Abstract
Diffusion Tensor Imaging (DTI) is a Magnetic Resonance Imaging method for measuring water diffusion in vivo. One powerful DTI contrast is fractional anisotropy (FA). FA reflects the strength of water’s diffusion directional preference and is a primary metric for neuronal fiber tracking. As with other DTI contrasts, FA measurements are obscured by the well established presence of bias. DTI bias has been challenging to assess because it is a multi-variable problem including SNR, six tensor parameters, and the DTI collection and processing method used. SIMEX is a modern statistical technique that estimates bias by tracking measurement error as a function of added noise. Here, we use SIMEX to assess bias in FA measurements and show the method provides; i) accurate FA bias estimates, ii) representation of FA bias that is data set specific and accessible to non-statisticians, and iii) a first time possibility for incorporation of bias into DTI data analysis.
Keywords: DTI, FA, bias, SIMEX, parameter estimation, bias correction, diffusion, tensor, imaging
1 Introduction
Radiological practice appears to be on the cusp of reliably and routinely using imaging biomarkers as surrogates for traditional tissue biopsy (e.g., virtual biopsy). One promising magnetic resonance imaging (MRI) technique, diffusion tensor imaging (DTI) provides unique, non-invasive contrasts sensitive to tissue micro-architecture, which can be correlated with cellular organization – for example, as seen through histological analysis [1]. However, quantification of observed data is an estimation process and numeric values must be viewed in light of their distributional properties. Extensive research has been invested in characterizing the impacts of noise and study design on DTI contrasts [2–4] and it is well recognized that both lead to changes in precision (variance) as well as accuracy (bias) of diffusion derived measures. Much progress has been made on variability analysis in DTI using Monte Carlo and boot-strap techniques [5], but quantitative post hoc assessment of the bias in diffusion estimated parameters has remained elusive.
Bias is explicitly defined as the difference between the expectation value of an experimentally observed measure, E(Θobs), and the true value of that measure, Θtruth; bias = E(Θobs) − Θtruth. Therefore, bias causes the statistical average of a measured DTI contrast to converge on the wrong value. Bias in DTI is a multi-variable function including imaging noise, field inhomogeneity, DTI experimental parameters, and the underlying biological diffusion parameters. No analytic equation for bias in DTI is known and no published method to account for bias in empirical DTI data has been tried. Herein, we present a statistical approach to estimate the expected level of bias in DTI contrasts on a voxel-wise basis.
In this initial evaluation, we focus on quantifying the bias in the commonly studied and important DTI contrast fractional anisotropy (FA) as measured using the traditional log-linear minimum mean-squared error (LLMMSE) tensor estimation framework. Significant bias in FA has been well documented both experimentally and through computational studies [2, 4, 6]. Within a clinical and research SNR range (~ 15:1 – 40:1), FA bias is generally a monotonic decreasing function, with lower FA values tending to have larger bias than higher FA values.
To estimate FA bias, we test the simulation extrapolation method (SIMEX) which emerged from the statistical literature as a way to compensate for bias due to additive noise within a regression framework [7, 8]. However, it can also be viewed as a general procedure to extrapolate the degree of bias in an estimate given noisy data by studying the sensitivity of the model fitting procedure when additional synthetic noise is added. Two key criterion of SIMEX are (i) the bias as a function of SNR is monotonic and smooth and (ii) the noise level of the observed data is well understood. Fortunately, (i) is generally holds and for (ii), noise estimation has been well-studied in the DTI literature and several robust methods exist to estimate the noise field given empirical data [9–11].
This manuscript is organized as follows. First, we briefly review the diffusion tensor formulation and describe how the SIMEX bias estimation can be applied to bias estimation for FA. Second, we present a simulation based on empirical data which enables knowledge of the true bias level for comparison with the SIMEX estimated bias. Third, we present single and multi-voxel demonstrations of SIMEX estimation of bias in FA. We close with a brief critique and discussion of potential applications for this approach and avenues for continuing research.
