Abstract
Microcalcification is a hallmark of breast cancer and a key diagnostic feature for mammography. We recently described the first robust animal model of breast cancer microcalcification. In this study, we hypothesized that high-resolution computed tomography (CT) could potentially detect the genesis of a single microcalcification in vivo and quantify its growth over time. Using a commercial CT scanner, we systematically optimized acquisition and reconstruction parameters. Two ray-tracing image reconstruction algorithms were tested, a voxel-driven “fast” cone beam algorithm (FCBA) and a detector-driven “exact” cone beam algorithm (ECBA). By optimizing acquisition and reconstruction parameters, we were able to achieve a resolution of 104 µm full-width at half maximum (FWHM). At an optimal detector sampling frequency, ECBA provided a 28 µm (21%) FWHM improvement in resolution over FCBA. In vitro, we were able to image a single 300 µm by 100 µm hydroxyapatite crystal. In a syngeneic rat model of breast cancer, we were able to detect the genesis of a single microcalcification in vivo and follow its growth longitudinally over weeks. Taken together, this study provides an in vivo “gold standard” for the development of calcification-specific contrast agents and a model system for studying the mechanism of breast cancer microcalcification.
Keywords: Breast Cancer, Microcalcifications, Hydroxyapatite, Micro-CT, Reconstruction Algorithms
INTRODUCTION
Breast cancer is a common malignancy, accounting for 26% of all new cancer cases in women.1 The 192,370 new cases of breast cancer diagnosed in the US in 2009 will ultimately result in an estimated 40,170 deaths.1 Since survival is a strong function of disease stage at diagnosis, early detection is of paramount importance.
Despite multiple technical limitations, x-ray mammography presently remains the screening approach of choice. A key diagnostic feature for mammography, seen in 30% to 50% of women with biopsy-confirmed breast cancer, is microcalcification.2 Whereas benign calcifications are typically composed of calcium oxalate, malignant calcifications are typically composed of hydroxyapatite (HA) deposited in specific patterns.3 HA microcalcifications range in size from 100 µm single crystals to clusters up to centimeters in diameter.4
To overcome the limitations of mammography, our group is developing calcification-specific diagnostic agents that have high sensitivity and high specificity for the HA calcium salt over all other calcium salts present in the human body.5–7 Since this approach requires a robust and reproducible animal model, we have created three different models of HA breast cancer microcalficiation.6,8,9 The newest model requires only a single intraperitoneal (IP) injection of recombinant bone morphogenetic protein-2 (rBMP-2) at four days after tumor cell implantation, and produces microcalcifications in 100% of animals in a dose- and time-controllable fashion.9
Armed with this animal model, the missing technology for diagnostic agent development is an in vivo “gold standard” to which newer HA-specific diagnostic agents can be compared. High-resolution computed tomography (CT) has the potential to provide such an internal gold standard, especially if it can be optimized to detect the genesis of a single breast cancer microcalcification and quantify its growth over time. Moreover, high-resolution CT imaging would permit tumor biologists to study the mechanism of breast cancer microcalcification by separating the processes of calcium salt nucleation from crystal growth.
High-resolution CT scanners for small animal imaging have been studied since the 1990s and have played a major role in molecular imaging.10–18 We hypothesized that optimization of a commercially available CT scanner might permit imaging and quantitation of single breast cancer microcalcifications in an animal model.
MATERIALS AND METHODS
High-Resolution CT Scanner
CT images were acquired on a NanoSPECT/CT™ imaging system (Bioscan, Washington, DC) equipped with an 8 W micro-focus x-ray source (Thermo, Scotts Valley, CA). It has a tungsten target at 40° with a cone beam x-ray and a 130 µm thick beryllium window to reduce the fluence of the low-energy window. The x-ray flux at 90 kVp is 27 Rem/min. The size of the focal spot is less than 9 µm at 8 W and less than 7 µm at 4 W. The system is capable of a range of tube voltages from 20 to 90 kVp, with a maximum tube current of 200 µA. The CT detection system consists of four complementary metal-oxide semiconductor (CMOS) sensors (Rad-icon, Sunnyvale, CA), each with a pixel array of 256 × 512 pixels (1024 × 2048 total), and a 48-µm pixel pitch, creating a total active area of 49.9 cm × 98.3 cm. The signal from each sensor is digitized to 12 bits. The acquisition software used was Nucline (Mediso, Budapest, Hungary).
