Abstract
Mosaic trisomy 17 is rare with only 28 cases reported and the clinical presentation is highly variable. The diagnosis is most commonly made by prenatal karyotype and in most cases is followed by a normal postnatal karyotype on blood lymphocytes. We present two cases of mosaic trisomy 17 diagnosed prenatally, with follow up in multiple tissues at birth. In the first case, trisomy 17 was identified in all amniocytes, and at birth standard results of chromosome analysis in peripheral blood were normal, but mosaic trisomy 17 was identified (50–75%) in skin fibroblasts by genome-wide SNP array analysis. This patient presented with minor anomalies, congenital heart disease, asymmetry, intestinal malrotation and died on day 9 of life. In the second patient amniocentesis after ultrasound finding of tetralogy of Fallot showed mosaic trisomy 17. Postnatally, results of a SNP array were normal in blood, buccal mucosa and skin. It is possible that the cardiac defect is related to trisomy 17 in key tissues during heart development, although at birth the aneuploidy could not be identified in tissues that are routinely analyzed for diagnosis. These cases add to our understanding of mosaic trisomy 17, highlighting the failure to diagnose this aneuploidy in peripheral blood.
Keywords: Mosaic trisomy 17, SNP microarray analysis, tissue specific mosaicism
INTRODUCTION
Aneuploidy is a common finding in spontaneously aborted fetuses and is less common among liveborns [Hassold, 1982]. The frequency of monosomy and trisomy for each of the chromosomes has been studied in abortuses and liveborns. Only full trisomies for autosomes 13, 18, and 21 are considered survivable to birth. Individuals with trisomy for chromosomes 8, 9, 14, 20 and 22 are usually survivable in the mosaic form while trisomy (mosaic or full) of the other autosomes is virtually never seen in late gestation or at birth. Full trisomy 17 has been seen in 0.1% of spontaneously aborted fetuses [Hassold et al., 1996], but never in a live born infant, consistent with lethality in utero. Mosaic trisomy 17 (mosaic T17) has been identified but is among the rarest of the aneuploidies identified with only 28 cases reported to date [Hassold, 1982; Kalousek et al., 1987; Wilson et al., 1989; Welborn et al., 1990; Djalali et al., 1991; Butler et al., 1996; Shaffer et al., 1996; Hsu et al., 1997; Djalali et al., 1998; Butler, 1999; Genuardi et al., 1999; Lesca et al., 1999; Nassar et al., 2000; Collado et al., 2003; Terhal et al., 2004; Abrams et al., 2005; Utermann et al., 2006; Witters et al., 2007; Witters et al., 2008]. Most of these cases were detected prenatally during CVS or at amniocentesis; all of the cases had normal karyotypes postnatally in peripheral blood lymphocytes. Those with no trisomy 17 in any tissue studied postnatally were apparently normal at birth, while the remaining patients had multiple congenital anomalies and confirmed mosaic trisomy in skin and/or other tissues [Shaffer et al., 1996; Genuardi et al., 1999; Lesca et al., 1999; Terhal et al., 2004; Utermann et al., 2006; Witters et al., 2007; Witters et al., 2008]. The proportion of trisomy 17 varied from patient to patient and from tissue to tissue accompanied by a wide range of phenotypes. Interestingly, eight patients with mosaic trisomy 17 in skin or other tissue also had normal chromosomes in lymphocytes suggesting that trisomy 17 is not tolerated in blood or it is lost during the culturing process.
Recent advances in microarray technologies have increased the sensitivity of detecting rare mosaic trisomies by allowing direct DNA analysis from uncultured samples [Gunderson et al., 2005; Shen et al., 2005]. In addition to detecting lower levels of mosaicism, the genotyping information obtained with single nucleotide polymorphisms (SNPs) allows for analysis of genotypes which can confirm biparental inheritance, provide information about the mechanism of trisomy formation, and identify uniparental disomy that may result from trisomy rescue[Conlin et al., 2010]. Since cross-over events occur during meiosis I, prior to chromosome separation of homologs, genotyping information can be used to identify the timing of the non-disjunction event by inspection of genotypes for evidence of altered allelic ratios.
