Abstract
Human olfactory receptor, hOR17-210, is identified as a pseudogene in the human genome. Experimental data has shown however, that the gene product of frame-shifted, cloned hOR17-210 cDNA was able to bind an odorant-binding protein and is narrowly tuned for excitation by cyclic ketones. Supported by experimental results, we used the bioinformatics methods of sequence analysis (genome-wide and pair-wise), computational protein modeling and docking, to show that functionality in this receptor is retained due to sequence-structure features not previously observed in mammalian ORs. This receptor does not possess the first two transmembrane helical domains (of seven typically seen in GPCRs). It however, possesses an additional TM that has not been observed in other human olfactory receptors. By incorporating these novel structural features, we created two putative models for this receptor. We also docked odor ligands that were experimentally showntobind hOR17-210. We show how and why structural modifications of OR17-210 do not hinder this receptor's functionality. Our studies reveal that novel gene rearrangements that result in sequence and structural diversity may have a bearing on OR and GPCR function and evolution.
Keywords: Olfactory receptors, Functional pseudogene, Computational modeling, Docking
Introduction
GTP-binding Protein Coupled Receptors (GPCRs) are proteins that traduce the cell membrane and are responsible for catalyzing or initiating a cellular response in the form of a signal transduction process following extracellular stimuli [1, 2]. GPCR function is wide and varied. GPCRs are ubiquitously found in mammals, plants and fungi [3]. Olfactory receptors (OR), which constitute the largest gene families in mammalian genomes [4] are believed to be rhodopsin-like GPCRs. They are structurally characterized by seven transmembrane helical regions that are connected by three extracellular and three intracellular loops, an extracellular N-terminus and an intracellular C-terminus. Earlier experimental observations have shown that there exists a many-many binding/activating relationship between ORs and odors. One odor may bind and activate more than one OR, while an OR might be activated by more than one odor [4–15]. The olfactory system can differentiate odorous molecules based on structural and chemical diversity and concentration. Olfactory receptors' (OR) interactions with odor ligands are widely accepted as the first specific step in the early events leading to olfactory perception. Research has also suggested that the interaction of odor ligands with the binding region of an OR [16, 17] may cause the receptor to evolve from a structurally inactive to active state.
After the publication of the first draft of the human genome, several groups, working independently, identified the human olfactory repertoire [18–21]. As other mammalian genomes became available, the OR repertoires of these species were also identified [22–24]. These genomic ORs were identified as either putatively functional or nonfunctional and pseudogenic. Initially, more than 60% of the human olfactory receptor genes were flagged as pseudogenes, while that number for the mouse OR repertoire stands at less than a third [23, 24]. As additional analysis is being carried out, the number of mammalian functional receptors however, is being constantly revised [25, 26]. There is evidence that primate evolution is marked by loss of olfactory functionality, as evidenced by a greater percentage of functional ORs in the evolutionary parent than the daughter [27].
OR gene, hOR17-210 was genomically identified as pseudogenic (OR1E3P in the HUGO and HORDE (Human Olfactory Receptor Database Exploratorium housed at http://bioportal.weizmann.ac.il/HORDE/) databases). This genomic pseudogene sequence was identified earlier as a possible functional pseudogene possessing a two-nucleotide frame shift [28]. A cDNA clone of this frame-shifted sequence was subsequently shown by Matarazzo, Clot-Faybesse and others to successfully initiate a G-protein mediated signal transduction cascade in the presence of a mixture of odorants (especially cyclic ketone compounds [29]) commonly perceived by humans. The researchers co-expressed two human ORs (OR17-209 and OR17-210 [the subject of this work]) with corresponding G-proteins in Sf9 cells using baculovirus vectors.
We used bioinformatics strategies to show that hOR17-210 receptor (the frame shifted, cDNA clone sequence mentioned above) protein has sequence-structure features that are atypical of previously studied ORs or GPCRs. Despite these differences, this receptor remains functional. We show that the regions in this protein that are responsible for G-protein coupling are not affected by the unusual sequence-structure attributes. OR17-209, which is shown to respond to esters [29], has predicted sequence structure correlates more typical of ORs.
The novel sequence-structural features mentioned above are: (1) hOR17-210 has only six transmembrane helical regions (TMs) instead of the typical seven. The first two TMs typically observed in models of ORs and GPCRs are missing. (2) While this presumably reduces the number of TMs to five, there exists an additional TM, which occurs after what is typically observed as the C-terminus in other ORs. The sequence for this TM is only found in two other ORs—a chimpanzee and a cow homolog, which themselves have additional unique structural features. (4) The amino acid sequence motifs for ORs that have been implicated in G-protein coupling and olfactory sensory neuron targeting [30], however, remain structurally and sequentially conserved. (5) Unlike mammalian ORs and GPCRs studied to date, the C-terminus is predicted to be extracellular.
We show how and why these structural modifications may not hinder the function of this OR. We created two putative computational models of this receptor. Our models incorporate the novel sequence-structural features for this OR. We also carried out computational docking studies using the preferred of the two models with selected odor ligands that are known to experimentally excite hOR17-210 [29].
Materials and methods
Sequence analysis and transmembrane domain prediction
Figure 1a shows the results of a comparative sequence analysis between the cDNA sequence functionally studied [29] and the pseudogene (OR1E3P) identified from the genome in the HORDE database [31]. The former is listed in GENBANK (http://www.ncbi.nih.gov/) under Accession Number AAC99555, the latter, Accession Number U53583. OR1E3P has a nucleotide sequence in a missing 5′ region located upstream from the cloned cDNA sequence. The missing region is as follows:
Fig. 1.

