Abstract
It is now understood that virtually all human cancer types are the result of the accumulation of both genetic and epigenetic changes. DNA methylation is a molecular modification of DNA that is crucial for normal development. Genes that are rich in CpG dinucleotides are usually not methylated in normal tissues, but are frequently hypermethylated in cancer. With the advent of high-throughput platforms, large-scale structure of genomic methylation patterns is available through genome-wide scans and tremendous amount of DNA methylation data have been recently generated. However, sophisticated statistical methods to handle complex DNA methylation data are very limited. Here we developed a likelihood based Uniform-Normal-mixture model to select differentially methylated loci between case and control groups using Illumina arrays. The idea is to model the data as three types of methylation loci, one unmethylated, one completely methylated, and one partially methylated. A three-component mixture model with two Uniform distributions and one truncated normal distribution was used to model the three types. The mixture probabilities and the mean of the normal distribution were used to make inference about differentially methylated loci. Through extensive simulation studies, we demonstrated the feasibility and power of the proposed method. An application to a recently published study on ovarian cancer identified several methylation loci that are missed by the existing method.
Keywords: DNA methylation, mixture model, case-control designs
Introduction
It is now understood that virtually all human cancer types are the result of the accumulation of both genetic and epigenetic changes [Jones and Baylin, 2002; Herman and Baylin, 2003; Feinberg and Tycko, 2004; Lund and Lohuizen, 2004; Baylin and Ohm, 2006; Egger et al., 2004; Kulis and Esteller, 2010; Kalari and Pfeifer, 2010; Kerkel et al., 2010]. Pathological epigenetic changes, that is, non-sequence-based alterations that are inherited are increasingly recognized as alternatives to mutations and chromosomal alterations in disrupting gene function [Egger et al., 2004]. DNA methylation, the addition of a methyl group to the 5’ position of cytosine in the context of a CpG dinucleotide, is a molecular modification of DNA that is crucial for normal development. Aberrant epigenetic mechanisms include global DNA hypomethylation, chromatin alterations, hypermethylation and hypomethylation of specific genes that influence the transcription of genes important to the cancer process. All of these can lead to aberrant activation of growth-promoting genes and aberrant silencing of tumor-suppressor genes [Feinberg and Tycko, 2004]. Genes that are rich in CpG dinucleotides are usually not methylated in normal tissues, but are frequently hypermethylated in cancer. This is often associated with gene silencing [Jones and Laird, 1999] and is an important mechanism for the inactivation of tumor suppressor genes. In more recent studies on DNA methylation and cancers, Eberth et al. [2010] found that CD44 may be a promising new epigenetic marker for diagnosis and a potential therapeutic target for the treatment of specific lymphoma subtypes as it is found to be epigenetically regulated in lymphoma and undergoes de novo methylation in distinct lymphoma subtypes; Bediaga et al. [2010] found that DNA methylation profiles may enable breast cancer subtype prediction; Christensen et al. [2010] found that breast cancer prognostic characteristics and risk-related exposures appear to be associated with gene-specific tumor methylation and overall methylation pattern; and Lugthart et al. [2011] found aberrant DNA hypermethylation in acute myeloid leukemia among many other studies. Apart from the importance in DNA methylation with cancer development, what makes DNA methylation even more interesting is, unlike genetic alterations, DNA methylation is reversible. Thus, to decode the human epigenome to understand DNA methylation alterations in tumorigenesis may help us with new therapeutic approaches.
DNA methylation has historically been studied in a locus-targeted manner. However, with the advent of high-throughput platforms, large-scale structure of genomic methylation patterns is available through genome-wide scans. Two high-throughput platforms that have been popularly used include the Illumina Infinium Human Methylation27 array and the Illumina GoldenGate array. Both arrays are based on genotyping bisulfite-converted DNA. DNA samples are treated with a methylation kit that converts unmethylated cytosines to uracils, whereas methylated cytosines are protected and remain cytosine. Therefore, whether the base at a given locus is converted or not provides information on its original methylation status. The results of the array, the methylation status of the interrogated CpG site is a sequence of β-values, one for each locus, calculated as the average of approximately 30 replicates (with approximately 30 beads per site per sample) of the quantity max(M, 0)/(max(U,0) + max(M,0) + 100). Here U is the fluorescent signal from an unmethylated allele on a single bead, M is that from a methylated allele. A maximum between signal intensity and 0 is chosen to compensate for negative signals due to background subtraction. The constant 100 is to regularize β-values when both M and U values are small [Bibikova et al., 2006]. This β-value ranges continuously from 0 (unmethylated) to 1 (completely methylated) and reflects the methylation level of each CpG site.