Reconstruction Algorithms
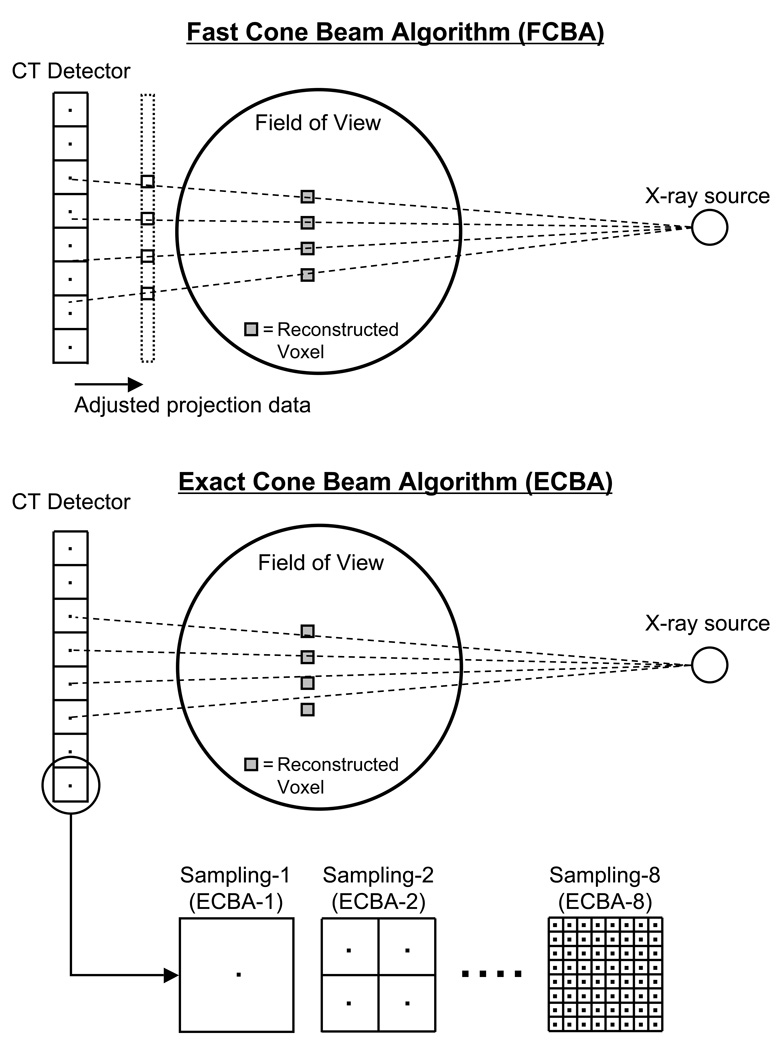
All scanned data were reconstructed using a modified Feldkamp ray-tracing that appropriately models the cone beam geometry19: the “fast” cone beam algorithm (FCBA) and the “exact” cone beam algorithm (ECBA). FCBA defines a ray from the source through each voxel in the reconstructed field of view (FOV). The value of the projection data to be back-projected is determined via interpolation of data on the x-ray detector. The number of voxels set in the image reconstruction volume determines the number of rays. ECBA defines rays from the pixels on the detector to the focal spot of the x-ray source. The value along the ray deposited in the voxel during back-projection is determined by the intersection of the ray and the voxel. The number of rays (sampling number) in ECBA depends on the number of pixels x (sampling number)2. Schematics for each algorithm are shown in Figure 1. Reconstruction was performed using these algorithms as implemented in InVivoScope post-processing software (Bioscan).
Figure 1. CT Reconstruction Algorithms.
Schematic of the fast cone beam algorithm (FCBA; top) and the exact cone beam algorithm (ECBA; bottom), as detailed in Materials and Methods. Shown are the ray tracings from X-ray source to detector, along with their relationship to reconstructed voxels. Also shown are the CT detector binnings (for ECBA) used to create various detector sampling frequencies.
Measurement of Maximum Resolution of Projected and Reconstructed Images
To measure the line spread function (LSF), a 12.7 µm diameter tungsten wire, oriented axially, was placed 10 mm transaxially from the center of the FOV in order to minimize any influence of potential center of rotation artifacts. A CT scan was performed at 45 kVp, 177 µA current, 3000 ms/projection, 360 projections/rotation, and a detector pixel size of 48 µm. The total acquisition time was 18 min/scan. The modulation transfer function (MTF) of the projected image was calculated by performing a Fourier transform of the LSF. The MTF was calculated by using the method developed by Melnyk et al.20 Spatial frequency was measured at 10% of the MTF. Resolution was calculated using the full-width at half maximum (FWHM) of the projected image. Projection data were reconstructed using the aforementioned FCBA and ECBA algorithms. The sampling number (i.e., sampling frequency, binning) of the ECBA was set to 1, 2, 4, 6, and 8 (i.e., ECBA-1, -2, -4, -6, and -8), corresponding to 1, 4, 16, 36, and 64 x-rays per detector pixel, respectively. A Shepp and Logan filter21 was used during the reconstruction process for both the FCBA and ECBA algorithms, and voxel size was set to 10 µm. The MTF and FWHM were obtained by using a method similar to the one used for the projection image.
To evaluate the dependence of CT detector size on the resolution of the reconstructed image, the tungsten wire was scanned using detector pixel sizes of 48, 96, 192, and 384 µm. Additionally, to evaluate the dependence on voxel size, projection data obtained with a pixel size of 48 µm were reconstructed by using either FCBA or ECBA with sampling numbers of 1, 2, 4, 6, and 8, with voxel size set to 10, 50, 100, 200, and 400 µm, respectively. The FWHM was measured on each projected and reconstructed image.
Measurement of Image Noise
A 21 mm diameter syringe phantom was filled with water and scanned to evaluate image noise. The phantom was placed in the center of the FOV. Image noise was evaluated by measuring the standard deviation (SD) of the averaged signal. A region of interest (ROI) with a diameter of 15 mm was set at the center of reconstructed image to measure SD. The reconstruction was performed by using either the FCBA or ECBA algorithm, again with sampling numbers of 1, 2, 4, 6, and 8 and a Shepp and Logan filter. The following tests were performed:
Effect of CT detector pixel size: CT scans were performed at 45 kVp, 177 µA current, 3000 ms/projection, and 360 projections/rotation. The pixel size of the detector was set to either 48, 96, 192, or 384 µm. Projection data were reconstructed with a voxel size of 100 µm. The SD as a function of the pixel size of the detector was measured.
Effect of exposure time: CT scans were performed at 45 kVp, 177 µA, 360 projections/rotation, and a 48 µm pixel size. Exposure time was set to 400, 1000, 1500, or 3000 ms/projection. Projection data were reconstructed by setting the voxel size to 100 µm. The SD as a function of the exposure time was measured.