(a) Results of the sequence alignment between OR17-210 cDNA found in GENBANK Accession number (AAC99555) and OR1E3P genomic DNA found in the HORDE database. The functional region in OR17-210 begins from nucleotide 170. This is caused by a two-residue frame shift in the genomic DNA. The sequence of the functional protein also contains an added region beginning from nucleotide number 977. This orphan TM7′ and the extracellular C-termini are contained in this region. The frame shift results in a stop codon beyond nucleotide 1030, (b) results of the translation of the genomic DNA for OR1E3P as found in the HORDE database. The presence of stop codons indicates that the receptor is pseudogenic as listed and (c) results of the translation of the genomic DNA of OR1E3P as found in the HORDE database, but following a 2-nucleotide frame shift. This sequence contains the orphan TM observed in GENBANK entry ACC99555. This sequence was also used in the experimental functional studies [29]
ATGATGAAGA AGAACCAAAC CATGATCTCA GAGTTCCTGC TCCTGGGCCT TCCATCCAAC CTGAGCAGCA GAATCTGTTC TATGCCTTGT TCTTGGCCGT GTATCTTACC ACCCTCCTGG GGAACCTCCT CGTCATTGTC CTCATTCGAC TGGACTCCCA CCTCCAC
On the other hand, the sequence used in our informatics-based work and which was shown to be functional possesses the following additional base pairs at the 3′ terminus.
TAGTAGGTGTAGTAAAGTTGATAATGAAA TATCACTCTAAATCAGTGG CTTAA
When the genomic gene sequences is translated using the TRANSLATE tool available through Swissprot's Expasy web site (http://ca.expasy.org/tools/dna.html), OR1E3P (genomic OR17-210) contained several stop codons (denoted by /) after the first 132 residues (Fig. 1b). The TRANSLATE program also translates a given nucleotide sequence in three frames. A two-nucleotide frame shift yielded the same peptide sequence as AAC99555. Figure 1c shows this sequence that was used in our analysis and in the experimental functional studies [29].
As a prelude to creating our computational model, we used Hidden Markov Models to predict transmembrane helices in hOR17-210. Figure 2 highlights regions that are predicted as TMs by two transmembrane prediction algorithms: TMHMM [32] and HMMTOP [33] TM. Both were identified as the best α-helical TM prediction programs in an analysis of over ten such programs [34]. The figure shows that both programs agree in their identification of only six TMs and an extra-cellular C-terminus (red circles).
We carried out a sequence comparison of hOR17-210 with rat OR I7, Olr266 in Genbank (Accession Number P23270, Fig. 3). OR I7, having been among the first cloned and identified ORs is well characterized, both experimentally [35] and computationally-structurally [36, 37]. We use rat I7 here to represent ORs with structural features that are typical of GPCRs. The I7 TM regions are highlighted as predicted by both TMHMM and HMMTOP. The figure shows that I7 has TM1 and 2. Both of these TMs are missing in hOR17-210; the latter has an additional predicted TM after the C-terminus. We denote this additional TM as TM7′. Interestingly, the region of TM2 correctly predicted as a helical TM region for rat I7 is not predicted as a TM in hOR17-210 despite the apparent sequence similarities.
We carried out a comprehensive BLAST search for hOR17-210 against GENBANK. Sequence identity was found between hOR17-210 and its predicted chimpanzee and cow OR homologs. Figure 4 shows the results of the alignment. The TM regions are highlighted. All three sequences possess the TM7′ region. The notable difference between the three sequences is that the predicted chimpanzee and cow homologs lack OR17-210's frame shift mutation and so have seven intact TMs, i.e., they possess what is typically TM1. And just as in hOR17-210, the cow and chimpanzee homologs are missing the typically observed TM2.
Fig. 2.
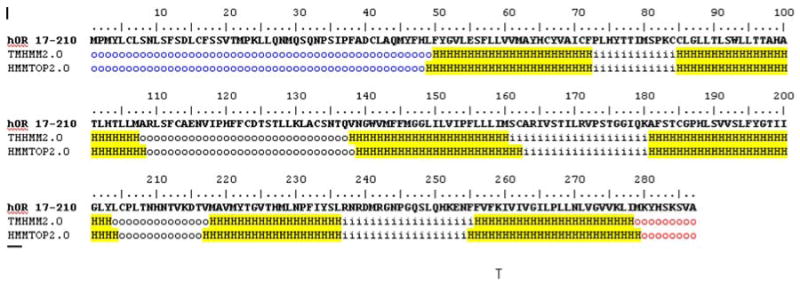
Figure shows the results of Hidden Markov Model predictions of termini, intra and extra-cellular loops and helices in hOR17-210. The yellow highlighted regions show the predicted TM helices. The blue colored regions indicate the extra-cellular N-termini, which contains the MPMY polypeptide motif. This typically marks the beginning of TM2 in most Ors. The last helical region is the orphan TM7′ region. The red colored region shows that the C-terminus for this protein is extracellular
Fig. 3.

Figure shows the results of Hidden Markov Model predictions of termini, intra and extra-cellular loops and helices in hOR17-210. The yellow highlighted regions show the predicted TM helices. The blue colored regions indicate the extra-cellular N-termini, which contains the MPMY polypeptide motif. This typically marks the beginning of TM2 in most Ors. The last helical region is the orphan TM7′ region. The red colored region shows that the C-terminus for this protein is extracellular
Fig. 4.

Figure shows the sequence alignment if hOR17-210 with its chimpanzee and cow homologs, other ORs that show the orphaned TM7′. Each OR shows atypical features of the polypeptide region beginning with PMY not being a TM. Additionally, the chimpanzee and cow OR gene products show the existence of TM1. Residues highlighted in yellow are predicted transmembrane helical regions
Constructing a structural model of hOR17-210
We created two computational, structural models of hOR17-210 (Fig. 5a, b). Our modeling strategies incorporated the new structural features discussed above. In addition to using homology modeling strategies [38] to create our preliminary model, we tested a new paradigm for rationalizing the hydrophobic nature of the inside of the receptor (Eq. 1).
Fig. 5.