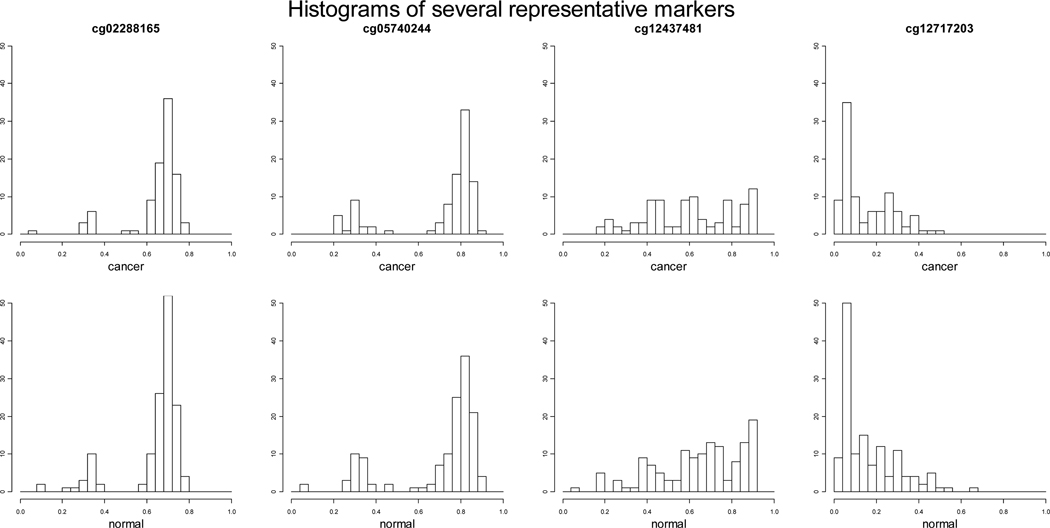
Recently tremendous amounts of DNA methylation data have been generated from high-throughput DNA methylation platforms. However, sophisticated statistical methods to handle complex DNA methylation data, especially data measured with proportions from popularly used commercial platforms such as Illumina, are either nonexistent or very limited. Currently, to select differentially methylated loci, researchers mainly apply either parametric methods such as regression-based methods or t-test or nonparametric methods such as rank sum test. Some research has been done on tumor type classifications using DNA methlyation data. Siegmund et al. [2004] introduced a Bernoulli-lognormal mixture model for classifying DNA methylation data generated using MethyLight. MethyLight results in percentages of methylated reference (PMR) and frequently contains an “excess” of zeros. Houseman et al. [2008] proposed a beta-mixture model to classify different tissue types using Illumina arrays. Most recently, Kuan et al. [2010] investigated issues in quality control steps. We noticed from many studies that β-values generated by BeadStudio with Illumina arrays usually have a heavy tail close to zero which represents unmethylated and a bump close to one which represents completely methylated. Figure 1 displays histograms of DNA methylation measures of several representative markers in cancer and normal groups using the data from the United Kingdom Ovarian Cancer Population Study (UKOPS) [Teschendorff et al., 2010]. Existing methods do not take this unique feature into account. Although DNA methylation measures generated by BeadStudio are continuous measures between 0 and 1, on the molecular level, some loci are unmethylated, some are completely methylated, and some are hemi-methylated, i.e., the cytosine is only methylated in one strand but not in the other (Human Molecular Genetics, 3rd Edition). In this paper, we developed a likelihood based Uniform-Normal-mixture model to select differentially methylated loci between case and control groups. The idea is to model the data as three types of methylation loci, one unmethylated, one completely methylated, and one partially methylated. A three-component mixture model with two Uniform distributions and one truncated normal distribution was used to model the three types. The mixture probabilities and the mean of the normal distribution were used to make inference about differentially methylated loci. Through extensive simulation studies, we demonstrated the feasibility and power of the proposed method. We further applied the proposed method to the United Kingdom Ovarian Cancer Population Study to select differentially methylated loci between ovarian cancer cases and age-matched healthy controls using Illumina Infinium Human Methylation27 Beadchip [Teschendorff et al., 2010] and identified some methylation loci that are missed by the existing method.