Effect of voxel size on the reconstructed image: Projection data were scanned with the following parameters: 45 kVp, 177 µA current, 3000 ms/projection, 360 projections/rotation, and a detector pixel size of 48 µm. Projection data were reconstructed by setting the voxel size to 10, 50, 100, 200, or 400 µm. The SD as a function of exposure time was measured and the images were evaluated visually.
Imaging of a Single HA Crystal
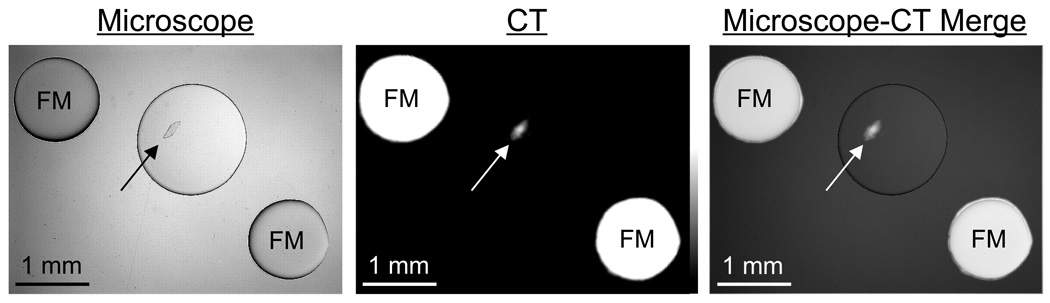
A single HA crystal (catalog #391947, Calbiochem, La Jolla, CA) was scanned with the following parameters: 45 kVp, 177 µA current, 3000 ms/projection, 360 projections/rotation, and a detector pixel size of 48 µm. The crystal was placed on an acrylic sheet with a thickness of 0.762 mm and immobilized with a drop of Fluoromount-G (Southern Biotech, Birmingham, AL). Spots of iodine-containing contrast (Renografin-60, Bracco Diagnostics Inc., Princeton, NJ) were used on the slide as fiducial markers. CT reconstruction was performed using the optimal parameters determined in preliminary studies (described below). Immediately after CT acquisition, a brightfield microscopy image was obtained and merged with the maximal intensity project (MIP) of the reconstructed CT image.
Syngeneic Rodent Model of Breast Cancer Microcalcifications
Animal experiments were conducted under the supervision of a protocol approved by the Institutional Animal Care and Use Committee. Female Fischer 344 rats were obtained from Taconic Farms (Germantown, NY). At the time of tumor cell inoculation, rats averaged 7 to 9 weeks of age and weighed 130 ± 20 g. R3230 syngeneic breast cancer cells were grown in DMEM (Mediatech, Inc., Manassas, VA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin, harvested, and suspended in DMEM for tumor cell implantation. Rats were anesthetized with isoflurane (2% in oxygen) and approximately 2 × 107 cells in 0.3 mL DMEM were injected subcutaneously into the mammary fat pad. rBMP-2 was prepared as described in detail previously.3 One hundred µg of rBMP-2 was administered as a single IP injection to four rats bearing R3230 breast tumors 4 days following tumor cell implantation.3
In Vivo and Ex Vivo Imaging of Rat Breast Cancer Microcalcifications
Animals were anesthetized with isoflurane (2% in oxygen) for imaging. Breast cancer-bearing rats were scanned daily by high-resolution CT beginning 12 d after rBMP-2 injection using a reconstruction voxel size of 50 µm. For both in vivo and ex vivo measurements, scan parameters were 45 kVp, 177 µA current, 3000 ms/projection, 360 projections/rotation, and a detector pixel size of 48 µm, and reconstruction was performed using the optimal parameters determined in preliminary studies (described below). Rats were sacrificed 33 d post-administration of rBMP-2, and the tumor was excised and scanned ex vivo using varying reconstruction voxel sizes of 50, 100, 200, and 400 µm.
RESULTS
Maximum Obtainable Resolution of Projected and Reconstructed Images
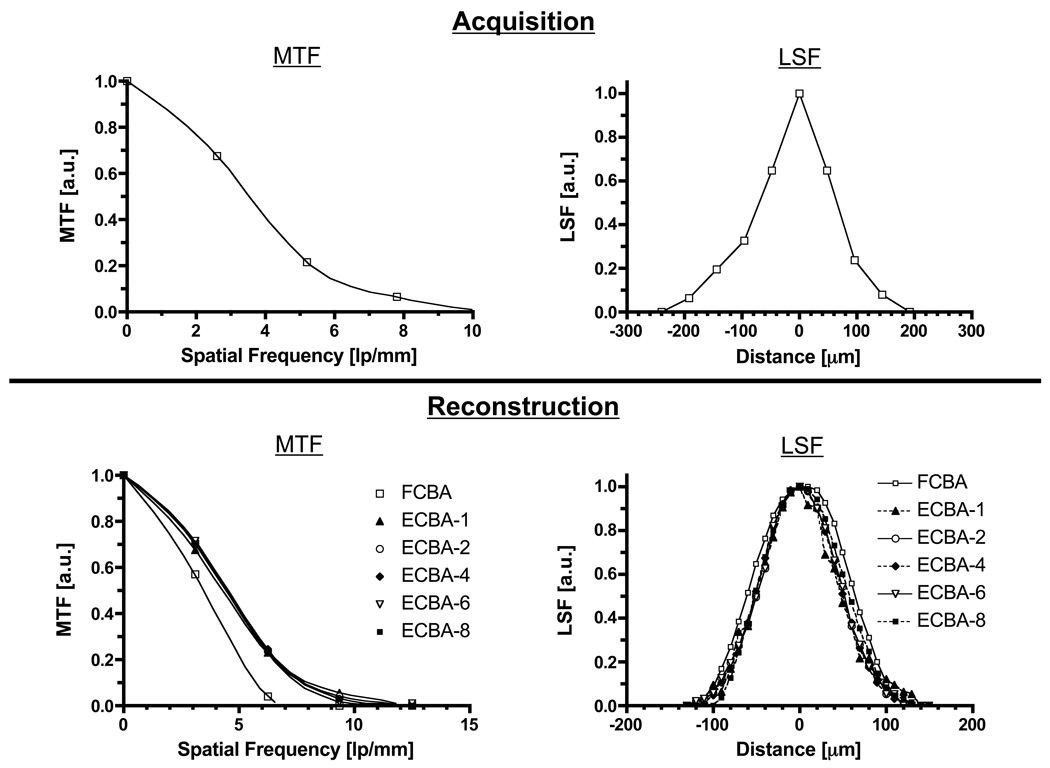
As shown in Figure 2 (top), the MTF curve of the projected image obtained using a 12.7 µm diameter tungsten wire exhibited a spatial frequency of 7.20 lp/mm at 10% of peak MTF. The FWHM of the projected image was 126 µm.