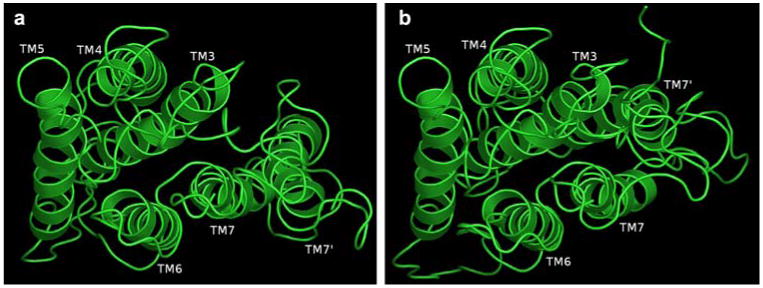
(a) Figure shows a structural model for OR17-210 with the TM7′ occupying the position typically occupied by TM1 in rhodopsin-like GPCRs and (b) figure shows a structural model for OR17-210 with the TM7′ occupying the position typically occupied by TM2 in rhodopsin-like GPCRs
One must recognize here that in using homology modeling to create our models, we do not use sequence homology, but structural homology. Indeed, the sequence identity between olfactory receptors (and most GPCRs) and rhodopsin does not justify the use of sequence homology. In most cases, the sequence identity is less than 20% with sequence homology not rising above 50%.
We used PRALINE [39] (http://zeus.cs.vu.nl/programs/pralinewww/example/prepro_iter/) sequence comparison software to align the sequences of OR17-210 and bovine rhodopsin. Figure 6 illustrates the sequence comparison with a color range depicting the level of conservation between aligned amino acids. The sequence identity is approximately 15% and the sequence homology is a little more than 25% (the sequence homology using other software is as high as 45%, depending on the assignment of gap extensions and penalties). The figure shows that both the identity and the homology suffer (more than usual) because of the missing TM1 region in OR17-210. In semi-empirical modeling methods for ORs, as in our modeling protocol, using the structure of bovine rhodopsin as a template, the use of structural homology (as opposed to sequence homology) is not unprecedented [14, 37, 39–48]. The assumption here is that since a GPCR has seven transmembrane regions, so will an OR. The helical regions are independently identified using helix prediction methods, or, as we have done here, using Hidden Markov Models. The predicted helical regions are then matched with the helical regions from the rhodopsin structure (which serves as a template) prior to homology modeling.
Fig. 6.
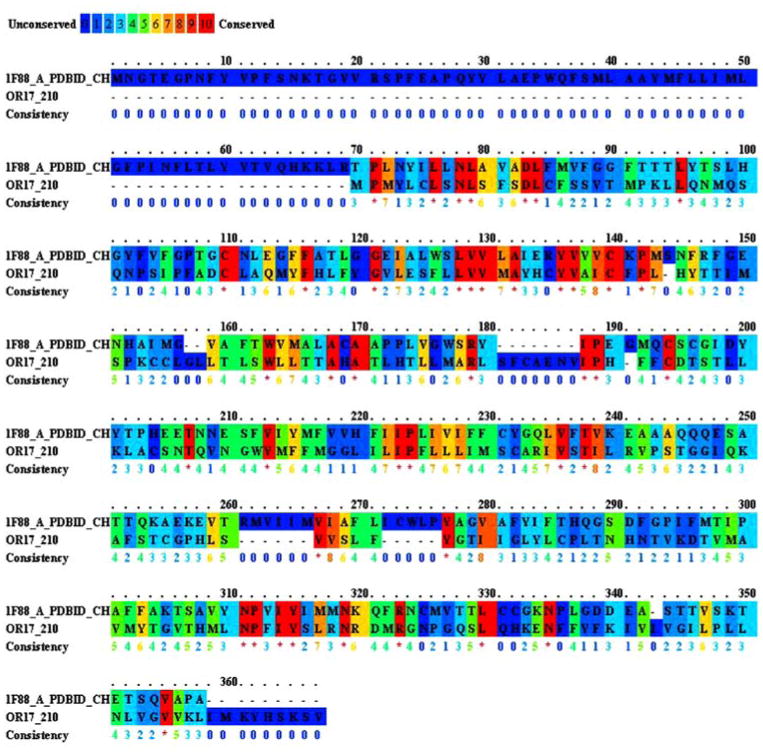
Figure shows a sequence alignment between bovine rhodopsin and OR17-210. The software PRALINE was used to perform the sequence alignment. The figure legend indicates a range from blue (unconserved) to red (identity). The figure indicates that the sequence identify is 15% and the sequence homology is approximately 25%
Our modeling protocol was as follows:
TM helical regions were predicted by using Hidden Markov Models through the programs: TMHMM2.0 [49] and HMMTOP2.0 [33].
Due to the missing TMs 1 and 2 and the addition of an orphaned TM7′, the secondary structures of helices in positions 3, 4, 5, 6 and 7 were refined by alignment of the first five regions of hOR17-210 against the previously predicted secondary structure of rat OR-I7. Each predicted OR helix was aligned to the center of the homologous helix in the rhodopsin structure, with no gaps. Two variants of helical packing were explored: in the first variant, the final orphaned helix of hOR17-210 was aligned against helix 2 in rhodopsin; in the second variant, the last (orphaned) helix of hOR17-210 was aligned against helix 1 in rhodopsin.
3D models were generated using the homology modeling program, Modeler 8v2 [38]. We used the highest resolution structure (2.2 Å) of dark-adapted bovine rhodopsin [50] (Protein Data Bank ID: 1U19) as a template. For the helical construction, each predicted OR helix was sequentially aligned to the center of the homologous helix in the rhodopsin structure, with no gaps. Two variants of helical packing were explored: in the first, the final orphaned helix of hOR17-210 was aligned against TM2 in rhodopsin; in the second variant, the last (orphaned) helix of hOR17-210 was aligned against TM1 in rhodopsin.
Each helix each structural model was minimized with typical α-helix H-bond distance constraints using the consistent valence force field (CVFF) and conjugate gradient algorithm in Accelrys Discover suite of programs (http://www.accelrys.com/products/insight/). The helices were individually submerged in water during the energy minimization step to relax helical features specific to rhodopsin.
