Figure 2.

Iron release from FeNhTF and the R143A FeNhTF mutant. (A) Iron release progress curve from FeNhTF (black curve) fit to the A → B → C → D model (red line). Residuals shown in green. (B) Iron release progress curve from the R143A FeNhTF mutant (black curve) fit to the A → B → C model (red line). Residuals shown in green. (C) Overlay of the iron release progress curves of FeNhTF and the R143A FeNhTF mutant. Note the loss of the initial fast event from the R143A mutant. All samples (375 nM in 300 mM KCl) were rapidly mixed with 200 mM MES, pH 5.6, 300 mM KCl, and 8 mM EDTA and excited with 280 nm light, and the emission was monitored using a 320 nm cut-on filter.
Proof of Iron Removal from the R143A Mutant in Each Background
Direct evidence that iron removal from the N-lobe is complete comes from analysis of the samples on urea gels (Supporting Information Figure 1). Migration of hTF species on urea gels depends on both charge and shape (39, 41); for the R143A mutant (in either the Fe2hTF or FeNhTF background) the loss of a surface-exposed positive charge results in a difference in the migration pattern compared to the respective control. Nevertheless, it is evident that the R143A mutant is iron-loaded at pH 7.4 in both backgrounds; furthermore, it is clear that the R143A mutants lack iron at pH 5.6 since there is no band corresponding to the monoferric N-lobe in either the Fe2hTF or FeNhTF background (Supporting Information Figure 1).
Kinetic Analysis of FeNhTF and the R143A FeNhTF Mutant as a Function of Increasing Concentration of KCl
As described in the introduction, there is a complex relationship between iron release and anion binding to specific site(s) on hTF. As reported previously (7), the effect of increasing salt is the most dramatic and obvious in the case of the rate constant for iron release from the N-lobe of FeNhTF. To investigate the effect of salt on the kinetics, we carried out titration experiments with the control, FeNhTF, in comparison to the R143A FeNhTF mutant (Figure 3A). In the case of the control, we note a 4-fold increase in the rate constant (k1) for iron release at 600 mM in comparison to 50 mM KCl (Figure 3A, inset). A more modest 1.6-fold increase is observed for one of the two conformational changes (k2) as a function of salt with no change in the final conformational change (k3) (Figure 3A, inset). In marked contrast, the kinetic analysis of the R143A FeNhTF mutant yielded only two rate constants, neither of which was sensitive to salt concentration, a strong indication that Arg143 constitutes a KISAB site.
Figure 3.
(A) Salt effect on rate constants from FeNhTF and the R143A FeNhTF mutant. Rate constants (k1 (black), k2 (white), and k3 (gray)) at pH 5.6 as a function of KCl concentration are shown for the FeNhTF control (inset) and the R143A FeNhTF mutant. Except for the salt concentration, the conditions are exactly as indicated in the legend to Figure 2. (B) Salt effect on rate constants from isolated N-lobe and the R143AN-lobemutant. Rate constants (k1 (black), k2 (white), and k3 (gray)) at pH 5.6 as a function of KCl concentration are shown for the N-lobe control (inset) and the R143A N-lobe mutant. Except for the salt concentration, the conditions are exactly as indicated in the legend to Figure 2. Note that the rate constants are plotted on the same scale to provide a direct comparison between the R143A mutant and the control.
Kinetic Analysis of the Isolated N-Lobe of hTF and the R143A N-Lobe Mutant as a Function of Increasing Concentration of KCl
To further investigate the effect of salt on iron release, we placed the R143A mutation into a plasmid expressing the His-tagged N-lobe (lacking a C-lobe). The results are presented in Figure 3B. In contrast to both the Fe2hTF and FeNhTF, mutation of Arg143 in the isolated N-lobe does not lead to loss of a kinetic rate constant as compared to the control. Similar to the FeNhTF construct, the isolated N-lobe of hTF fits well to the A → B → C → D model, where k1 reports iron release followed by two conformational changes (k2 and k3) (7). In the case of the N-lobe control, the first two rate constants show sensitivity to the concentration of KCl: a 1.5-fold increase in the rate constant (k1) for iron release at 300 mM in comparison to 50 mM KCl and a 1.9-fold increase for the first conformational change (k2) as a function of salt with no change in the final conformational change (k3) (Figure 3B, inset). As reported previously the rate constant for iron release actually decreases in the presence of 600 mM KCl (7). In contrast, the rate constant for iron release (k1) is significantly lower in the R143A mutant (>2-fold at each salt concentration). None of the three rate constants for theR143A N-lobe are sensitive to the concentration of salt (Figure 3B), with the fastest rate presumably corresponding to iron release.