Figure 2. Maximum Resolution of CT Detector and Reconstructed Images.
The modulation-transfer functions (MTF; left) and line-spread functions (LSF; right) as measured during acquisition (top) and reconstruction (bottom) using a 12.7 µm diameter tungsten wire. CT acquisition parameters were: 45 kVp, 3000 ms/projection, and 360 projections/rotation. ECBA reconstruction sampling number is defined in Figure 1.
The MTF and LSF curves of reconstructed images with a reconstruction voxel size of 10 µm, obtained using different reconstruction algorithms and sampling (i.e., detector binning) numbers, are shown in Figure 2 (bottom). The resolution of FCBA showed significant degradation compared to ECBA. The spatial frequencies of FCBA and ECBA with sampling numbers of 1, 2, 4, 6, and 8 were 5.90, 8.60, 8.41, 8.38, 8.33, and 8.17 lp/mm, respectively. FWHMs of reconstructed images obtained using FCBA and ECBA with sampling numbers of 1, 2, 4, 6, and 8 were 132, 95, 96, 98, 99, and 99 µm, respectively.
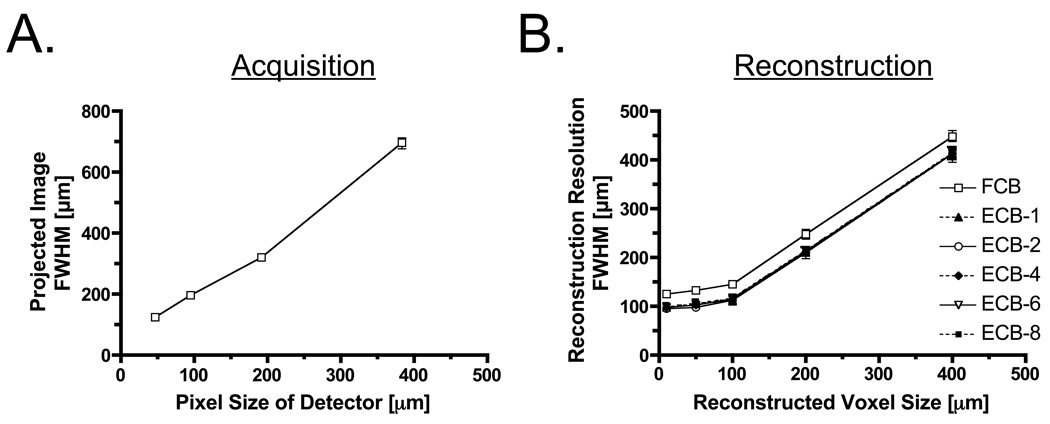
FWHM of projected and reconstructed images as a function of detector pixel size and reconstruction voxel size are shown in Figure 3. When the pixel size of the detector becomes small, FWHM is dramatically improved. The resolution with a pixel size of 48 µm was improved by 568 µm over that with a pixel size of 400 µm (Figure 3, left). FWHMs of reconstructed images as a function of reconstruction voxel size are shown in Figure 3 (right). FWHMs of ECBA-based reconstructions suggest higher resolution compared to FCBA. This observation also occurs when the voxel size is smaller than 100 µm, albeit with a lower slope. FWHMs of reconstructed images at a reconstruction voxel size of 50 µm obtained using FCBA and ECBA with sampling numbers of 1, 2, 4, 6 and 8 were 132, 103, 98, 104, 104 and 105 µm, respectively. Using ECBA-6, the FWHM were improved by 28 µm (21%) compared to FCBA.
Figure 3. Full-Width Half-Maxima (FWHM) of Projected and Reconstructed Images.
- FWHM of projected image as a function of detector pixel size.
- FWHM of reconstructed image as a function of voxel size. ECBA reconstruction sampling number is defined in Figure 1.