The hydrophobic moments at each residue around a helix were calculated using the following expression:
| (1) |
where, μθ is the effective aggregate hydrophobicity at each point around a helical wheel computed by summing the arc contributions to the hydrophobicity moment on that residue from all other points along the helical wheel for a given TM. In order to establish the correct frame of reference with respect to the entire helical assembly for hydrophobic moments derived from this algorithm, these were initially calculated for the TMs in rhodopsin and subsequently mapped to the actual rotational orientations within the helical bundle. We observed that in rhodopsin, that the largest Θ valued residues in TM 1, 3, 4, and 7 pointed toward the binding pocket; for TM2, the largest Θ pointed away from the binding region; and for TMs 5 and 6, the largest aggregate hydrophobic moments pointed toward each other. The hydrophobicities for hOR17-210 were computed and the TM helices were rotated and oriented using this rationale.
After helix construction and rotation, the helices were used as the input template into the Modeller software for ab initio assignment of the intra- and extracellular loop residues. The resulting structure was then rigorously minimized using the Accelrys Discover program by constraining only the motion of the alpha-carbon atoms of the protein in order to maintain the integrity of the transmembrane helices.
Of the two models created, our preferred model is one where TM7′ is positioned in place of TM2. As shown in Fig. 4, the protein sequences in the chimpanzee and cow homologs both have strongly predicted TM1s and therefore, in order to maintain the helical bundle, TM7′ would have to be placed in the position typically occupied by TM2.
Ligand docking
We docked eight ligands: beta-ionone, d- and l-camphor, 2-and 6-undecanone, heptanal, decanal, nonanol and nonanone (ligand positional parameters and those for the two model variants are available from corresponding author) in the binding pocket of our preferred model of hOR17-210—with the TM7′ homology-modeled in the place of missing TM2. Figure 7a (ringed ligands) and b (straight chain ligands) shows the results of computational docking. Of these ligands, experimentally, the cyclic ketones show strong responses, the straight chain ketones show weak responses and the alcohols show no response at any concentration (Personal Communication—CR). These docked ligands vary in length of carbon chain and functional groups (aldehyde, ketone, alcohol and ring structures). Ligand models were constructed using the InsightII suite of software (http://www.accelrys.com/insight/).
Fig. 7.
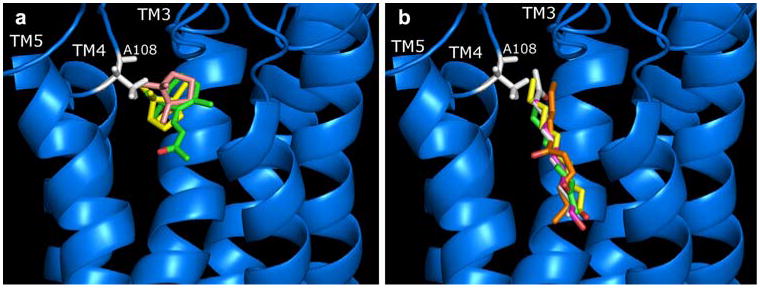
(a) Figure shows the docking of three ligands with ring structures: beta ionone (green), D-camphor (yellow) and L-camphor (pink). The figure shows the proximity of the docked ligands to ALA108 in white. The binding is expectedly in the region bound by TMs 3 (1), 4 (2), 5(3) and 6(4). The TM identifiers are numbers typical of ORs and GPCRs. The numbers in parentheses are TM numbers for hOR17-210 and (b) figure shows the docking of five ligands straight chains: decanol (yellow), nonanone(green), nonanol (pink), 2-undecanone (orange) and 6-undecanone (white). The figure shows the proximity of the docked ligands to ALA108 in white. The binding is expectedly in the region bound by TMs 3 (1), 4 (2), 5(3) and 6(4). The TM identifiers are numbers typical of ORs and GPCRs. The numbers in parentheses are TM numbers for hOR17-210
Ligand conformational energies were minimized using the Discover module in InsightII. We added hydrogen atoms to our I7 OR model [37] to create a system of pH 7.0. Atomic charges were assigned using Consistent Valence Force Field (CVFF). We used DOCK [51, 52] to identify the ideal binding configurations of the ligands in the binding pocket [37] of the 17–210 human olfactory receptor model. Using every atom in the OR model as input for the DMS (Dot Molecular Surface) program [53], we calculated a solvent accessible molecular surface-area for the I7 model; and DOCK's SPHGEN (SPHere GENerator) module identified cavity site-points in the receptor. We discarded spheres that represented cavities on the intracellular side of the receptor; these spheres were structurally “below” the TM3 and TM4 crossover plane in the model. The GRID module in DOCK was used to generate force fields and interaction parameters to compute intermolecular binding. DOCK used spheres that were retained to compute spatial restraints based on van der Waals interactions. Flexible_Ligand, a module in DOCK, allowed the modification of torsion angles in the ligand. Figure 7a and b shows the docked ligands in the receptor model.
Results
The helical regions and the internal and external locations of the intra and extracellular loops and the N- and C-termini are highlighted in the Fig. 3, which compares the sequences of hOR17-210 and rat OR I7, one of the first ORs cloned [54], functionally analyzed [35] and modeled [36, 37]. The highlighted regions in the I7 sequence in Fig. 3 are representative of what is typically known about ORs. Figure 2 shows that both HMMTOP and TMHMM transmembrane prediction programs agree that for hOR17-210: (1) the region beginning with PMY—is not a TM, although it is predicted to be the second TM in most ORs; (2) the region we identify as the orphaned TM, TM7′ exists and (3) the C-terminus of the receptor must be atypically extracellular. TM7′ in OR17-210 extends past the C-terminus of ORI7. The amino acids of the C-terminus in I7 and the final intracellular loop (between TM7 and 7′) of OR17-210 are conserved.