Here we report on post-natal SNP microarray analysis on several tissues from 2 patients identified prenatally with trisomy17. The first case demonstrated full trisomy 17 on amniocentesis, mosaic trisomy 17 in skin fibroblasts and was born with multiple congenital anomalies. The second case demonstrated mosaic trisomy 17 on amniocentesis, yet no evidence for trisomy 17 was found in three tissues sampled post-natally.
CLINICAL REPORT
Patient 1
The 36-year-old mother presented for follow-up evaluation at 25 weeks of gestation after a finding of trisomy 17 on amniocentesis. Trisomy 17 was detected in all 15 colonies from three cultures. Ultrasound evaluation (at 25 weeks and 4 days) demonstrated relatively short long bones (right humerus consistent with a gestational age of 15 weeks 5 days and right tibia and fibula consistent with gestational age less than 15 weeks), hypertelorism, micrognathia, hypoplasia of the inferior vermis, right talipes with toe anomalies, absent fourth sacral vertebra and right kidney, and a two vessel cord. Fetal echocardiogram was consistent with a single ventricle and truncus arteriosus, type 1A. There were no previous pregnancy losses and the family history was unremarkable except for 2 maternal brothers with mitral valve prolapse.
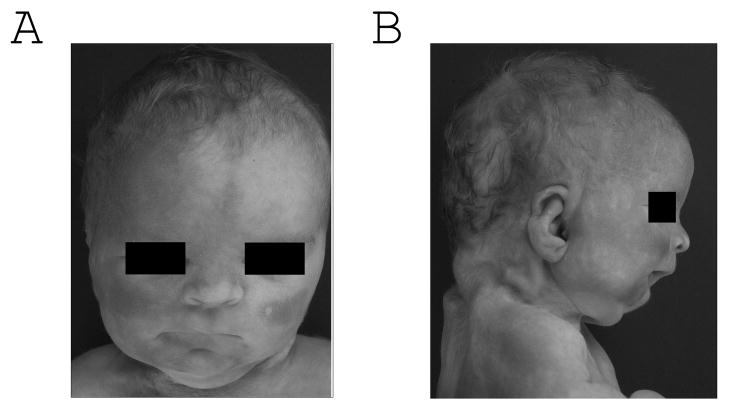
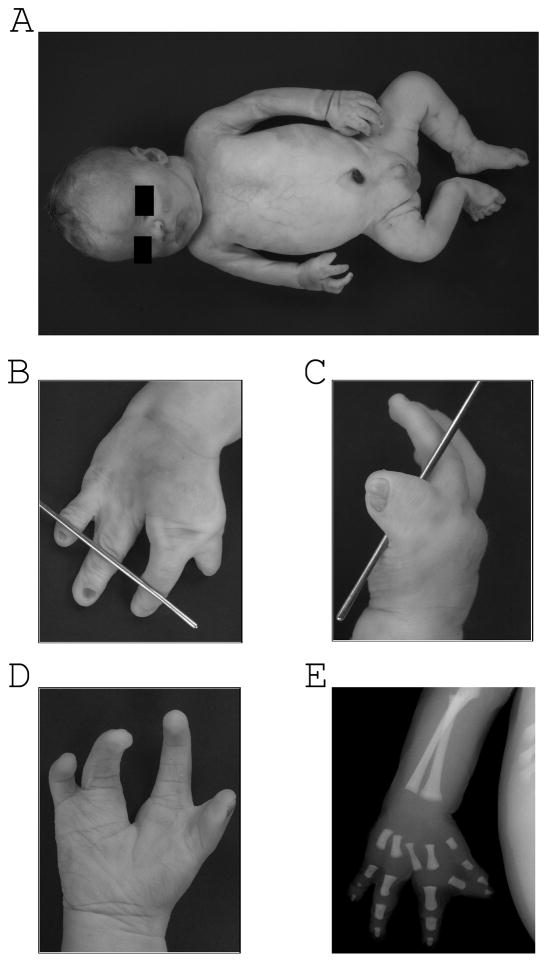
The infant was delivered at 36 weeks and survived for 9 days. After birth, cytogenetic and molecular analyses were carried out at an outside lab to detect mosaic trisomy 17. No abnormalities were found by conventional cytogenetics, carried out on peripheral blood lymphocytes and a BAC array on DNA extracted from buccal mucosa. At autopsy the cardiac abnormality was found to be a single ventricle with a truncus arteriosus, communis type I and a 4-cuspid valve. Brain findings were consistent with hypoplasia of the inferior cerebellar vermis. A right pelvic kidney was found with ipsilateral absence of the vas deferens and hypoplasia of seminal vesicle and ectopic ureterovesical orifices. Asymmetric anomalies were also noted in the thyroid gland (right lobe smaller than left) and salivary glands (right submandibular gland smaller than left). In addition, intestinal malrotation was identified with a Meckel’s diverticulum and an ileal duplication cyst. Bilateral unilobar lungs were also identified. Facial features included: frontal bossing, deep set eyes, hypertelorism and micrognathia (Fig 1). The limbs were asymmetric with the right side being smaller than the left, right foot talipes, and right hand abnormalities (Fig. 2). Skin samples from the right and left arms were collected for SNP array analysis.