Measurement of Image Noise
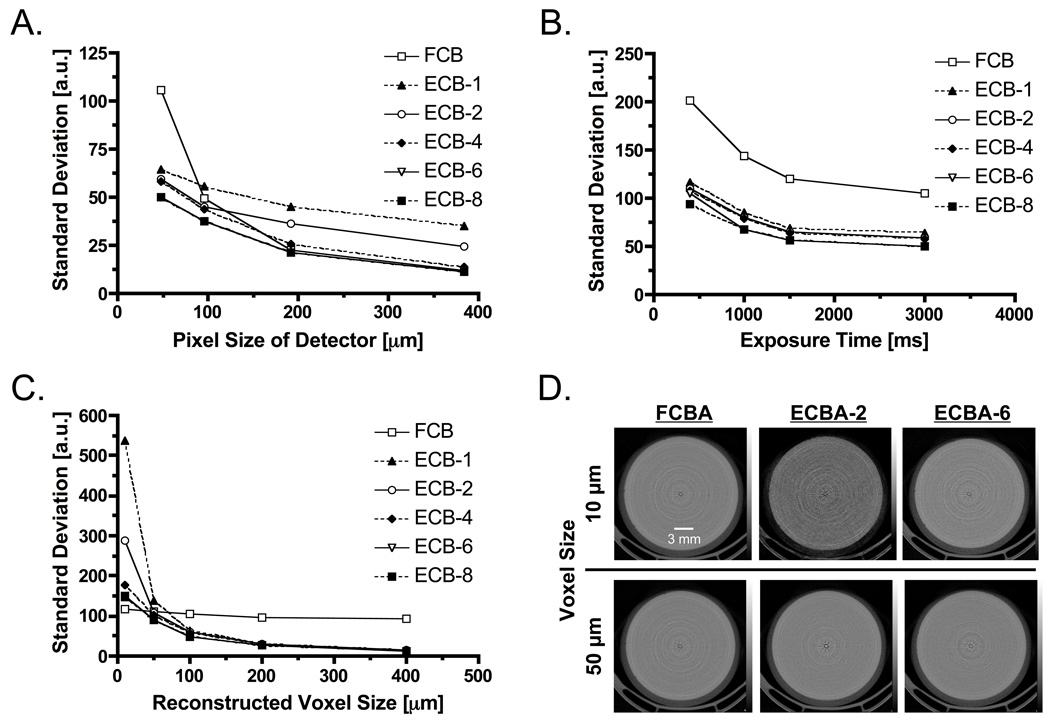
The correlation between the SD of image noise and the detector pixel size is shown in Figure 4A. With both reconstruction algorithms, there is an inverse relationship between detector pixel size and SD. Specifically, SD of FCBA at a detector pixel size of 48 µm was significantly higher than SD of ECBA. SD decreased by 52.7% when using ECBA-6 or ECBA-8. Figure 4B shows SD as a function of exposure time. As expected, when exposure time increases, SD decreases. SD was lowest at 3000 ms/projection. Figure 4C shows SD as a function of the reconstructed voxel size. SD was independent of voxel size for FCBA but dependent on voxel size for ECBA. When voxel size was greater than 50 µm, SD for ECBA (with the exception of ECBA-1) was smaller than SD for FCBA. SDs of ECBA-6 and -8 decreased by 20% and 85% at voxel sizes of 50 µm and 400 µm, respectively, compared to FCBA. Conversely, when voxel size was smaller than 50 µm, FCBA showed the smallest SD. Reconstructed phantom images using reconstruction voxel sizes of 10 and 50 µm are shown in Figure 4D. The results of visual evaluation of the phantom images are in agreement with the results shown in Figure 4C.
Figure 4. Analysis of Image Noise.
- The relationship between standard deviation of image noise and detector pixel size as a function of reconstruction algorithm.
- The relationship between standard deviation of image noise and exposure time as a function of reconstruction algorithm.
- The relationship between standard deviation of image noise and reconstructed voxel size as a function of reconstruction algorithm.
- Reconstructed phantom images obtained using 10 µm (top) or 50 µm (bottom) reconstruction voxels, as a function of reconstruction algorithm.
Based on this study, we selected a detector pixel size of 48 µm, an exposure time of 3000 ms/projection, the ECBA-6 reconstruction algorithm, and a reconstruction voxel size of 50 µm in order to achieve the highest possible resolution and image quality for in vivo experiments.
In Vitro Imaging of Single HA Crystals
Single HA crystals were imaged sequentially by high-resolution CT and brightfield microscopy. A single crystal with a diameter of approximately 300 µm by 100 µm is seen clearly in the reconstructed CT image, which co-registered perfectly with the optical microscopy image (Figure 5 and Supplementary Video).
Figure 5. CT Imaging of a Single Hydroxyapatite Crystal.
Brightfield microscopy image (left), CT image (middle), and a merge of the two (right). Arrows indicate single HA crystal measuring 300 µm × 100 µm. CT parameters included: 45 kVp, 177 µA current, 3000 ms/projection, 360 projections/rotation, and a detector pixel size of 48 µm. FM = Iodine-containing fiducial markers. See also Supplementary Video.