2 Theory
2.1 Diffusion Tensor Imaging
In DTI, magnetic gradients (g) are used to label the position of water at the start and end of an experiment. Given the random thermal motions of water, the labels add incoherently and result in an attenuated, diffusion weighted image (dwi). In general, the greater probabilistic movement, the greater the signal attenuation. In the tensor model, the dwi will be a function of the unweighted signal intensity, So, diffusion direction and magnitude (encoded in diffusion tensor D), the experimental timing and gradient strength (encoded in parameter b), and the direction of the gradient applied (encoded in g). For a single voxel we write
| (1) |
Here, j indexes the gradient directions. . For convenience, we define the observed data for a single voxel, X⇀obs;, as a vector with elements xi, representing dwij for j = 1,2, … m, and So.
| (2) |
D is a 3 × 3 symmetric matrix who’s Eigenvalues, λ1, λ2, and λ3, describe the three principal axis of diffusion. Herein, we consider the FA of the tensor which is the normalized standard deviation of the Eigenvalues and reflects the relative degree of orientation (i.e., 0: no orientation preference = isotropic diffusion, 1: infinite direction preference = 1−D diffusion).
| (3) |
2.2 SIMEX applied to DTI
The premise of SIMEX is simple. Bias can be understood by adding noise to data in incremental amounts and measuring the resulting contrast. The trend in the contrast with added noise should enable prediction of the contrast with ‘removed’ noise. For simplicity, the following description assumes a single DTI experiment is performed (e.g. single subject, one DTI dataset) and describes the single-voxel case, though the analysis can be extended to multiple experiments and multiple voxels.
Let a truth data set be described by zero experimental noise,
| (4) |
where σE is the standard deviation of the experimental noise. The function T mapping X⇀truth to FA determines the calculated ground truth FA value, FAtruth.
| (5) |
The observed data of an actual experiment contains experimental noise and is described by,
| (6) |
Here, Z⇀ is a vector of m+1 random drawings from a standard normal distribution. The resulting observed FA from the experiment is then,
| (7) |
In SIMEX, synthetic normally distributed noise with standard deviation ω1/2σE is added to X⇀obs, and the new simulated data is written as,
| (8) |
Where M.C. stands for ‘Monte Carlo’ (M.C. replaces ‘b’ and ω replaces ‘λ’ as defined in the SIMEX literature [8]). The simulated FA is then described by,
| (9) |
The variance of the simulated data as a function of ω is
| (10) |
A key observation is to note that at the value ω = −1, (which cannot be simulated) the variance goes to 0. For repeated Monte Carlo simulations of FAM.C.(ω), a mean value can be calculated, , and the trend approximated by fitting with a standard non-linear equation, [8],
| (11) |
The FA at zero noise can then be estimated by solving eq. 11 for = −1,
| (12) |
The bias in the data can then be estimated,
| (13) |
The true bias can also be calculated if the truth data is known,
| (14) |
3 Methods and Results
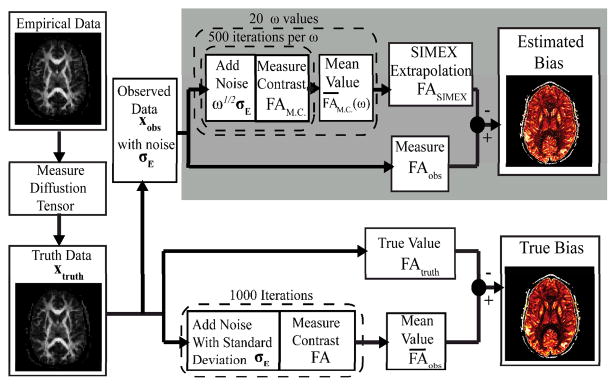
The steps used here to evaluate SIMEX on FA are summarized in the flow chart of Fig. 1 and exampled by individual voxels in Fig. 2. Unless otherwise stated, all processing and analysis was performed in Matlab 2010 (Mathworks, Natick, MA). Diffusion tensor estimates were calculated by fitting the diffusion model (eq.1) to the data using a simple LLMMSE. In all cases, noise was added to data using a Rician distribution, which is approximately Gaussian for SNR > 5:1[12].
Fig. 1.
Flow chart of steps to create SIMEX Estimated Bias and True Bias maps. Variables refer to terms defined explicitly by equations in the Theory section.