Figure 1.
A and B. Patient 1. Note hypertelorism, deep set eyes, micrognathia, and broad forehead.
Figure 2.
A. Patient 1 body asymmetry. B, C, D. Findings of split hand on the right and the corresponding radiograph E.
Patient 2
A 31-year-old G2P1 woman presented following an ultrasound study at 21 weeks of gestation. The ultrasound demonstrated tetralogy of Fallot (TOF). On echo he was also found to have moderate pulmonary vein stenosis. Cells from a subsequent amniocentesis studied in a commercial laboratory showed trisomy 17 in 4/17 cells (23%). At birth the patient’s head circumference was at the 25th, length and weight at the 50th centile. The infant had three fontanelles, a normal appearance with exception of mild micrognathia, a wide 1–2 sandal gap on the right, mild clinodactyly of the 5th digit, and normal dermatoglyphics (ulnar loops). Ears were normally formed and placed, inter-nipple distance to chest circumference ratio was 40%. He was found to have normal male genitalia with bilaterally descended testes. On MRI he was found to have ventriculomegaly consistent with hydrocephalus. The child is now 1 year of age and post surgical correction of TOF and ventricular-peritoneal (VP) shunt placement. Psychomotor development is normal thus far; however, there is a concern for mild speech delay.
MATERIAL AND METHODS
SNP microarray karyotyping
DNA was extracted directly from uncultured peripheral blood, buccal mucosa as well as from cultured fibroblasts in the standard manner. The quality of the DNA was monitored by analysis of OD260/OD280 and OD260/OD230 ratios. OD260/OD280 values were between 1.8 and 2.0 and OD260/OD230 ratios were > 2.0. Samples were genotyped using the Illumina Quad610 array and the Illumina BeadStation. The samples were whole genome-amplified, fragmented, hybridized, fluorescently tagged and scanned, as per standard protocols [Gunderson et al., 2005; Shen et al., 2005]. Quality was monitored using call rate (> 98%) and standard deviation of the LogR Ratio (< 0.3). All copy number alterations were visually inspected and manually detected using BeadStudio software, in combination with a CNV detection tool [Gai et al., 2010]. Calculations of mosaicism were performed as described previously [Conlin et al., 2010].