Kinetic Analysis of the hTF Mutants in the Presence of the sTFR
As mentioned in the introduction, binding of hTF to its receptor has a significant impact on iron release, reversing the order such that iron comes out of the C-lobe first and the N-lobe second. It was therefore of interest to determine the effect of the TFR on the kinetics of the four N-lobe mutants. In the present study, we find that the rate constants for iron release from the C-lobe (k1C) of all of the mutants in complex with the sTFR in the Fe2hTF background are very similar to the control, Fe2hTF/sTFR complex (Supporting Information Table 1A). Likewise, when in complex with the sTFR, iron release from the N-lobe (k2N) of the four mutants is similar to the control. As detailed below, these results are not surprising because the main effect of the TFR is on the C-lobe, which in the complex is the first lobe to release iron.
Analysis of iron release from the four mutants in the FeNhTF background in complex with the sTFR also reveals no significant differences in the rate constants for conformational change (k1) or iron release (k2) (Supporting Information Table 1B). Although there are some differences in the rate constants of the mutants, they fall within or close to the standard deviation and overlap the values for the control. Because the N-lobe alone does not bind to the TFR, it could not be evaluated in its presence (7).
DISCUSSION
Alanine scanning of four residues (Pro142, Arg143, Lys144, and Pro145) residing in a loop in the NII subdomain reveals their importance in the mechanism of iron release from the N-lobe. Significant findings include definitive identification of Arg143 as a KISAB site as indicated by the following observations: (1) the curves of the R143A mutant in the Fe2hTF and FeNhTF constructs are clearly missing a rapid kinetic event (Figures 1C and 2C); (2) the absence of Arg143 results in a substantial decrease in the rate constant for iron release from the N-lobe (which may be indistinguishable from the rate constant for iron release from the C-lobe in the Fe2hTF background or from conformational changes in the N-lobe in the FeNhTF background over the same time frame used for the controls) (Tables 1 and 2); and (3) the anion effect is absent from the R143A FeNhTF mutant and from the isolated N-lobe containing the R143A mutation (Figure 3).
Mutation of Lys144 to alanine results in a small, but consistent, increase in the rate constant for iron release from the N-lobe (Tables 1 and 2), indicating that Lys144 normally slows the process and, additionally, that it is not a KISAB site. Iron release from the N-lobe of the K144A mutant remains sensitive to salt (data not shown). We suggest that the absence of Lys144 might increase access of chloride ions to Arg143, resulting in an enhancement of iron release relative to the control. Although both Arg143 and Lys144 were suggested as a potential KISAB sites in a modeling study (42), our data clearly show that, unlike Arg143, Lys144 does not meet the criteria for this designation.
Mutation of the two proline residues (P142A and P145A) within the loop marginally slows the rate of iron release from the Fe2hTF constructs (Table 1) but has a somewhat greater effect (~2-fold slower) on iron release from the constructs in the FeNhTF background (Table 2). Clearly, the substitution of an alanine does not provide the unique properties of proline residues. As detailed previously (43), this region of the NII subdomain is relatively mobile due to its proximity to α-helices 5 and 11 and a disulfide bond in hTF formed betweenCys137 and Cys331. The opening of the cleft involves the pivoting of α-helix 5 (residues 129–135) on α-helix 11 (residues 317–328). As noted, the Pro-Arg-Lys-Pro sequence is completely conserved in rabbit and pig TF which bind to human TFR and is not conserved in bovine TF, chicken ovotransferrin, or human lactoferrin which do not bind to the human TFR.