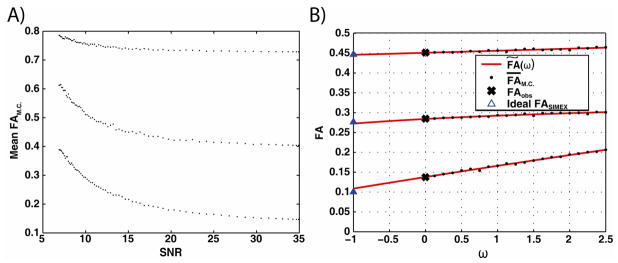
Fig. 2.
(A) Exploratory SIMEX plots of simulated FAM.C. as a function of SNR for three voxels from the simulated observed data. 1000 Monte Carlo simulations were averaged for each . The FAobs is the point at the highest SNR value (SNR = 35:1). An upward bias with decreasing SNR is observed for all three voxels. (B) FAM.C.(ω) and the extrapolation fit for three voxels in the observed data set. The x-axis for extrapolation is ω ~1/SNR and the fit is extrapolated to ω = −1. The ideal FASIMEX (blue triangles) represents the extrapolated FASIMEX value if the true bias was correctly estimated.
Empirical Data
Empirical data (KKI2009-33-DTI) was downloaded from the Multi-Modal MRI Reproducibility study available online at www.nitrc.org [13]. Full collection details are provided in the reference. Briefly, DTI data was collected using a multi-slice, single-shot, echo planar imaging (EPI) sequence. Thirty two gradient directions were used with b = 700 s/mm2 (m = 32, b = 700, eq. 1 & 2). Five So images were collected and averaged into a single So. The resulting images consisted of sixty-five transverse slices with a field of view = 212 × 212 reconstructed to 256 × 256.
Creating Noiseless Ground Truth (eq. 4)
The 32 dwi images were registered to the single So image using FLIRT affine registration (FMRIB, Oxford, UK). For spatial consistency, the resulting affine transformation matrix was applied to the gradient table. A single axial slice of interest was selected and diffusion tensor estimates (D) for that slice were calculated. Using the So image, the estimated diffusion tensor, and the given g and b as inputs to eq. 1, dwi images were simulated. The So image and the 32 noiseless dwi images make the noiseless ground truth dataset, X⇀truth.
Creating Observed Data (eq. 6)
To create the observed data, X⇀obs, noise was added to the ground truth data set such that SNR = 35:1. The signal strength was estimated from the average signal intensity of the masked So image. Noise, σE = signal/35, was added to the ground truth data to create the 32 dwi images and the single So image.
Calculating the Estimated Bias (eq. 13)
To calculate FASIMEX, first a plot of needed to be constructed. 500 Monte Carlo simulations for 20 different ω values were performed and calculated from the average. Each simulation consisted of a noise adding step to create XM.C. (eq. 8), followed by a calculation of FAM.C.. Example for three voxels are shown in Fig. 2A. was fit using the empirical non-linear equation (eq. 11). The fit equation was extrapolated to ω = −1, producing FASIMEX (Fig. 2B). The FAobs from eq. 13 was calculated directly from diffusion tensor fits to the observed data set, X⇀obs. The difference of FASIMEX and FAobs was taken and the Estimated Bias calculated.
Calculating True Bias (eq. 14)
The average of 1000 iterations was used to estimate the expected value of FAobs, . Each simulation consisted of first adding noise (SNR = 35:1) to X⇀truth followed by calculation of FA. FAtruth was calculated directly from X⇀truth. The True Bias was calculated from the difference between and FAtruth. Exploratory M.C. simulations applying SIMEX to different SNR levels of X⇀obs for a single ground truth voxel are shown in Fig. 3. The True Bias and Estimated Bias are compared in Fig. 4. The results shown in Fig. 4 were typical for multiple test trials (data not shown).
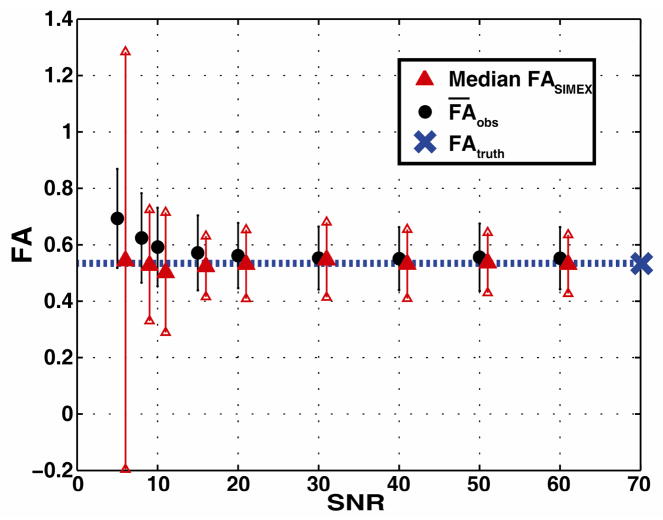
Fig. 3.