Figure 1.
Histograms of DNA methylation measures of several representative markers in cancer and normal groups from the ovarian cancer data.
Method
For each DNA methylation marker, let yi,i = 1,…,n denote independent observations of β-values representing DNA methylation levels, where n is the number of subjects in one group. Let τ1 and τ2 be two threshold values (0 < τ1 < 0.5 < τ2 <1) conditional on which we have two Uniform distributions representing two components, U[0,τ1] for unmethylated and U[τ2,1] for completely methylated. The third component is a truncated normal component for those partially methylated loci. Under a Uniform-Normal-mixture model, where yi ’s are assumed to be a mixture of three different methylation patterns, let zi, i = 1,…,n be the latent indicator variable for each observation yi that determines the component from which the observation originates, where
We consider {yi, zil} as the “complete data”. Here zi = (zi1,zi2,zi3)' follows a 3-category multinomial distribution with probabilities π = (π1,π2,π3)', and πl is the prior distribution for zi and π1 + π2 + π3 = 1. Thus, we have
where N[0,1](yi | μ,σ2) is the truncated normal density truncated at 0 and 1. The likelihood of observing n subjects is thus:
Here θ = (τ1,τ2,π1,π2,μ,σ) are the unknown parameters representing “mixture” proportions and the parameters of the Uniform and normal distributions.
At a specific methylation locus k, assuming two different sets of parameters for case and control groups, parameters are θ1 = (τ1,τ2,π1,π2,μ,σ) for the case group and for the control group, where τ1, and τ2 are the same for both groups. The joint likelihood of observing n1 independent cases and n2 independent controls at DNA methylation locus k is L(y | θ) = L(yi | θ1)L(yj | θ2). In estimating the parameters, we apply a numerical estimation using the profile likelihood coupled with an Expectation-Maximization (EM) algorithm [Dempster et al., 1977]. The likelihood of (τ1,τ2) is defined for each fixed (τ1,τ2) (0 < τ1 < 0.5 < τ2 <1) by maximizing L(τ1,τ2,π1,π2,μ,σ) over (π1,π2,μ,σ) and maximizing using the EM algorithm. That is, . The maximum likelihood estimators of (τ1,τ2) are defined by maximizing the profile likelihood, that is, (τ̂1,τ̂2) = argmax Lp(τ1,τ2). The EM algorithm is applied to estimate the log-likelihood and obtain the maximum likelihood estimates (MLE) of the parameters (π1,π2,μ,σ) for the case group and MLE of the parameters for the control group separately, with the E-step and the M-step updated iteratively. Using the case group as an example, to get the MLE of the parameters (π1,π2,μ,σ) for the case group, the EM algorithm proceeds as follows:
Step 1: Start with a set of initial best guesses of the parameters ;
- Step 2 (E-step): Given the current estimates of the parameters , compute the posterior probabilities for all cases i = 1,…,n1, and l = 1,2,3:
- Step 3: (M-step): Update the parameters :
Step 4: Repeat the E-step and the M-step until convergence.
To test the null hypothesis that locus k is not differentially methylated between case and control groups, we test , and μ = μ'. That is, we compare the mixture probabilities and mean of the normal distribution between case and control groups. The corresponding alternative hypothesis is H1 : not all π1's,π2's, and μ's are the same. We use the Likelihood Ratio Test (LRT) with 3 degrees of freedom (dfs) to test H0. The LRT statistic will be compared to a χ2 distribution with 3 dfs to assess significance. To obtain the MLE of the parameters under H0, the same profile likelihood coupled with the EM algorithm is applied, while the steps of the EM algorithm are changed. The joint likelihood of observing n1 independent cases and n2 independent controls at DNA methylation locus j under H0 is:
The EM algorithm to get the MLE of the parameters (π1,π2,μ,σ,σ') proceeds as follows:
Step 1: Start with a set of initial best guesses of the parameters ;
- Step 2 (E-step): Given the current estimates of the parameters , compute the posterior probabilities for all cases i = 1,…,n1, and controls j = 1,…,n2, and l = 1,2,3:
- Step 3: (M-step): Update the parameters :
Step 4: Repeat the E-step and the M-step until convergence.