RESULTS
Patient 1
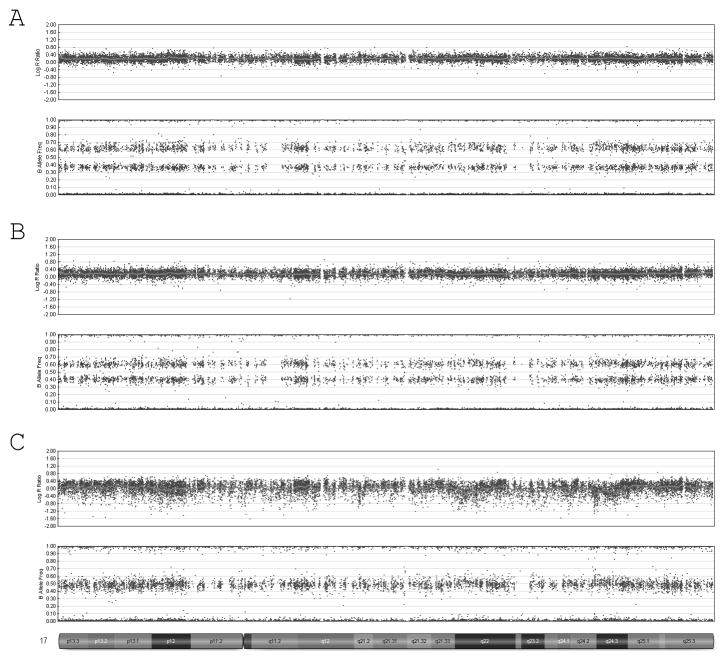
Skin fibroblasts were obtained from the right and left side of the patient and DNA was extracted for analysis on the genome wide SNP array in our laboratory. Both samples demonstrated an elevated logR ratio and split in the B-allele frequency consistent with mosaic trisomy 17 (Fig. 3A,B). The logR ratio and B-allele frequencies for each sample were then used to estimate the level of mosaicism with in-house algorithms. These algorithms look at the average of the logR intensity for a given chromosome as well as the distribution of B-allele frequencies then correlate them to standard curves generated for control mosaics. Consistent with a mosaic trisomy for both samples, the average logR ratio of all probes along chromosome 17 was between 0 and 0.5. The spread in the B-allele frequencies was slightly more informative in determining the mosaic percentages and showed the sample from the right side had 75% mosaic trisomy 17 while the sample from the left side had a lower level near 50%. The genotyping pattern in each sample was also analyzed and found to be consistent with only two haplotypes present in the trisomic cell line for chromosome 17. This suggests that the trisomy 17 resulted from a post-zygotic nondisjunction or a meiosis II nondisjunction in the absence of crossing over [Conlin et al., 2010].
Figure 3.
SNP microarray results: SNP array results for case 1 (A and B) showing 75% trisomy 17 in right sided fibroblast (A) and 50% trisomy 17 in left sided fibroblasts (B). Mosaic trisomy 17 could not be detected in fibroblasts from case 2 (C).
Patient 2
Postnatally, DNA was extracted from skin, buccal mucosa and peripheral blood, and analyzed using a genome-wide SNP array (Illumina Quad610 beadchip). Analysis of the LogR ratio and B-allele frequency from all three samples from this child was consistent with two copies of chromosome 17, with no evidence for trisomy mosaicism. The genotypes at each locus were compared between the samples from all three tissues, and all tissues had identical genotypes, with a pattern of alleles consistent with biparental inheritance, with no evidence for either uniparental disomy, or altered combinations of chromosome 17 alleles secondary to resolution of the mosaicism (Fig. 3C).
DISCUSSION
We present two cases of mosaic trisomy 17 diagnosed at amniocentesis, with very different clinical and cytogenetic outcomes observed postnatally (Table I). In case 1, amniocentesis was carried out due to an increased risk identified on maternal serum screening and complete trisomy 17 was diagnosed at amniocentesis. Postnatal follow up studies showed 50–75% trisomy 17 in uncultured skin when analyzed by a genome-wide SNP array. The patient had multiple anomalies and died at 9 days of age. In case 2, amniocentesis was carried out after tetralogy of Fallot was diagnosed prenatally and cytogenetic analysis of cells cultured from amniotic fluid demonstrated mosaic trisomy 17 in 4/17 cells (23%). Postnatal studies documented a normal genome-wide SNP array in DNA from skin, buccal mucosa and peripheral blood. We estimate that this array will rule out mosaicism as low as 5–10% [Conlin et al., 2010]. Clinically, this patient had only mild micrognathia and a 1–2 sandal gap on one foot, in addition to the tetralogy of Fallot. Development is normal at age 3 months.