The hOR17-210 sequence begins with a MPMY polypeptide region. Typically, in ORs, the PMY sequence motif marks the beginning of TM2. HMMs for this functional hOR17-210 sequence predict however, that the region beginning with PMY (i.e., MPMYLCLSNLSFSDLC FSSVTM is not a helical TM and is extracellular). Experimentally, heterologous expression of a FLAG (DYKDDDDK polypeptide sequence) tagged receptor in insect cells confirmed an extracellular N-terminus [29]. This leads us to conclude that hOR17-210 is missing both the first and second TMs when compared to other mammalian ORs. The hOR17-210 transmembrane regions start from what would typically be TM3 in other ORs. The “MAYDRY” motif region, a highly conserved sequence within Class A GPCRs and known to be located at the intracellular end of this TM, has been shown to be essential for G-protein coupling and the initiation of signal transduction following ligand-binding [30, 55, 56]. In the case of hOR17-210, the “MAYDRY” sequence has undergone mutation to MAYHCY; since this TM must be oriented extracellular to intracellular in order for correct positioning of this conserved motif, it allows the polarity of the remaining TMs predicted by the HMM to be determined with certainty.
A sequence similarity search of the TM7′ peptide region “FVFKIVIVGILPLLNLVGVVKLI,” returns only two matches. The first is a chimpanzee OR (GENBANK Accession Number XP_523775), which is homologous to hOR17-210; the second is a cow OR (GENBANK Accession Number XP_872923). HMM of these OR sequences predicts that, in addition to the presence of TM7′, the polypeptide regions beginning with PMY are also not TM helices. One major difference is that these two orthologs do, however, possess intact TM1 helices (Fig. 4).
The constrained polarity of the final TM (TM7′) causes the C-terminus of OR17-210 to become extracellular. Extracellular C-termini have been predicted in Drosophila odorant receptors [57]. The sequence region RNRDMRGNPGQSLQHKENFF is the third intracellular loop in hOR17-210 (between TM7 and TM7′). We carried out a BLAST search using the above sequence of this loop, focusing the search to return only sequences for olfactory receptors. From over 2,500 results, the “RNRDMRG” region is strongly conserved (greater than 70% identity and 100% positive matches, where R is often replaced by K) in the C-termini of most ORs (and is possibly involved in GPCR-G-protein interactions). hOR17-210 functionality is therefore, not affected by an extra-cellular C-terminus or the lack of a TM (the absence of TMs 1 and 2).
Figure 6a and b indicates that all the docked odorous ligands are clustered in the same spatial region bound by the first four TMs (3, 4, 5 and 6) of hOR17-210. The numbers in parenthesis indicate the TM numbers for typical ORs. An inspection of the interior of the receptor, which is modeled using the hydrophobicities determined using Eq. 1 indicate that there are no strongly polar residues pointing into the binding pocket, except His48 on TM1. This residue however, is greater than 10 Å away from the nearest ligand atom. The nearest distances from every side-chain atom within the receptor's binding pocket to each atom of the docked ligands were calculated. The closest distance (between 0.8 and 1.5 Å) was for Ala108, specifically between ligand atoms and the methyl hydrogens in Ala108. Some of the interactions can be considered to be electrostatic in nature because they are between the ligand carbonyl oxygen and the Ala108 hydrogen atoms. Possible interactive distances were also observed between ligands and Phe122 and Cys123. These residues however, belong to the first extracellular loop, which in our model dips into the binding pocket. The contributions of these residues however, cannot be ascertained because of the dynamic nature of loop conformations.
Discussion
We created computational structural models for two possible variants to account for the atypical nature of the hOR17-210 (Fig. 5a, b). Such a model, based on structural template matching (the sequence homology between I7 and rhodopsin is less than 40%) [8, 13] may however, introduce rhodopsin structure-specific biases into the model. Biases include differences in lengths of loops [58] and kinks [59, 60] in TM helical domains. We attempted to limit the intra-helical biases by allowing each helix to structure-energetically relax in an aqueous medium individually before assembling the TM domains.
The first step in any GPCR modeling study is the identification of the TM regions. TM helices presumably protect the interior of the binding pocket from the surrounding lipid bilayer, while at the same time, ensuring that the signal-traducing structural features of the receptor are properly positioned inside and outside the cell.
We aligned the first five TMs of hOR17-210 cloned sequence in the positions occupied by TMs 3, 4, 5, 6 and 7 of typical ORs, respectively. In each variant, TM7′ occupied the positions typically occupied by TM1 and TM2, respectively. Our modeling strategy ensured that sequentially conserved (and possibly functionally implicated) regions were positioned as found in typical ORs. TM7′ in the two variants was positioned to maintain the structural integrity of the TM scaffold while protecting the interior of the OR and the odor ligand. We have indicated earlier that orthologs of hOR17-210 exist in cow and chimpanzee. Evidence of sequence predicted as TM7′ and absence of TM2 in observed in only these three mammalian ORs. During hOR17-210 modeling, when presented with a choice of placing TM7′ in the position of TM1 or TM2, we posit that since the cow and chimpanzee orthologs retained TM1 and were missing only the typical TM2, the orphaned TM7′ would favorably occupy in the position of TM2 (Fig. 5b). For olfactory function to persist, the main GPCR scaffold needs to be maintained.
Katada et al. [61] have shown that the C-termini of ORs are involved in G-protein interactions. Our BLAST results have shown that the third intra-cellular loop shows strong sequence homology with several hundred vertebrate ORs, especially in the “RNRDMRG” sequence motif, which is invariably in the C-terminus, specifically, in the region where the seventh TM ends and the C-terminus begins. A few of the more than 900 results also show homology in the rest of this intra-cellular loop and the homolgous ORs are always in the C-terminus. This indicates to us that if a certain motif of amino acids interacts with the G-protein, then this motif is present in OR17-210 intracellularly. We conclude therefore that OR function is not hindered because of the presence of an intracellular third loop.
An extracellular C-terminus for mammalian ORs has also not been experimentally shown. The presence of an orphaned TM puts the C-terminus extracellularly. This is confirmed by two TM prediction programs. Experiments involving the attachment of a poly-Histidine tag to the end of the OR would confirm the extracellular C-terminus for hOR17-210.