Significantly, and in contrast to the other three residues comprising the loop, the side chain of Arg143 extends away from the hTF/TFR interface (32, 34) (Figure 4). In the crystal structures of the apo- and iron-containing N-lobe, the guanidinium moiety of Arg143 is in proximity (~14 Å) to Trp128, which along with Trp264 completely accounts for the increase in the fluorescent signal upon iron release from the N-lobe of hTF (40). We therefore considered the possibility that our results might be explained by a direct effect of Arg143 on Trp128, such that the report of the pH-induced change when the Arg is changed to Ala is lost. However, as shown in Supporting Information Figure 2, the positioning of residues Arg143 and Trp128 appears to preclude any direct interaction and additionally undergoes no significant change with respect to each other as a result of the iron status of the N-lobe, making this alterative explanation unlikely.
Figure 4.
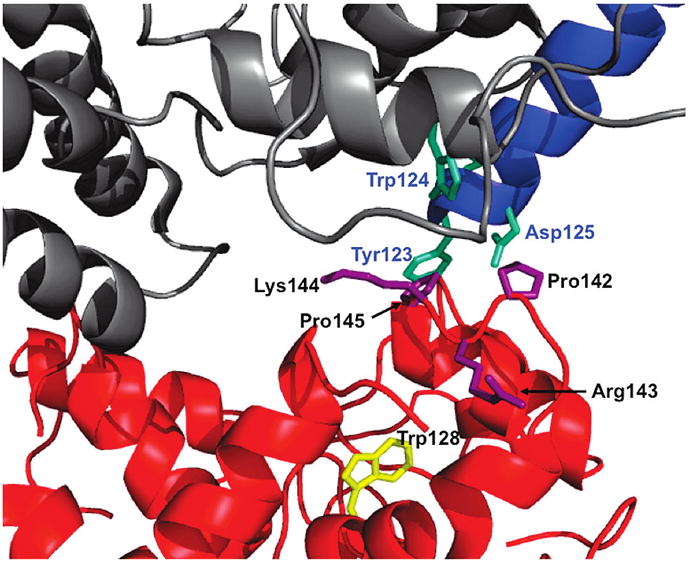
Close-up of the residues identified in the cryo-EM model (PDB: 1SUV) thought to be involved in the interaction between the N-lobe and the TFR: sTFR (gray), sTFR protease-like domain (blue), and N-lobe of hTF (red). Residues on the sTFR proposed to interact with the N-lobe (cyan). Residues in the NII subdomain proposed to interact with the TFR (purple). A contributing factor to the increase in intrinsic tryptophan fluorescence upon the removal of iron from the N-lobe, Trp128 is highlighted (yellow). Arrows used for clarity. Figure generated using Pymol (48).
Simple inorganic anions have multiple roles in hTF which differ according to the conformation of the lobes (open/closed) and the pH. We suggest that an authentic KISAB site should exert its maximal effects on a closed iron-containing lobe at low pH and should assist and/or promote iron release by inducing a conformational change. In the present study, this is precisely what is observed for Arg143. Similarly, Lys569 in the C-lobe of hTF was identified as a KISAB site (44). At pH 5.6, the K569Q mutation in a construct in which the C-lobe was selectively loaded with iron shows a reduced rate constant for iron release, with 10 mM PPi as the chelator, and also a muted salt effect in accord with Lys569 being a KISAB site. Conversely, at neutral pH, anion binding would ideally exert a negative or retarding effect on iron release. In the case of the N-lobe, this is observed such that chloride significantly retards iron release from the isolated N-lobe at neutral pH (31).
Further complicating the story are the numerous studies measuring binding of various anions to apo hTF (19, 24, 25). We suggest that once the iron is removed from the cleft by a chelator, anions are able to bind to newly exposed residues and to stabilize the open conformation. This certainly is true in the case of the N-lobe in which the anion binding sites were identified by titration of apo N-lobe (and single point mutants of the N-lobe) with sulfate (45). Residues that bind to sulfate included Lys206 and Lys296 (the two residues comprising the dilysine trigger crucial to efficient iron release from the N-lobe), as well as Arg124 and Tyr188 which are bound to the synergistic anion and to iron, respectively. Because these residues are buried in the closed iron-bound N-lobe, anions may not exert their full effect on them until the iron is removed and the cleft begins to open. This implies that they would have a limited role as allosteric anion binding sites which directly drive iron release.