Table I.
Overview of cytogenetic findings
| Amniocentesis | Buccal | Lymphocytes/cordocentesis | Fibroblasts | |
|---|---|---|---|---|
| Case 1 | 47, XX, +17[15] | Normal BAC array | 46, XX | Right side: ~75% +17 Left side: ~50% +17 |
| Case 2 | 47, XY, +17 [4]/46, XY[13] | Normal SNP array | Normal SNP array | Normal SNP Array |
Mosaic T17 is among the rarer trisomies seen in liveborn infants. Most of the reported cases were initially identified through prenatal amniocentesis or CVS and resulted in normal, healthy infants suggesting that the trisomy was confined to extra-embryonic tissues. The nine cases with confirmed mosaic trisomy 17 in other tissues all presented with congenital anomalies, although the phenotypes reported were variable. Considering that different mosaic proportions of trisomy 17 were found in various tissues, the clinical variability may be due to tissue-specific mosaicism. We add two additional cases of mosaic trisomy 17 that were identified in amniocytes and presented with variable manifestations.
The first case presented here showed a number of features consistent with the physical findings described in patients diagnosed with mosaic trisomy 17 (summarized in Table II). These findings include facial dysmorphia (broad forehead, hypertelorism, and micro/retrognathia), extremity asymmetry, congenital heart defects, and intestinal malrotation. Consistent with the presence of various malformations, mosaic trisomy 17 appeared to be widely distributed and was confirmed in skin fibroblasts. Using the genotyping information we were able to confirm that only two haplotypes were present at each of the loci on chromosome 17. The absence of a third haplotype at any of the loci allows us to rule out a meiosis I non-disjunction event as the cause of the trisomy. Given that the trisomy could only result from a meiosis II nondisjunction in the absence of crossing over and there are several hot spots for crossing over on chromosome 17[Cheung et al., 2007], the error more likely occurred post-zygotically. An error in mitosis also rules out the possibility of uniparental disomy in the “normal” cells since there was no trisomy rescue event. Several of the patients in the literature also show evidence of post-zygotic non-disjunction (Table II, Patients 1, 4 and 7) [Shaffer et al., 1996; Terhal et al., 2004; Utermann et al., 2006]. Each of these patients had confirmed mosaic trisomy 17 in skin fibroblasts with similar clinical findings including asymmetry of the limbs. We were able to study tissue from both sides in Patient 1 demonstrating different percentages of trisomy 17 mosaicism (50% - left and 75% - right) which presumably explains the asymmetry, although it may also represent sampling bias.
Table II.
Findings in the eight cases in the literature with physical descriptions and amniocentesis results and our two patients.