Identifying the active and inactive states for an OR and elucidating its role in olfaction at a molecular level necessitates an experimental determination of its protein structure, in addition to knowledge of its odorant repertoire. There is currently, no experimentally derived structure of an olfactory receptor. This lack of a structure engenders the assumption that GPCRs are structurally similar to rhodopsin. Every computational study of olfactory receptors and other GPCRs uses a rhodopsin structure (the X-ray derived structure with the highest resolution [50] as used in our modeling here or a lower resolution electron diffraction structure [62]). Modifications to remove rhodopsin-specific biases as detailed in the Materials and Methods section begin from this point.
Computational modeling, docking and simulation studies [14, 36, 37, 48] have shown that the OR binding region is on the extracellular side—a pocket that is created by side chains belonging to TM regions 3, 4 5 and 6—which is confirmed by our docking results. Our docking results indicate that the ALA108 is the only residue in the binding pocket that is within Van der Waals distances with the ligands. These results are preliminary at best. Since all the odors docked in the same general area of the binding region, we believe that the binding region is apt for odor-OR interactions. We have previously shown that dynamic simulation of odorous molecules in the olfactory receptor binding pocket provide instances of interactions with key amino acid residues in the binding pocket [36]. These interactions are however, not always observed as a result of static docking. The time and computational effort required to complete the dynamic simulation of all odors identified that excite hOR17-210 strongly make it the subject of another paper. Certainly, site-directed mutagenesis results would provide us with better starting points in our docking and simulation studies. These results unfortunately, do not exist for this receptor.
Our standard model building protocols differ slightly from those previously established [37, 47, 48], by independently predicting the TM regions followed by removing of rhodopsin structure specific biases. In the case of hOR17-210, we are breaking new ground because we have identified and attempt to model a novel TM (TM7′) that has not been previously observed. Once TM7′ was placed in position of TM2 (our preferred structure), it was subject to the same TM-modeling protocol as other TMs in our model. Further validation of the novel method we introduced with Eq. 1 of calculating hydrophobic moments for determining TM rotations would also be aided by future experimental work followed by fine tuning of our modeling strategies.
Future work with experimental functional analyses following key point mutations would aid in identifying the role of binding pocket residues in ligand interactions. Simulating the dynamic motion of ligand in the OR binding pocket where its interactions with key residues in the binding pocket can vary over the time period of the simulation would be useful to identify if other residues are involved in the ligand–OR interaction. Also, computational docking shows that ligands tested (Fig. 6a, b) are clustered within a single region, we can only surmise from the docking results that this region is the preferred binding region for this OR.
Conclusion
This informatics-based study, supported by experimental results, identifies an OR possessing atypical sequence-structure features while still maintaining olfactory functionality. The human olfactory repertoire reveals that the ratio of functional ORs to pseudogenes is 1:2 (http://senselab.med.yale.edu/senselab/ORDB/humanOR.html). This number has been revised recently and will likely be further revised as more information becomes available and more genome-level experiments are carried out.
Evolutionarily, hOR17-210 could occupy a position of transition between functional and pseudogenic ORs. This receptor is a possible illustration of how loss in OR function may occur, namely, through mutations that create sequences unfavorable for transmembrane helical assembly. Our study, we hope, will cause researchers to reassess the sequence-structure–function correlates in olfactory receptors, and also the necessity to incorporate structural features in the classification of ORs and GPCRs.
We have used bioinformatics methods to show how and why a receptor appearing pseudogenic in several portions of the population can be functional in others. Functionality was confirmed by measuring the experimental, varying excitatory responses to odor ligands with different of functional groups. While hOR17-210 appears to lack the first two TMs typically observed in ORs, it possesses an additional TM present in only two other non-human olfactory receptors in all of GenBank. This TM, named TM7′, may preserve olfactory function within this OR (when functional) by maintaining the TM structure, thus protecting the binding odor ligand. The intracellular positions of regions identified as possibly responsible for olfactory function, due to their highly conserved nature, are preserved. hOR17-210 possibly straddles the point in mammalian OR evolution where loss of function occurs.
Acknowledgments
This work was generously supported by Grant 2 P01 DC 004732-05 from the National Institute for Deafness and Communicative Disorders, National Institutes of Health. P. L. was supported as a part-time undergraduate researcher at the Department of Neurobiology, Yale University School of Medicine. This work is also supported by a Faculty Development Grant (CC) at the University of Alabama at Birmingham.
Abbreviations
- OR
Olfactory Receptors
- GPCR
GTP-binding Protein Coupled Receptors
- HUGO
Human Genome Organization
- HORDE
Human Olfactory Receptor Database Exploratorium
- SPHGEN
Sphere Generator
- CVFF
Consistent Valence Force Field
- DMS
Dot Molecular Surface
Contributor Information
Peter C. Lai, Division of Natural Science, Mathematics, and Computing, Bard College at Simon's Rock, Great Barrington, MA, USA
Gautam Bahl, Department of Radiology, Wayne State University/Detroit, Medical Center, Detroit, MI, USA.
Maryse Gremigni, Laboratoire de Neuroglycobiologie, UMR 6149 CNRS, Université de Provence, Pole 3C, 3 Pl. V.Hugo, 13331 Marseille Cedex 3, France.
Valery Matarazzo, Laboratoire de Neuroglycobiologie, UMR 6149 CNRS, Université de Provence, Pole 3C, 3 Pl. V.Hugo, 13331 Marseille Cedex 3, France.
Olivier Clot-Faybesse, Laboratoire de Neuroglycobiologie, UMR 6149 CNRS, Université de Provence, Pole 3C, 3 Pl. V.Hugo, 13331 Marseille Cedex 3, France.
Catherine Ronin, Laboratoire de Neuroglycobiologie, UMR 6149 CNRS, Université de Provence, Pole 3C, 3 Pl. V.Hugo, 13331 Marseille Cedex 3, France.