In Vivo and Ex Vivo Imaging of Breast Cancer Microcalcifications
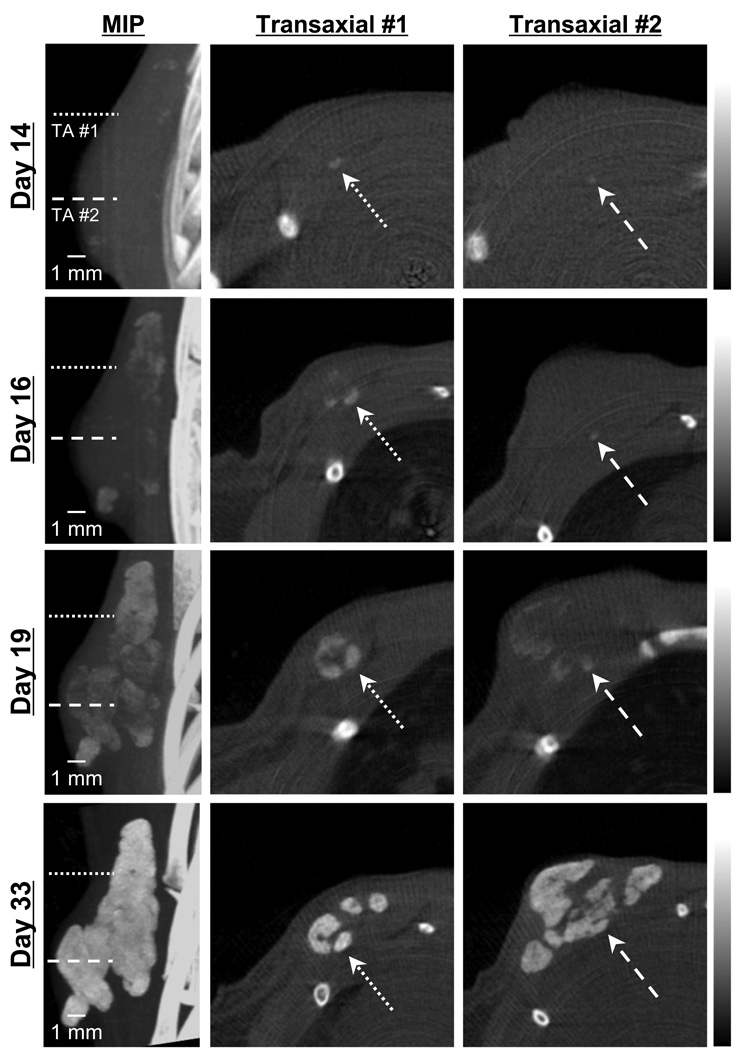
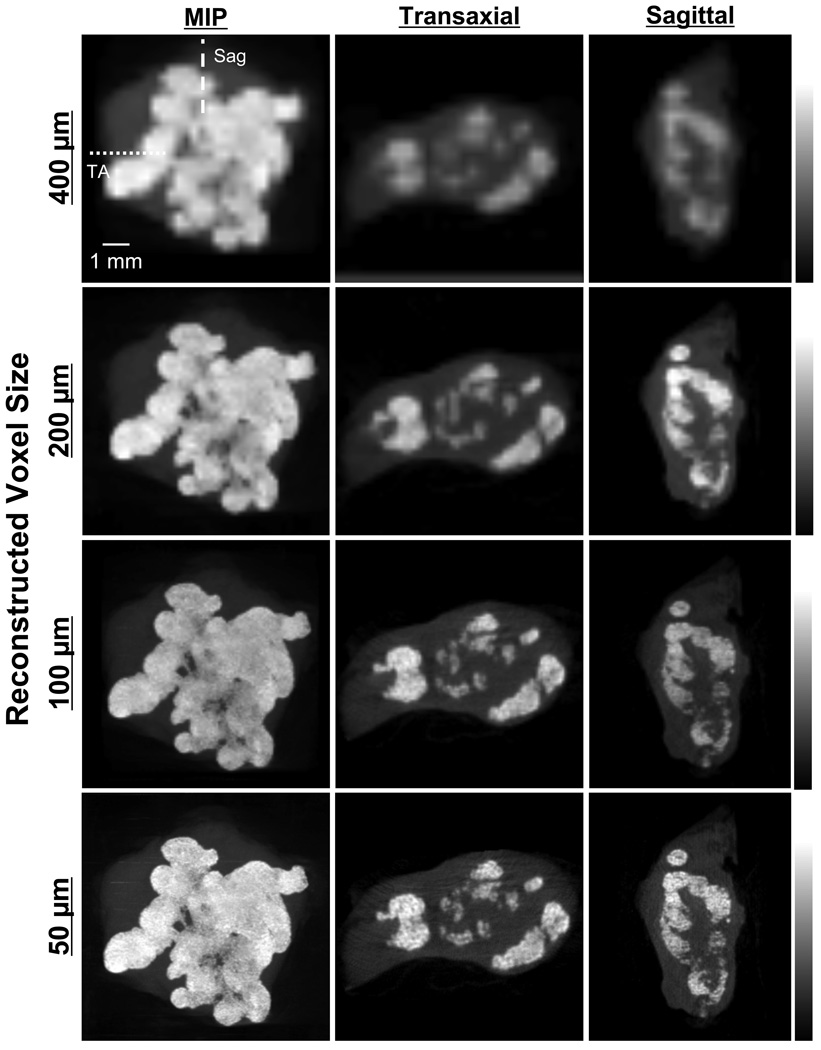
Longitudinal monitoring of syngeneic breast cancer in rat revealed the first evidence of tumor microcalcification 14 d post-injection with rBMP-2. As shown in a representative animal (Figure 6), the first two detectable calcifications, presumably single crystals or extremely small clusters, had dimensions of 300 µm and 100 µm in diameter. After initial nucleation, sites of microcalcification expanded rapidly over the next 19 d, with high-resolution CT being capable of following individual clusters and quantifying tumor volume and calcification volume over time.9 The effect of reconstructed voxel size on image quality (see also Figures 3 right and 4C) is shown in Figure 7 for an excised tumor. As reconstructed voxel size was reduced from 400 µm to 50 µm, image quality improved steadily, albeit at the expense of reconstruction time, which increased from 30 min for 400 µm voxels to 180 min for 50 µm voxels.
Figure 6. In Vivo Imaging of the Genesis and Growth of Breast Cancer Microcalcifications over Time.