We reiterate that, in our studies, loss of the nonsynergistic anion binding site at position 143 is observed to slow the rate of iron release in the absence of the TFR. In the native protein, anion binding to Arg143 normally induces a conformational change which promotes iron release from the N-lobe prior to the C-lobe. However, in the presence of the sTFR the rate constant for iron release from the N-lobe is roughly equivalent in all four mutants (Supporting Information Table 1); in all cases the sTFR decreases the rate of iron release from the N-lobe of Fe2hTF 16-fold and increases the rate of iron release from the C-lobe ~7-fold (7). The question is whether, as previously suggested, one can conclude that the effects of anions and of the TFR on iron release are independent (30). This conclusion, based on analysis of iron release to pyrophosphate from the C-lobe of FeChTF bound to placental-derived TFR, yielded a single rate constant ascribed to iron release. Unfortunately, the many experimental differences in our current work and this previous study do not allow a direct comparison. More studies will be needed to completely resolve this issue.
Our analysis and more precise definition of a KISAB site have allowed us to positively identify the first such site in the N-lobe of hTF. It is possible (and likely) that other KISAB sites exist to influence the kinetic process of iron release from hTF. It is significant that the concentration of anion has the greatest effect on iron release from the N-lobe because it is the first lobe to release iron (in the absence of the TFR). An interesting observation is that the R143A N-lobe mutant does not show a “missing” rate but instead a slower rate of iron release which is salt insensitive. This indicates that the rate that appears to be “missing” (or very slow and overlapping with the other rate constants) occurs only when a C-lobe (with or without iron) is present. The results are consistent with the often observed cooperativity between the two lobes. As reported previously (7, 39), iron release from the N-lobe is affected by the iron status of the C-lobe (but not vice versa). While anion binding to the KISAB sites has a significant influence on the rates of iron exchange between hTF and chelators at pH 5.6 and 7.4, binding to the TFR clearly dominates iron release from the N-lobe at pH 5.6. The present study highlights the complexities of this system and helps to explain previous conflicting results. Specific experimental conditions including TF constructs employed, pH, salt, and chelators must be considered. Generalizations are not easily made.
The TF family is characterized by the very significant conformational changes which occur when the two lobes open and close. Specifically in hTF rotations of 59.4° and 49.5° are required to open the N- and C-lobes, respectively (46). The current work describes what a KISAB site looks like in the N-lobe of hTF in solution at pH 5.6. However, we need to point out that it is unlikely that hTF in the circulation would ever encounter the lower pH found within the endosome. In the bigger picture, i.e., when hTF is bound to the TFR, the physiological significance and/or importance of the identified KISAB site in iron release from the N-lobe is less clear. In spite of this caveat the elucidation of this site remains very interesting as a noteworthy example of protein dynamics. The increasing awareness of proteins as ensembles of conformations seems very relevant (47). The magnitude of the effect of mutation of this single arginine residue to alanine is substantial, indicating a very real effect of anion binding on a specific local region that has more global consequences.
Supplementary Material
Footnotes
This work was supported by USPHS Grant R01 DK21739 (A.B.M.) and R37-GM-20194 from the National Institute of General Medical Sciences (to N.D.C.). Support for A.N.S. came from Hemostasis and Thrombosis Training Grant 5T32HL007594, issued to Dr. Kenneth G. Mann at The University of Vermont by the National Heart, Lung, and Blood Institute.
Abbreviations: TF, serum transferrin; hTF, human serum transferrin; Fe2hTF, recombinant diferric hTF that contains an N-terminal hexa-His tag and is nonglycosylated; FeNhTF, recombinant monoferric N-lobe hTF (mutations Y426F and Y517F preclude iron binding in the C-lobe) that contains an N-terminal hexa-His tag and is nonglycosylated; P142A, R143A, K144A, and P145A refer to single point mutants of hTF placed in the Fe2hTF background or into the FeNhTF background as specified; TFR, transferrin receptor; sTFR, glycosylated soluble portion of the transferrin receptor (residues 121–760) expressed as a recombinant entity that contains an N-terminal hexa-His tag; BHK, baby hamster kidney cells; KISAB site, kinetically significant anion binding site; EPR, electron paramagnetic resonance spectroscopy; NMR, nuclear magnetic resonance spectroscopy.