Simulated outcomes for SIMEX at several SNR values. values for each SNR value (circles) were calculated starting from a voxel in the simulated truth data. The values are shown with the standard deviation (error bars) from 1000 simulations run at each SNR value. 100 observations at each SNR value were randomly chosen for SIMEX. The median FASIMEX values (triangles) are shown with the standard deviation(error bars). The median value was chosen due to the decreased robustness of only 100 simulations.
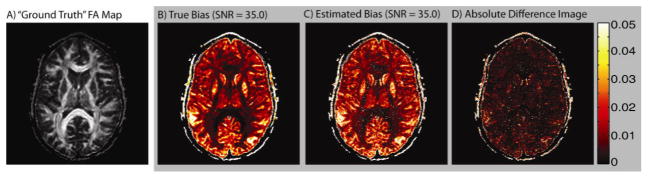
Fig. 4.
(A) The ‘ground truth’ FA map (FAtruth) of the slice selected for data analysis. (B) The True Bias map for an SNR of 35:1 is compared to (C) the absolute value of the Estimated Bias at SNR = 35:1. The absolute value difference map was calculated from the raw bias data (not the difference in the absolute values). For a significant majority of voxels the True Bias is positive and the Estimated Bias is greater than the True Bias.
4 Discussion
Potential systematic differences in distributional properties of tensor-derived contrasts pose serious hazards for statistical analysis. Understanding “statistical artifacts” in DTI has long been an essential challenge in proper interpretation of DTI contrasts [14]. Routine approaches for interpreting contrasts (e.g., regions of interest, voxel-based morphometry, tract-based spatial statistics) hinge upon the assumption of equivalent (or zero) bias between populations. If this assumption is violated then the inferences (and p-values) may be suspect. In any empirical experiment, there will be variations in bias due to subject characteristics, protocols, scanner stability, hardware/software changes, etc. Without a method to estimate bias in measures derived from individual empirical datasets, evaluation of any change requires an extensive simulation study based on a range of potential anatomical models.
With the proposed SIMEX approach, it is possible to estimate bias in contrasts on a voxel-by-voxel level given the empirical data. SIMEX proved a sensitive method for detecting FA bias as evidenced by its success on the high SNR (and therefore relatively low bias) dataset used here (Fig. 4). Although bias may be negligible for some individual cases at high SNR values (such as exampled for high SNR values in Fig. 3), bias may become increasingly important at lower SNR values or in hypothesis test settings where even a small bias can significantly inflate the Type-I error rate. To that end, the individual voxel test case (Fig. 3) suggests SIMEX to be a promising technique for empirical data collected at lower SNR levels (stable median FASIMEX values) as well as a reliable method for repeated use on individual datasets within grouped data (stable error bars). The instability of the SIMEX estimate for SNR ≤ 10:1 (Fig. 3) is likely to be a result of the non-monotonic behavior of bias at extremely low SNR values [6].
We note that at these early stages, we are not specifically recommending using the estimated bias measures to correct for voxel-wise bias. Optimization of user input SIMEX parameters (e.g. the approximation function in eq. 11 or the number of Monte Carlo iterations) and repeat testing on empirical data of various SNR levels is underway. Refinements may help decrease the variability in the SIMEX estimate (Fig. 3 red error bars) and mediate some of the isolated outliers seen in Fig. 4D. Additionally, methods for constructing error estimates of the measured SIMEX bias exist and have yet to be tested in the context of DTI [7, 8]. With continued future refinement and careful incorporation of robust/advanced tensor estimators it may be possible to produce unbiased contrast estimates without the disadvantages of direct maximum likelihood methods [15, 16].
In summary, a method for direct, empirical assessment of bias in tensor derived contrasts opens many possibilities for useful statistical assessments and improved analysis approaches. Further investigation is warranted into bias estimation with other tensor contrasts, using advanced tensor fitting methods, and examining quantitative tissue models beyond the tensor formulation.