High-resolution CT scanning of a syngeneic rat breast cancer at days 14, 16, 19, and 33 days post-injection with rBMP-2. Shown are the maximal intensity projection (MIP; left) and two transaxial (TA) slices (middle and right) whose positions are indicated with dotted and dashed lines on the MIP. Arrows indicate 300 µm (TA #1) and 100 µm (TA #2) microcalcifications first seen at day 14, which grew over time. CT acquisition parameters included: 45 kVp, 177 µA current, 3000 ms/projection, 360 projections/rotation, and a detector pixel size of 48 µm. Reconstruction parameters included the ECBA algorithm with a sampling number of 6 (ECBA-6) and a voxel size of 50 µm. Data are representative of n = 3 independent experiments.
Figure 7. Ex vivo Imaging of Breast Cancer Microcalcifications.
High-resolution, ex vivo CT scanning of a syngeneic rat breast tumor 33 d post-injection with rBMP-2. Shown are MIPs (left), and transaxial (TA) and sagittal (Sag) slices as indicated with dotted and dashed lines, respectively. CT acquisition parameters included: 45 kVp, 177 µA current, 3000 ms/projection, 360 projections/rotation, and a detector pixel size of 48 µm. Reconstruction parameters included the ECBA algorithm with a sampling number of 6 (ECBA-6). Reconstructed voxel size was varied from 400 µm (top row; instrument default) to 50 µm (bottom row). Data are representative of n = 3 independent experiments.
DISCUSSION
To the best of our knowledge, this is the first report of imaging single breast cancer microcalcifications in vivo using high-resolution CT. We purposely utilized a commercially available scanner for the study to demonstrate that systematic optimization of acquisition and reconstruction parameters can result in satisfactory imaging without specialized hardware or software. Nor did the in vivo results we presented employ correction for respiratory motion of the rib cage to which the breast tumor was adhered. Addition of such correction would be expected to improve image quality even further, towards that seen during ex vivo scanning (compare Figure 6 to Figure 7).
The detection of single breast cancer microcalcifications by high-resolution CT, and the ability to quantify calcification volume over time,9 have several implications. First, groups like ours who are interested in developing HA-specific contrast agents5–7 now have an in vivo gold-standard to which to compare sensitivity of various imaging modalities. For example, when developing a HA-specific radiotracer for positron emission tomography (PET), it is now possible to know the precise location of single HA crystals in a tumor, which in turn will permit measurement of the limit of detection of PET radiotracers as a function of PET voxel size and scanner sensitivity. This is particular important in PET, where voxel resolution will typically be ≈ 10-fold lower than CT. Second, the technology we describe (i.e., combination of animal model and high-resolution CT scanning) will permit biologists interested in the mechanism of breast cancer microcalcification to quantify calcification burden after modulation of gene expression. In a prior study, for example, we demonstrated that the effect of rBMP-2 was humoral, rather than local, in inducing microcalficiations.8 Finally, our technology provides a convenient method to test possible pharmacologic modulators of breast cancer microcalcification. For example, rBMP-2-like agents could be developed that increase the sensitivity of screening mammography by inducing microcalcification (i.e., increasing detectability) of otherwise occult malignancies.
Our study is complementary to ongoing clinical studies using dedicated clinical breast CT scanners pioneered by the Boone group at the University of California, Davis.22 In a study by Gong et al., breast calcifications on the order of 175 µm in diameter could be detected using their system.4 In our study, we evaluated the use of FCBA vs. ECBA reconstruction algorithms and found that the best tradeoff between resolution and image noise was found with ECBA and an optimal sampling frequency (i.e., detector binning). Resolution of ECBA improved by 28 µM (21%) of FWHM compared to FCBA. We believe this is due to the fact that projection data are interpolated using adjacent pixels using FCBA, which degrades resolution. In contrast, ECBA is reconstructed by binning each pixel on the detector into squares according to the sampling number. We suggest that ECBA is superior for high-resolution imaging of in vivo microcalcifications. Other parameters that appear to optimize high-resolution imaging include a detector pixel size of 48 µm and a reconstructed voxel size of less than 100 µm. Of note, the default reconstructed voxel size for rat imaging on the NanoSPECT/CT is 400 µm, which is far from optimal.
Since small pixel and voxel sizes are required to obtain high-resolution images, there is the risk that image noise might be increased in the process. We evaluated noise as a function of pixel size, exposure time, and reconstructed voxel size. Image noise of FCBA is significantly increased compared to ECBA, when detector pixel size is held constant at 48 µm. SD increases as pixels are interpolated to a larger size, therefore image noise was higher with FCBA compared to ECBA. We also evaluated the correlation between image noise and reconstructed voxel size. Since projected data is interpolated based on voxel size prior to back-projection, image noise of the FCBA was relatively independent of voxel size. In contrast, since projection data are not interpolated with ECBA, image noise depends on the voxel size. The image noise of the ECBA also depends on the relationship between detector pixel size and reconstructed voxel size. Specifically, when the reconstructed voxel size is smaller than the detector pixel size, image noise is increased dramatically. The number of rays used to reconstruct each voxel decreases when the pixel size of the detector is much greater than the reconstructed voxel size. Our data suggest that if the pixel size of the detector is larger than the voxel size, ECBA may help reduce image noise.
Acquisition and reconstruction times are important parameters. Marxen et al. reported a high-resolution CT scanner that had a 38 lp/mm improvement in resolution over our data.17 However, the acquisition of CT data took ≈ 2 h to image mice. Since small animals are stressed by anesthesia, minimization of scan time is important. In the optimized protocol we describe, the total scan time for visualizing single microcalcifications is only 18 min, thus providing both high-resolution and practicality.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eugenia Trabucchi for administrative assistance and Lorissa Moffitt and Mary McCarthy for editing. We gratefully acknowledge funding from the following sources: National Institutes of Health High-End Instrumentation Grant (S10-RR-023010) to J.V.F.; Lewis Family Foundation to J.V.F.; Foundation for Promotion of Cancer Research to K. I.; Tokyo Metropolitan University to K. I.; BIRD of Japan Science and Technology Agency to K. I. and H. F.