We test the K DNA methylation loci independently. To adjust for multiple comparisons, a False Discovery Rate (FDR) q-value [Storey and Tibshirani, 2003] of ≤ 0.05 as significant for the examination of K loci could be applied.
Results
In this section, using extensive simulation studies, the performance of the proposed method is compared to that of the two-sample t-test. Method that assumes a general beta distribution for proportion measures like DNA methylation measures has not been applied by researchers partly due to its lack of biologically meaningful interpretation in parameters. Therefore, only t-test is considered as an alternative method to the proposed method. The simulation procedures were repeated 10,000 times to evaluate the Type I error rates and 1,000 times to evaluate power.
Simulation Setup
In the simulation studies to evaluate Type I error rates and power, the total sample size was fixed at n1 = 250 cases and n2 = 250 controls. To mimic the distribution of β-values generated by BeadStudio with Illumina arrays, which usually have a heavy tail close to zero (unmethylated) and a bump close to one (completely methylated), we set τ1,τ2 at different values, τ1 = 0.1, τ2 = 0.9 and τ1 = 0.15, τ2 = 0.85. With fixed τ1,τ2, we considered different parameter settings with different mixture probabilities π1,π2, and π3, and different mean and standard deviation (μ,σ) for the truncated normal distribution N[0,1]. Therefore, within case and control groups, for each methylation measure, it has a probability π1 to be generated from a Uniform distribution U[0,τ1], a probability π3 to be generated from a Uniform distribution U[τ2,1], and a probability π2 to be generated from a truncated normal distribution N[0,1](μ,σ).
We chose different parameter settings such that case and control groups would be different in term of the percentage of unmethylated, the percentage of completely methylated, or the mean methylation level of partially methylated.
Type I error
In order to evaluate the Type I error rates for the proposed test, data were generated under the null hypothesis that a specific locus k is not differentially methylated between case and control groups, i.e., , and μ = μ'. The simulation procedure was repeated 10,000 times. Type I error rates of the proposed method and the two-sample t-test were then estimated by the proportion of times that the null hypothesis of locus k is not differentially methylated between case and control groups was rejected by the two methods, respectively.
Table 1 displays the Type I error rates to detect differentially methylated loci with the proposed method and the t-test under different parameters settings. The nominal Type I error rate of 0.05 was well controlled by both the proposed method and the t-test although the proposed method is a little more conservative than the t-test.
Table 1.
Type I error rates to detect differentially methylated loci at the 0.05 significance level with the proposed method and the two-sample t-test under different parameter settings*.
| Parameter settings | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| π1 | 0.3 | 0.4 | 0.4 | 0.5 | 0.4 | |
| π2 | 0.5 | 0.5 | 0.5 | 0.1 | 0.2 | |
| π3 | 0.2 | 0.1 | 0.1 | 0.4 | 0.4 | |
| μ | 0.3 | 0.2 | 0.3 | 0.3 | 0.2 | |
| σ | 0.1 | 0.1 | 0.2 | 0.2 | 0.1 | |
| τ1 = 0.10 τ2 = 0.90 |
t-test | 0.0491 | 0.0509 | 0.0518 | 0.0487 | 0.0474 |
| Proposed | 0.0504 | 0.0349 | 0.0460 | 0.0414 | 0.0441 | |
| τ1 = 0.15 τ2 = 0.85 |
t-test | 0.0517 | 0.0463 | 0.0500 | 0.0530 | 0.0478 |
| Proposed | 0.0554 | 0.0248 | 0.0387 | 0.0407 | 0.0431 | |
Total sample size was fixed at n1 = 250 cases and n2 = 250 controls; two nuisance parameters τ1,τ2 were fixed at τ1 = 0.1 and τ2 = 0.9 or τ1 = 0.15 and τ2 = 0.85; simulation procedure was repeated 10,000 times.
Power
To assess the performance of the proposed method, three scenarios of the parameter settings were considered. Table 2 displays power results to detect differentially methylated loci between case and control groups with the proposed method and the t-test under the three scenarios when τ1,τ2 are set at τ1 = 0.1, τ2 = 0.9. Power was assessed with 1,000 simulations.