SUPPORTING INFORMATION AVAILABLE
Rate constants for iron release from Fe2hTF/sTFR and FeNhTF/sTFR complexes and mutants thereof, urea gel analysis proving that iron is released from the R143Amutants (in both the Fe2hTF and FeNhTF backgrounds), and a model showing the proximity and orientation of Arg143 to Trp128. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Biol. 2001;33:940–959. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- 2.Aisen P, Leibman A, Zweier J. Stoichiometric and site characteristics of the binding of iron to human transferrin. J Biol Chem. 1978;253:1930–1937. [PubMed] [Google Scholar]
- 3.Hemmaplardh D, Morgan EH. The role of endocytosis in transferrin uptake by reticulocytes and bone marrow cells. Br J Haematol. 1977;36:85–96. doi: 10.1111/j.1365-2141.1977.tb05758.x. [DOI] [PubMed] [Google Scholar]
- 4.Morgan EH. Effect of pH and iron content of transferrin on its binding to reticulocyte receptors. Biochim Biophys Acta. 1983;762:498–502. doi: 10.1016/0167-4889(83)90052-6. [DOI] [PubMed] [Google Scholar]
- 5.Klausner RD, Ashwell G, van Renswoude J, Harford JB, Bridges KR. Binding of apotransferrin to K562 cells: explanation of the transferrin cycle. Proc Natl Acad Sci USA. 1983;80:2263–2266. doi: 10.1073/pnas.80.8.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc Natl Acad Sci USA. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne SL, Chasteen ND, Steere AN, Mason AB. The unique kinetics of iron release from transferrin: the role of receptor, lobe-lobe interactions, and salt at endosomal pH. J Mol Biol. 2010;396:130–140. doi: 10.1016/j.jmb.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlabach MR, Bates GW. The synergistic binding of anions and Fe(III) by transferrin. J Biol Chem. 1975;250:2182–2188. [PubMed] [Google Scholar]
- 9.Dubach J, Gaffney BJ, More K, Eaton GR, Eaton SS. Effect of the synergistic anion on electron paramagnetic resonance spectra of iron-transferrin anion complexes is consistent with bidentate binding of the anion. Biophys J. 1991;59:1091–1100. doi: 10.1016/S0006-3495(91)82324-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folajtar DA, Chasteen ND. Measurement of non-synergistic anion binding to transferrin by EPR difference spectroscopy. J Am Chem Soc. 1982;104:5775–5780. [Google Scholar]
- 11.Baker EN. Structure and reactivity of transferrins. Adv Inorg Chem. 1994;41:389–463. [Google Scholar]
- 12.Kretchmar SA, Raymond KN. Effects of ionic strength on iron removal from the monoferric transferrins. Inorg Chem. 1988;27:1436–1441. [Google Scholar]
- 13.Price EM, Gibson JF. Electron paramagnetic resonance evidence for a distinction between the two iron binding sites in transferrin and in conalbumin. J Biol Chem. 1972;247:8031–8035. [PubMed] [Google Scholar]
- 14.Chasteen ND. Transferrin: a perspective. Adv Inorg Biochem. 1983;5:201–233. [PubMed] [Google Scholar]
- 15.Thompson CP, McCarty BM, Chasteen ND. The effects of salts and amino group modification on the iron binding domains of transferrin. Biochim Biophys Acta. 1986;870:530–537. doi: 10.1016/0167-4838(86)90262-1. [DOI] [PubMed] [Google Scholar]
- 16.Grady JK, Mason AB, Woodworth RC, Chasteen ND. The effect of salt and site-directed mutations on the iron(III)-binding site of human serum transferrin as probed by EPR spectroscopy. Biochem J. 1995;309:403–410. doi: 10.1042/bj3090403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubal G, Mason AB, Patel SU, Sadler PJ, Woodworth RC. Oxalate- and Ga3+-induced structural changes in human serum transferrin and its recombinant N-lobe. 1H NMR detection of preferential C-lobe Ga3+ binding. Biochemistry. 1993;32:3387–3395. doi: 10.1021/bi00064a024. [DOI] [PubMed] [Google Scholar]