REFERENCES
- 1.American Cancer Society: Cancer facts and figures. 2010 www.cancer.org/downloads/STT/500809web.pdf.
- 2.Morgan MP, Cooke MM, McCarthy GM. Microcalcifications associated with breast cancer: an epiphenomenon or biologically significant feature of selected tumors? J Mammary Gland Biol Neoplasia. 2005;10:181–187. doi: 10.1007/s10911-005-5400-6. [DOI] [PubMed] [Google Scholar]
- 3.Haka AS, Shafer-Peltier KE, Fitzmaurice M, et al. Identifying microcalcifications in benign and malignant breast lesions by probing differences in their chemical composition using Raman spectroscopy. Cancer Res. 2002;62:5375–5380. [PubMed] [Google Scholar]
- 4.Gong X, Vedula AA, Glick SJ. Microcalcification detection using cone-beam CT mammography with a flat-panel imager. Phys Med Biol. 2004;49:2183–2195. doi: 10.1088/0031-9155/49/11/005. [DOI] [PubMed] [Google Scholar]
- 5.Bhushan KR, Misra P, Liu F, et al. Detection of breast cancer microcalcifications using a dual-modality SPECT/NIR fluorescent probe. J Am Chem Soc. 2008;130:17648–17649. doi: 10.1021/ja807099s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenkinski RE, Ahmed M, Zaheer A, et al. Near-infrared fluorescence imaging of microcalcification in an animal model of breast cancer. Acad Radiol. 2003;10:1159–1164. doi: 10.1016/s1076-6332(03)00253-8. [DOI] [PubMed] [Google Scholar]
- 7.Zaheer A, Lenkinski RE, Mahmood A, et al. In vivo near-infrared fluorescence imaging of osteoblastic activity. Nat Biotechnol. 2001;19:1148–1154. doi: 10.1038/nbt1201-1148. [DOI] [PubMed] [Google Scholar]
- 8.Liu F, Bloch N, Bhushan KR, et al. Humoral bone morphogenetic protein 2 is sufficient for inducing breast cancer microcalcification. Mol Imaging. 2008;7:175–186. [PMC free article] [PubMed] [Google Scholar]
- 9.Liu F, Misra P, Lunsford EP, et al. A dose- and time-controllable syngeneic animal model of breast cancer microcalcification. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0535-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulus MJ, Sari-Sarraf H, Gleason SS, et al. A new X-ray computed tomography system for laboratory mouse imaging. IEEE Trans Nucl Sci. 1999;46:558–564. [Google Scholar]
- 11.Guldberg RE, Lin AS, Coleman R, et al. Microcomputed tomography imaging of skeletal development and growth. Birth Defects Res C Embryo Today. 2004;72:250–259. doi: 10.1002/bdrc.20016. [DOI] [PubMed] [Google Scholar]
- 12.Ritman EL. Molecular imaging in small animals--roles for micro-CT. J Cell Biochem Suppl. 2002;39:116–124. doi: 10.1002/jcb.10415. [DOI] [PubMed] [Google Scholar]
- 13.Wyss C, Schaefer SC, Juillerat-Jeanneret L, et al. Molecular imaging by micro-CT: specific E-selectin imaging. Eur Radiol. 2009;19:2487–2494. doi: 10.1007/s00330-009-1434-2. [DOI] [PubMed] [Google Scholar]
- 14.Zhu S, Tian J, Yan G, et al. Cone Beam Micro-CT System for Small Animal Imaging and Performance Evaluation. Int J Biomed Imaging. 2009:960573. doi: 10.1155/2009/960573. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burghardt AJ, Kazakia GJ, Laib A, Majumdar S. Quantitative assessment of bone tissue mineralization with polychromatic micro-computed tomography. Calcif Tissue Int. 2008;83:129–138. doi: 10.1007/s00223-008-9158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du LY, Umoh J, Nikolov HN, et al. A quality assurance phantom for the performance evaluation of volumetric micro-CT systems. Phys Med Biol. 2007;52:7087–7108. doi: 10.1088/0031-9155/52/23/021. [DOI] [PubMed] [Google Scholar]
- 17.Marxen M, Thornton MM, Chiarot CB, et al. MicroCT scanner performance and considerations for vascular specimen imaging. Med Phys. 2004;31:305–313. doi: 10.1118/1.1637971. [DOI] [PubMed] [Google Scholar]
- 18.Seo Y, Hashimoto T, Nuki Y, Hasegawa BH. In vivo microCT imaging of rodent cerebral vasculature. Phys Med Biol. 2008;53:N99–N107. doi: 10.1088/0031-9155/53/7/N01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldkamp LA, Davis LC, Kress JW. Practical cone-beam algorithm. J. Opt. Soc. Am. A. 1984;1:612–619. [Google Scholar]
- 20.Melnyk R, DiBianca FA. Modeling and measurement of the detector presampling MTF of a variable resolution x-ray CT scanner. Med Phys. 2007;34:1062–1075. doi: 10.1118/1.2436977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shepp LA, Logan BF. The Fourier reconstruction of a head section. IEEE Trans Nucl Sci. 1974;21:21–43. [Google Scholar]
- 22.Lindfors KK, Boone JM, Nelson TR, et al. Dedicated breast CT: initial clinical experience. Radiology. 2008;246:725–733. doi: 10.1148/radiol.2463070410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.