Chiquito J. Crasto, Department of Neurobiology, Yale University School of Medicine, New Haven, CT 06516, USA; Yale Center for Medical Informatics, Yale University School of Medicine, New Haven, CT 06516, USA
References
- 1.Ji TH, Grossmann M, Ji I. G protein-coupled receptors. I. Diversity of receptor–ligand interactions. J Biol Chem. 1998;273:17299–17302. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- 2.Muller G. Towards 3D structures of G protein-coupled receptors: a multidisciplinary approach. Curr Med Chem. 2000;7:861–888. doi: 10.2174/0929867003374534. [DOI] [PubMed] [Google Scholar]
- 3.King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 4.Mombaerts P. Molecular biology of odorant receptors in vertebrates. Annu Rev Neurosci. 1999;22:487–509. doi: 10.1146/annurev.neuro.22.1.487. [DOI] [PubMed] [Google Scholar]
- 5.Abaffy T, Malhotra A, Luetje CW. The molecular basis for ligand specificity in a mouse olfactory receptor: a network of functionally important residues. J Biol Chem. 2007;282:1216–1224. doi: 10.1074/jbc.M609355200. [DOI] [PubMed] [Google Scholar]
- 6.Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruch RC, Rulli RD. Ligand binding specificity of a neutral l-amino acid olfactory receptor. Comp Biochem Physiol B. 1988;91:535–540. doi: 10.1016/0305-0491(88)90018-1. [DOI] [PubMed] [Google Scholar]
- 8.Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 10.Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr Opin Neurobiol. 2004;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Mombaerts P. Targeting olfaction. Curr Opin Neurobiol. 1996;6:481–486. doi: 10.1016/s0959-4388(96)80053-5. [DOI] [PubMed] [Google Scholar]
- 12.Oka Y, Omura M, Kataoka H, Touhara K. Olfactory receptor antagonism between odorants. EMBO J. 2004;23:120–126. doi: 10.1038/sj.emboj.7600032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothman A, Feinstein P, Hirota J, Mombaerts P. The promoter of the mouse odorant receptor gene M71. Mol Cell Neurosci. 2005;28:535–546. doi: 10.1016/j.mcn.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Touhara K. Odor discrimination by G protein-coupled olfactory receptors. Microsc Res Tech. 2002;58:135–141. doi: 10.1002/jemt.10131. [DOI] [PubMed] [Google Scholar]
- 15.Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc Natl Acad Sci USA. 1999;96:4040–4045. doi: 10.1073/pnas.96.7.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buck LB. Information coding in the vertebrate olfactory system. Annu Rev Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- 17.Shepherd GM. Toward a molecular basis for sensory perception. In: Gazzaniga MS, Bizzi E, editors. The cognitive neurosciences. MIT Press; Cambridge: 1995. pp. 105–137. [Google Scholar]
- 18.Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 19.Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci USA. 2004;101:2584–2589. doi: 10.1073/pnas.0307882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci USA. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zozulya S, Echeverri F, Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2:RESEARCH0018. doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quignon P, Giraud M, Rimbault M, Lavigne P, Tacher S, Morin E, Retout E, Valin AS, Lindblad-Toh K, Nicolas J, Galibert F. The dog and rat olfactory receptor repertoires. Genome Biol. 2005;6:R83. doi: 10.1186/gb-2005-6-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young JM, Shykind BM, Lane RP, Tonnes-Priddy L, Ross JA, Walker M, Williams EM, Trask BJ. Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 2003;4:R71. doi: 10.1186/gb-2003-4-11-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Firestein S. The olfactory receptor gene super-family of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 25.Niimura Y, Nei M. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene. 2005;346:13–21. doi: 10.1016/j.gene.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 26.Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D. Primate evolution of an olfactory receptor cluster: diversification by gene conversion and recent emergence of pseudogenes. Genomics. 1999;61:24–36. doi: 10.1006/geno.1999.5900. [DOI] [PubMed] [Google Scholar]
- 28.Menashe I, Man O, Lancet D, Gilad Y. Population differences in haplotype structure within a human olfactory receptor gene cluster. Hum Mol Genet. 2002;11:1381–1390. doi: 10.1093/hmg/11.12.1381. [DOI] [PubMed] [Google Scholar]
- 29.Matarazzo V, Clot-Faybesse O, Marcet B, Guiraudie-Capraz G, Atanasova B, Devauchelle G, Cerutti M, Etievant P, Ronin C. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem Senses. 2005;30:195–207. doi: 10.1093/chemse/bji015. [DOI] [PubMed] [Google Scholar]
- 30.Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314:657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- 31.Fuchs T, Glusman G, Horn-Saban S, Lancet D, Pilpel Y. The human olfactory subgenome: from sequence to structure and evolution. Hum Genet. 2001;108:1–13. doi: 10.1007/s004390000436. [DOI] [PubMed] [Google Scholar]
- 32.Visiers I, Ballesteros JA, Weinstein H. Three-dimensional representations of G protein-coupled receptor structures and mechanisms. Methods Enzymol. 2002;343:329–371. doi: 10.1016/s0076-6879(02)43145-x. [DOI] [PubMed] [Google Scholar]
- 33.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 34.Moller S, Vilo J, Croning MD. Prediction of the coupling specificity of G protein coupled receptors to their G proteins. Bioinformatics. 2001;17(Suppl 1):S174–S181. doi: 10.1093/bioinformatics/17.suppl_1.s174. [DOI] [PubMed] [Google Scholar]
- 35.Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci. 2000;3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- 36.Lai PC, Singer MS, Crasto CJ. Structural activation pathways from dynamic olfactory receptor–odorant interactions. Chem Senses. 2005;30:781–792. doi: 10.1093/chemse/bji070. [DOI] [PubMed] [Google Scholar]