- 18.Pecoraro VL, Harris WR, Carrano CJ, Raymond KN. Siderophilin metal coordination. Difference ultraviolet spectroscopy of di-, tri-, and tetravalent metal ions with ethylenebis-[(o-hydroxyphenyl)glycine] Biochemistry. 1981;20:7033–7039. doi: 10.1021/bi00527a040. [DOI] [PubMed] [Google Scholar]
- 19.Harris WR. Thermodynamics of anion binding to human serum transferrin. Biochemistry. 1985;24:7412–7418. doi: 10.1021/bi00346a057. [DOI] [PubMed] [Google Scholar]
- 20.Harris WR, Bali PK. Effects of anions on the removal of iron from transferrin by phosphonic acids and pyrophosphate. Inorg Chem. 1988;27:2687–2691. [Google Scholar]
- 21.Harris WR, Cafferty AM, Abdollahi S, Trankler K. Binding of monovalent anions to human serum transferrin. Biochim Biophys Acta. 1998;1383:197–210. doi: 10.1016/s0167-4838(97)00152-0. [DOI] [PubMed] [Google Scholar]
- 22.Harris WR, Nesset-Tollefson D, Stenback JZ, Mohamed-Hani N. Site selectivity in the binding of inorganic anions to serum transferrin. J Inorg Biochem. 1990;38:175–183. doi: 10.1016/0162-0134(90)84011-d. [DOI] [PubMed] [Google Scholar]
- 23.Giannetti AM, Halbrooks PJ, Mason AB, Vogt TM, Enns CA, Bjorkman PJ. The molecular mechanism for receptor-stimulated iron release from the plasma iron transport protein transferrin. Structure. 2005;13:1613–1623. doi: 10.1016/j.str.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Bali PK, Harris WR. Site-specific rate constants for iron removal from diferric transferrin by nitrilotris(methylene-phosphonic acid) and pyrophosphate. Arch Biochem Biophys. 1990;281:251–256. doi: 10.1016/0003-9861(90)90440-a. [DOI] [PubMed] [Google Scholar]
- 25.Harris WR, Cafferty AM, Trankler K, Maxwell A, MacGillivray RTA. Thermodynamic studies on anion binding to apotransferrin and to recombinant transferrin N-lobe half molecules. Biochim Biophys Acta. 1999;1430:269–280. doi: 10.1016/s0167-4838(99)00007-2. [DOI] [PubMed] [Google Scholar]
- 26.Sun H, Li H, Sadler PJ. Chem Rev. Vol. 99. 1999. Transferrin as a metal ion mediator; pp. 2817–2842. [DOI] [PubMed] [Google Scholar]
- 27.Marques HM, Egan TJ, Pattrick G. The non-reductive removal of iron from human serum N-terminal monoferric transferrin by pyrophosphate. S Afr J Sci. 1990;86:21–24. [Google Scholar]
- 28.Marques HM, Watson DL, Egan TJ. Kinetics of iron removal from human serum monoferric transferrins by citrate. Inorg Chem. 1991;30:3758–3762. [Google Scholar]
- 29.Egan TJ, Ross DC, Purves LR, Adams PA. Mechanism of iron release from human serum C-terminal monoferric transferrin to pyrophosphate: kinetic discrimination between alternative mechanisms. Inorg Chem. 1992;31:1994–1998. [Google Scholar]
- 30.Egan TJ, Zak O, Aisen P. The anion requirement for iron release from transferrin is preserved in the receptor-transferrin complex. Biochemistry. 1993;32:8162–8167. doi: 10.1021/bi00083a016. [DOI] [PubMed] [Google Scholar]
- 31.He QY, Mason AB, Nguyen V, MacGillivray RTA, Woodworth RC. The chloride effect is related to anion binding in determining the rate of iron release from the human transferrin N-lobe. Biochem J. 2000;350:909–915. [PMC free article] [PubMed] [Google Scholar]