Summary of Physical Findings
| Shaffer et al. (1996) | Lesca et al. (1999) | Genuardi et al. (1999) case 3 | Terhal et al. (2004) | Utermann et al. (2006) case 1 | Utermann et al. (2006) case 3 | Utermann et al. (2006) case 4 | Witters et al. (2007) | Case 1 | Case 2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Amniocentesis | n.t. | 14% | 8% | 63 – 67% | 30% | 40% | ~33% | 13 – 50% | 100% | 23% |
| Buccal | n.t. | n.t. | n.t. | n.t. | n.t. | <1% | 0% | 4% | 0% | 0% |
| Lymphocytes/cordocentesis | 0% | 0% | n.t. | 0% | n.t. | 0% | 0% | 0% | 0% | 0% |
| Bladder cells | n.t. | n.t. | n.t. | n.t. | n.t. | n.t. | 66% | 16% | n.t. | n.t. |
| Fibroblasts | 22% | 76–96% | 5% | 25–72% | 9% | 0–70% | 60% | n.t. | 50–75% | 0% |
| Ventriculomegaly | U | mild | U | U | U | U | U | U | U | U |
| Cerebellar abnormalities | U | U | U | Y | Y | U | Y | U | Y | U |
| Facial asymmetry | U | U | U | U | U | Y | U | U | Y | U |
| Single palmar crease | U | Y | U | U | U | Y | U | U | Y | U |
| Hearing Loss | U | Y (unilateral) | U | U | U | U | Y | U | U | U |
| IUGR | U | U | U | U | Y | U | U | U | U | U |
| Hypotonia | U | Y | U | U | U | Imp | U | U | U | U |
| Developmental delay/Intellectual Disability | U | U | U | Y | U | Y | Y | U | U | U |
| Post-natal growth retardation | U | U | U | Y | U | Y | Y | U | U | U |
| Asymmetry lower limbs | U | U | U | Y | U | U | Y | U | Y | U |
| Asymmetry upper limbs | U | U | U | U | U | Post-axial polydactyly | U | U | Y | U |
| Intestinal malrotation | U | U | U | U | U | U | Y | U | Y | U |
| Retrognatia/Micrognathia | U | Y | U | U | Y | Y | Y | U | Y | Y |
| Broad forehead | U | U | U | Y | U | U | Y | U | Y | U |
| Structural heart defect | U | U | U | U | VSD | VSD, ASD | AV stenosis; hypoplastic aortic isthmus | U | Single ventricle, truncus arteriosum | TOF |
| Electrical heart abnormality | U | Long QT | U | U | U | U | Super-ventricular tachycardia | U | U | U |
| Two vessel cord | U | U | U | U | U | Y | U | U | Y | U |
| Hypertelorism | U | U | U | Y | Y | U | U | U | Y | U |
| Age at report | 8 y 8 m | 2.5 mos | Term (21w) | 7 y 2 m | Term (16w) | 2 y | 9 y | 18 mos | 9 d | 3 mos |
| Origin of nondisjunction | Paternal post-zygotic | n.d. | n.d. | Maternal Post-zygotic | Maternal Meiosis I | n.d | Paternal post-zygotic | n.d. | Post-zygotic | n.d. |
n.d.: not determined, n.t.: not tested, U: not reported, Y: Yes, Imp: improving, VSD: Ventricular septal defect, ASD: Atrial septal defect, TOF: Tetralogy of Fallot, Term: terminated
It is noteworthy that standard postnatal testing did not show a genomic alteration although mosaicism had been diagnosed by amniocentesis in the second patient who presented with a severe congenital heart defect (TOF). The patient was generally non-dysmorphic with normal neonatal growth dermatoglyphics, but had three fontanelles, mild micrognathia, and a unilateral 1–2 sandal gap. This patient represents the first report of an infant with clinical manifestations yet no detectable trisomy 17 on postnatally analyzed tissue. Since the SNP array is able to rule out mosaic trisomy as low as 5–10%, the data suggests that either really low levels of trisomy are present, the trisomy may reside in other tissues that were not tested (cardiac tissue) or the heart defect is unrelated to the prenatal finding of mosaic trisomy 17.
Although both cases described here had amniocentesis finding consistent with full or mosaic trisomy 17, as well as congenital heart defects, only one had SNP findings consistent with this diagnosis. Both cases had normal karyotypes from lymphocytes, as have all cases reported to date, again confirming the recommendation that follow-up analysis be performed on skin fibroblasts for infants with a pre-natal diagnosis of trisomy 17. Since the SNP array is performed on DNA extracted from uncultured blood, these cases also suggest that the absence of the trisomic cell line in the blood is not a culture artifact and is, in fact, due to a growth disadvantage in vivo. Several other mosaic cytogenetic abnormalities such as mosaic trisomy 20 and Pallister-Killian syndrome have also shown deficient growth of the abnormal lymphocytes compared to the normal cells.