- 37.Singer MS. Analysis of the molecular basis for octanal interactions in the expressed rat 17 olfactory receptor. Chem Senses. 2000;25:155–165. doi: 10.1093/chemse/25.2.155. [DOI] [PubMed] [Google Scholar]
- 38.John B, Sali A. Comparative protein structure modeling by iterative alignment, model building and model assessment. Nucleic Acids Res. 2003;31:3982–3992. doi: 10.1093/nar/gkg460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pirovano W, Feenstra KA, Heringa J. PRALINETM: a strategy for improved multiple alignment of transmembrane proteins. Bioinformatics. 2008;24:492–497. doi: 10.1093/bioinformatics/btm636. [DOI] [PubMed] [Google Scholar]
- 40.Floriano WB, Hall S, Vaidehi N, Kim U, Drayna D, Goddard WA., III Modeling the human PTC bitter-taste receptor interactions with bitter tastants. J Mol Model. 2006;12:931–941. doi: 10.1007/s00894-006-0102-6. [DOI] [PubMed] [Google Scholar]
- 41.Floriano WB, Vaidehi N, Goddard WA., III Making sense of olfaction through predictions of the 3-D structure and function of olfactory receptors. Chem Senses. 2004;29:269–290. doi: 10.1093/chemse/bjh030. [DOI] [PubMed] [Google Scholar]
- 42.Floriano WB, Vaidehi N, Goddard WA, Singer MS, III, Shepherd GM. Molecular mechanisms underlying differential odor responses of a mouse olfactory receptor. Proc Natl Acad Sci USA. 2000;97:10712–10716. doi: 10.1073/pnas.97.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Floriano WB, Vaidehi N, Zamanakos G, Goddard WA., III HierVLS hierarchical docking protocol for virtual ligand screening of large-molecule databases. J Med Chem. 2004;47:56–71. doi: 10.1021/jm030271v. [DOI] [PubMed] [Google Scholar]
- 44.Freddolino PL, Kalani MY, Vaidehi N, Floriano WB, Hall SE, Trabanino RJ, Kam VW, Goddard WA., III Predicted 3D structure for the human beta 2 adrenergic receptor and its binding site for agonists and antagonists. Proc Natl Acad Sci USA. 2004;101:2736–2741. doi: 10.1073/pnas.0308751101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall SE, Floriano WB, Vaidehi N, Goddard WA., III Predicted 3-D structures for mouse I7 and rat I7 olfactory receptors and comparison of predicted odor recognition profiles with experiment. Chem Senses. 2004;29:595–616. doi: 10.1093/chemse/bjh063. [DOI] [PubMed] [Google Scholar]
- 46.Man O, Gilad Y, Lancet D. Prediction of the odorant binding site of olfactory receptor proteins by human–mouse comparisons. Protein Sci. 2004;13:240–254. doi: 10.1110/ps.03296404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singer MS, Shepherd GM. Molecular modeling of ligand– receptor interactions in the OR5 olfactory receptor. Neuroreport. 1994;5:1297–1300. doi: 10.1097/00001756-199406020-00036. [DOI] [PubMed] [Google Scholar]
- 48.Vaidehi N, Floriano WB, Trabanino R, Hall SE, Freddolino P, Choi EJ, Zamanakos G, Goddard WA., III Prediction of structure and function of G protein-coupled receptors. Proc Natl Acad Sci USA. 2002;99:12622–12627. doi: 10.1073/pnas.122357199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melen K, Krogh A, von Heijne G. Reliability measures for membrane protein topology prediction algorithms. J Mol Biol. 2003;327:735–744. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- 50.Okada T. X-ray crystallographic studies for ligand–protein interaction changes in rhodopsin. Biochem Soc Trans. 2004;32:738–741. doi: 10.1042/BST0320738. [DOI] [PubMed] [Google Scholar]
- 51.Gschwend DA, Good AC, Kuntz ID. Molecular docking towards drug discovery. J Mol Recognit. 1996;9:175–186. doi: 10.1002/(sici)1099-1352(199603)9:2<175::aid-jmr260>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 52.Gschwend DA, Kuntz ID. Orientational sampling and rigid-body minimization in molecular docking revisited: on-the-fly optimization and degeneracy removal. J Comput Aided Mol Des. 1996;10:123–132. doi: 10.1007/BF00402820. [DOI] [PubMed] [Google Scholar]
- 53.Richards FM. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 54.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 55.Burger M, Burger JA, Hoch RC, Oades Z, Takamori H, Schraufstatter IU. Point mutation causing constitutive signaling of CXCR2 leads to transforming activity similar to Kaposi's sarcoma herpesvirus-G protein-coupled receptor. J Immunol. 1999;163:2017–2022. [PubMed] [Google Scholar]
- 56.Scheer A, Fanelli F, Costa T, De Benedetti PG, Cotecchia S. Constitutively active mutants of the alpha 1B-adrenergic receptor: role of highly conserved polar amino acids in receptor activation. EMBO J. 1996;15:3566–3578. [PMC free article] [PubMed] [Google Scholar]
- 57.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otaki JM, Firestein S. Length analyses of mammalian G-protein-coupled receptors. J Theor Biol. 2001;211:77–100. doi: 10.1006/jtbi.2001.2272. [DOI] [PubMed] [Google Scholar]
- 59.Olender T, Feldmesser E, Atarot T, Eisenstein M, Lancet D. The olfactory receptor universe—from whole genome analysis to structure and evolution. Genet Mol Res. 2004;3:545–553. [PubMed] [Google Scholar]
- 60.Yohannan S, Faham S, Yang D, Whitelegge JP, Bowie JU. The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors. Proc Natl Acad Sci USA. 2004;101:959–963. doi: 10.1073/pnas.0306077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katada S, Touhara K. A molecular basis for odorant recognition: olfactory receptor pharmacology. Nippon Yakurigaku Zasshi. 2004;124:201–209. doi: 10.1254/fpj.124.201. [DOI] [PubMed] [Google Scholar]
- 62.Krebs A, Villa C, Edwards PC, Schertler GF. Characterisation of an improved two-dimensional p22121 crystal from bovine rhodopsin. J Mol Biol. 1998;282:991–1003. doi: 10.1006/jmbi.1998.2070. [DOI] [PubMed] [Google Scholar]