- 32.Mason AB, Byrne SL, Everse SJ, Roberts SE, Chasteen ND, Smith VC, MacGillivray RT, Kandemir B, Bou-Abdallah F. A loop in the N-lobe of human serum transferrin is critical for binding to the transferrin receptor as revealed by mutagenesis, isothermal titration calorimetry, and epitope mapping. J Mol Recognit. 2009;22:521–529. doi: 10.1002/jmr.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giannetti AM, Snow PM, Zak O, Bjorkman PJ. Mechanism for multiple ligand recognition by the human transferrin receptor. PLoS Biol. 2003;1:341–350. doi: 10.1371/journal.pbio.0000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- 35.Mason AB, Halbrooks PJ, Larouche JR, Briggs SK, Moffett ML, Ramsey JE, Connolly SA, Smith VC, MacGillivray RTA. Expression, purification, and characterization of authentic monoferric and apo-human serum transferrins. Protein Expression Purif. 2004;36:318–326. doi: 10.1016/j.pep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Byrne SL, Leverence R, Klein JS, Giannetti AM, Smith VC, MacGillivray RTA, Kaltashov IA, Mason AB. Effect of glycosylation on the function of a soluble, recombinant form of the transferrin receptor. Biochemistry. 2006;45:6663–6673. doi: 10.1021/bi0600695. [DOI] [PubMed] [Google Scholar]
- 37.Mason AB, He QY, Adams TE, Gumerov DR, Kaltashov IA, Nguyen V, MacGillivray RTA. Expression, purification, and characterization of recombinant nonglycosylated human serum transferrin containing a C-terminal hexahistidine tag. Protein Expression Purif. 2001;23:142–150. doi: 10.1006/prep.2001.1480. [DOI] [PubMed] [Google Scholar]
- 38.James NG, Mason AB. Protocol to determine accurate absorption coefficients for iron containing transferrins. Anal Biochem. 2008;378:202–207. doi: 10.1016/j.ab.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byrne SL, Mason AB. Human serum transferrin: a tale of two lobes. Urea gel and steady state fluorescence analysis of recombinant transferrins as a function of pH, time, and the soluble portion of the transferrin receptor. J Biol Inorg Chem. 2009;14:771–781. doi: 10.1007/s00775-009-0491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James NG, Berger CL, Byrne SL, Smith VC, MacGillivray RT, Mason AB. Intrinsic fluorescence reports a global conformational change in the N-lobe of human serum transferrin following iron release. Biochemistry. 2007;46:10603–10611. doi: 10.1021/bi602425c. [DOI] [PubMed] [Google Scholar]
- 41.Makey DG, Seal US. The detection of four molecular forms of human transferrin during the iron binding process. Biochim Biophys Acta. 1976;453:250–256. doi: 10.1016/0005-2795(76)90270-1. [DOI] [PubMed] [Google Scholar]
- 42.Amin EA, Harris WR, Welsh WJ. Identification of possible kinetically significant anion-binding sites in human serum transferrin using molecular modeling strategies. Biopolymers. 2004;73:205–215. doi: 10.1002/bip.10551. [DOI] [PubMed] [Google Scholar]
- 43.Wally J, Buchanan SK. A structural comparison of human serum transferrin and human lactoferrin. Biometals. 2007;20:249–262. doi: 10.1007/s10534-006-9062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zak O, Tam B, MacGillivray RTA, Aisen P. A kinetically active site in the C-lobe of human transferrin. Biochemistry. 1997;36:11036–11043. doi: 10.1021/bi970628v. [DOI] [PubMed] [Google Scholar]
- 45.He QY, Mason AB, Templeton DM. Molecular and Cellular Iron Transport. Marcel Dekker; New York: 2002. Molecular aspects of release of iron from transferrins; pp. 95–123. [Google Scholar]
- 46.Wally J, Halbrooks PJ, Vonrhein C, Rould MA, Everse SJ, Mason AB, Buchanan SK. The crystal structure of iron-free human serum transferrin provides insight into inter-lobe communication and receptor binding. J Biol Chem. 2006;281:24934–24944. doi: 10.1074/jbc.M604592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilser VJ. Biochemistry. An ensemble view of allostery. Science. 2010;327:653–654. doi: 10.1126/science.1186121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