Both a normal outcome and multiple congenital anomalies (MCA) are possible with a pre-natal finding of mosaic trisomy 17, making it difficult to predict the outcome in the absence of other clinical findings. From the cases reported in the literature, there is no correlation between the proportion of cells containing trisomy 17 at amniocentesis and the presence of trisomy 17 detected postnatally (Table II). Phenotypic correlation is further complicated by the uncertainty of specific proportions of trisomy 17 in various tissues. While the clinical outcome of a prenatal finding of trisomy 17 is difficult to predict, if there is a high level of suspicion, especially based on physical findings or amniocentesis results, analysis of fibroblasts or direct DNA assays are necessary for a more definitive chromosomal diagnosis.
Acknowledgments
The authors would like to thank the patients and their families for their participation. K.A.C. and L.C. are supported by a Medical Genetics Research Training Grant, 5-T32-GM-008638-11, to the University of Pennsylvania. R.D. was supported by Award number T32NS007413 from the National Institutes of Neurological Disorders and Stroke (NINDS). The content is the sole responsibility of the authors and does not necessarily represent the official views of the NINDS of the National Institutes of Health. We are grateful for support of these studies from The Children’s Hospital of Philadelphia.
References
- Abrams DJ, Augustyn AM, Geier MR. Prenatally diagnosed mosaic trisomy 17: a case report with two-year follow-up. Prenat Diagn. 2005;25:968–969. doi: 10.1002/pd.1236. [DOI] [PubMed] [Google Scholar]
- Butler MG. Trisomy 17 mosaicism in a four-year seven-month-old white girl: follow-up report. Prenat Diagn. 1999;19:689. doi: 10.1002/(sici)1097-0223(199907)19:7<689::aid-pd592>3.0.co;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Neu RL, Mitchell K. Trisomy 17 detected in amniotic fluid cells but not in newborn infant. Am J Med Genet. 1996;65:247–248. doi: 10.1002/ajmg.1320650402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Burdick JT, Hirschmann D, Morley M. Polymorphic variation in human meiotic recombination. Am J Hum Genet. 2007;80:526–530. doi: 10.1086/512131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado FK, Fisher AJ, Bombard AT. Counseling patients with trisomy 17 mosaicism found at genetic amniocentesis. Prenat Diagn. 2003;23:948–950. doi: 10.1002/pd.714. [DOI] [PubMed] [Google Scholar]
- Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, Zackai EH, Deardorff MA, Krantz ID, Hakonarson H, Spinner NB. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19:1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djalali M, Barbi G, Grab D. Mosaic trisomy 17 in amniotic fluid cells not confirmed in the newborn. Prenat Diagn. 1991;11:399–402. doi: 10.1002/pd.1970110610. [DOI] [PubMed] [Google Scholar]
- Djalali M, Barbi G, Mueller-Navia J, Schneider M, Tettenborn U, Trautmann U, Ulmer R, Wolf M, Vogel W. Further observations of true mosaic trisomy 17 ascertained in amniotic fluid cell cultures. Prenat Diagn. 1998;18:1191–1194. doi: 10.1002/(sici)1097-0223(199811)18:11<1191::aid-pd417>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Gai X, Perin JC, Murphy K, O’Hara R, D’Arcy M, Wenocur A, Xie HM, Rappaport EF, Shaikh TH, White PS. CNV Workshop: an integrated platform for high-throughput copy number variation discovery and clinical diagnostics. BMC Bioinformatics. 2010;11:74. doi: 10.1186/1471-2105-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuardi M, Tozzi C, Pomponi MG, Stagni ML, Della Monica M, Scarano G, Calvieri F, Torrisi L, Neri G. Mosaic trisomy 17 in amniocytes: phenotypic outcome, tissue distribution, and uniparental disomy studies. Eur J Hum Genet. 1999;7:421–426. doi: 10.1038/sj.ejhg.5200333. [DOI] [PubMed] [Google Scholar]
- Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- Hassold T. Mosaic trisomies in human spontaneous abortions. Hum Genet. 1982;61:31–5. doi: 10.1007/BF00291327. [DOI] [PubMed] [Google Scholar]
- Hassold T, Abruzzo M, Adkins K, Griffin D, Merrill M, Millie E, Saker D, Shen J, Zaragoza M. Human aneuploidy: incidence, origin, and etiology. Environ Mol Mutagen. 1996;28:167–175. doi: 10.1002/(SICI)1098-2280(1996)28:3<167::AID-EM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Hsu LY, Yu MT, Neu RL, Van Dyke DL, Benn PA, Bradshaw CL, Shaffer LG, Higgins RR, Khodr GS, Morton CC, Wang H, Brothman AR, Chadwick D, Disteche CM, Jenkins LS, Kalousek DK, Pantzar TJ, Wyatt P. Rare trisomy mosaicism diagnosed in amniocytes, involving an autosome other than chromosomes 13, 18, 20, and 21: karyotype/phenotype correlations. Prenat Diagn. 1997;17:201–242. doi: 10.1002/(sici)1097-0223(199703)17:3<201::aid-pd56>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kalousek DK, Dill FJ, Pantzar T, McGillivray BC, Yong SL, Wilson RD. Confined chorionic mosaicism in prenatal diagnosis. Hum Genet. 1987;77:163–167. doi: 10.1007/BF00272385. [DOI] [PubMed] [Google Scholar]
- Lesca G, Boggio D, Bellec V, Magaud JP, Till M. Trisomy 17 mosaicism in amniotic fluid cells not found at birth in blood but present in skin fibroblasts. Prenat Diagn. 1999;19:263–265. doi: 10.1002/(sici)1097-0223(199903)19:3<263::aid-pd506>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Nassar AH, Chakhtoura N, Martin D. Update on prenatal diagnosis of true mosaic trisomy 17 in amniocyte cultures. Prenat Diagn. 2000;20:521–522. doi: 10.1002/1097-0223(200006)20:6<521::aid-pd843>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, McCaskill C, Hersh JH, Greenberg F, Lupski JR. A clinical and molecular study of mosaicism for trisomy 17. Hum Genet. 1996;97:69–72. doi: 10.1007/BF00218835. [DOI] [PubMed] [Google Scholar]
- Shen R, Fan JB, Campbell D, Chang W, Chen J, Doucet D, Yeakley J, Bibikova M, Wickham Garcia E, McBride C, Steemers F, Garcia F, Kermani BG, Gunderson K, Oliphant A. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Terhal P, Sakkers R, Hochstenbach R, Madan K, Rabelink G, Sinke R, Giltay J. Cerebellar hypoplasia, zonular cataract, and peripheral neuropathy in trisomy 17 mosaicism. Am J Med Genet A. 2004;130:410–414. doi: 10.1002/ajmg.a.30124. [DOI] [PubMed] [Google Scholar]
- Utermann B, Riegel M, Leistritz D, Karall T, Wisser J, Meisner L, Fauth C, Baldinger R, Johnson J, Erdel M, Taralczak M, Pauli RM, Baumer A, Schinzel A, Kotzot D. Pre- and postnatal findings in trisomy 17 mosaicism. Am J Med Genet A. 2006;140:1628–1636. doi: 10.1002/ajmg.a.31319. [DOI] [PubMed] [Google Scholar]
- Welborn JL, Lewis JP. Analysis of mosaic states in amniotic fluid using the in-situ colony technique. Clin Genet. 1990;38:14–20. doi: 10.1111/j.1399-0004.1990.tb03542.x. [DOI] [PubMed] [Google Scholar]
- Wilson MG, Lin MS, Fujimoto A, Herbert W, Kaplan FM. Chromosome mosaicism in 6,000 amniocenteses. Am J Med Genet. 1989;32:506–513. doi: 10.1002/ajmg.1320320417. [DOI] [PubMed] [Google Scholar]
- Witters I, Cannie M, Fryns JP. Prenatal diagnosis of trisomy 17 mosaicism. Prenat Diagn. 2007;27:677–678. doi: 10.1002/pd.1749. [DOI] [PubMed] [Google Scholar]
- Witters I, Fryns JP. Follow-up of a child with trisomy 17 mosaicism. Prenat Diagn. 2008;28:1080. doi: 10.1002/pd.2090. [DOI] [PubMed] [Google Scholar]