Acknowledgments
This work was supported in part by NIH/NINDS 1R01NS056307 and NIH/NINDS 1R21NS064534.
References
- 1.Le Bihan D, van Zijl P. From the diffusion coefficient to the diffusion tensor. NMR Biomed. 2002;15:431–434. doi: 10.1002/nbm.798. [DOI] [PubMed] [Google Scholar]
- 2.Farrell JA, Landman BA, Jones CK, Smith SA, Prince JL, van Zijl PC, Mori S. Effects of signal-to-noise ratio on the accuracy and reproducibility of diffusion tensor imaging-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5 T. J Magn Reson Imaging. 2007;26:756–767. doi: 10.1002/jmri.21053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landman BA, Farrell JA, Jones CK, Smith SA, Prince JL, Mori S. Effects of diffusion weighting schemes on the reproducibility of DTI-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5T. Neuroimage. 2007;36:1123–1138. doi: 10.1016/j.neuroimage.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 4.Jones DK, Basser PJ. “Squashing peanuts and smashing pumpkins”: How noise distorts diffusion-weighted MR data. Magnetic Resonance in Medicine. 2004;52:979–993. doi: 10.1002/mrm.20283. [DOI] [PubMed] [Google Scholar]
- 5.Whitcher B, Tuch DS, Wisco JJ, Sorensen AG, Wang L. Using the wild bootstrap to quantify uncertainty in diffusion tensor imaging. Hum Brain Mapp. 2008;29:346–362. doi: 10.1002/hbm.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landman BA, Farrell JA, Huang H, Prince JL, Mori S. Diffusion tensor imaging at low SNR: nonmonotonic behaviors of tensor contrasts. Magn Reson Imaging. 2008 doi: 10.1016/j.mri.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll R, Kuchenho H, Lombard F, Stefanski L. Asymptotics for the SIMEX estimator in structural measurement error models. Journal of the American Statistical Association. 1996;91:242–250. [Google Scholar]
- 8.Cook J, Stefanski L. Simulation-extrapolation estimation in parametric measurement error models. Journal of the American Statistical Association. 1994;89 [Google Scholar]
- 9.Landman BA, Bazin PL, Smith SA, Prince JL. Robust estimation of spatially variable noise fields. Magn Reson Med. 2009;62:500–509. doi: 10.1002/mrm.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajan J, Poot D, Juntu J, Sijbers J. Noise measurement from magnitude MRI using local estimates of variance and skewness. Phys Med Biol. 2010;55:N441–449. doi: 10.1088/0031-9155/55/16/N02. [DOI] [PubMed] [Google Scholar]
- 11.Aja-Fernandez S, Tristan-Vega A, Alberola-Lopez C. Noise estimation in single- and multiple-coil magnetic resonance data based on statistical models. Magn Reson Imaging. 2009;27:1397–1409. doi: 10.1016/j.mri.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 12.Gudbjartsson H, Patz S. The Rician distribution of noisy MRI data. Magn Reson Med. 1995;34:910–914. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landman B, Huang A, Gifford A, Vikram D, Lim I, Farrell J, Bogovic J, Hua J, Chen M, Jarso S, Smith S, Joel S, Mori S, Pekar J, Barker P, Prince J, van Zijl P. Multi-Parametric Neuroimaging Reproducibility: A 3T Resource Study. Neuroimage. 2011;4:2854–2866. doi: 10.1016/j.neuroimage.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basser PJ, Pajevic S. Statistical artifacts in diffusion tensor MRI (DT-MRI) caused by background noise. Magn Reson Med. 2000;44:41–50. doi: 10.1002/1522-2594(200007)44:1<41::aid-mrm8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 15.Landman BA, Bazin P-L, Prince JL. Robust Diffusion Tensor Estimation by Maximizing Rician Likelihood. International Society for Magnetic Resonance in Medicine. 2007 doi: 10.1109/iccv.2007.4409140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson JL. Maximum a posteriori estimation of diffusion tensor parameters using a Rician noise model: why, how and but. Neuroimage. 2008;42:1340–1356. doi: 10.1016/j.neuroimage.2008.05.053. [DOI] [PubMed] [Google Scholar]