Table 2.
Power to detect differentially methylated loci with the proposed method and the two-sample t-test under different parameter settings.
| §Scenario 1: | Parameter Settings |
1 | 2 | 3 | 4 | ||||
| Case | Control | Case | Control | Case | Control | Case | Control | ||
| π1 | 0.2 | 0.3 | 0.4 | 0.4 | 0.2 | 0.3 | 0.3 | 0.4 | |
| π2 | 0.5 | 0.4 | 0.5 | 0.4 | 0.5 | 0.4 | 0.3 | 0.3 | |
| π3 | 0.3 | 0.3 | 0.1 | 0.2 | 0.3 | 0.3 | 0.4 | 0.3 | |
| μ | 0.3 | 0.4 | 0.3 | 0.2 | 0.3 | 0.25 | 0.3 | 0.4 | |
| σ | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | |
| Sample Mean* | 0.445 | 0.461 | 0.265 | 0.292 | 0.445 | 0.401 | 0.486 | 0.425 | |
| t-test | 0.088 | 0.160 | 0.271 | 0.418 | |||||
| Proposed | 1.0 | 1.0 | 0.979 | 1.0 | |||||
| §Scenario 2: | Parameter Settings |
1 | 2 | 3 | 4 | ||||
| Case | Control | Case | Control | Case | Control | Case | Control | ||
| π1 | 0.4 | 0.4 | 0.3 | 0.2 | 0.3 | 0.3 | 0.4 | 0.4 | |
| π2 | 0.2 | 0.2 | 0.5 | 0.6 | 0.5 | 0.5 | 0.4 | 0.3 | |
| π3 | 0.4 | 0.4 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.3 | |
| μ | 0.4 | 0.3 | 0.3 | 0.3 | 0.3 | 0.2 | 0.3 | 0.3 | |
| σ | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | |
| Sample Mean* | 0.481 | 0.465 | 0.354 | 0.380 | 0.355 | 0.307 | 0.330 | 0.395 | |
| t-test | 0.073 | 0.151 | 0.380 | 0.544 | |||||
| Proposed | 0.272 | 0.564 | 1.0 | 0.703 | |||||
| §Scenario 3: | Parameter Settings |
1 | 2 | 3 | 4 | ||||
| Case | Control | Case | Control | Case | Control | Case | Control | ||
| π1 | 0.3 | 0.2 | 0.3 | 0.3 | 0.5 | 0.1 | 0.4 | 0.3 | |
| π2 | 0.5 | 0.5 | 0.5 | 0.5 | 0.3 | 0.7 | 0.5 | 0.4 | |
| π3 | 0.2 | 0.3 | 0.2 | 0.2 | 0.2 | 0.2 | 0.1 | 0.3 | |
| μ | 0.4 | 0.4 | 0.4 | 0.2 | 0.3 | 0.3 | 0.3 | 0.2 | |
| σ | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | |
| Sample Mean* | 0.405 | 0.496 | 0.404 | 0.309 | 0.304 | 0.406 | 0.265 | 0.383 | |
| t-test | 0.866 | 0.897 | 0.940 | 0.977 | |||||
| Proposed | 0.769 | 1.0 | 1.0 | 1.0 | |||||
Total sample size was fixed at n1 = 250 cancer cases and n2 = 250 normal controls; two nuisance parameters τ1,τ2 were fixed at τ1 = 0.1 and τ2 = 0.9; simulation procedure was repeated 1,000 times.
Sample mean over 1,000 simulated data set with 250 cases, and 250 controls.
Scenario 1: parameter settings where 1) there are differences in both the mixture probabilities πl 's and the mean μ of the normal distribution component between case group and control group, 2) but the difference between the overall means of the methylation percentage of the case group and the control group is small.
Scenario 2: parameter settings where 1) there are difference in either the mixture probabilities πl 's or the mean μ of the normal distribution component between case group and control group, 2) but the difference between the overall means of the methylation percentage of the case group and the control group is small.
Scenario 3: the parameter settings where 1) there is difference in both/either the mixture probabilities πl 's and/or the mean μ of the normal distribution component between case group and control group, 2) and the difference between the overall means of the methylation percentage of the case group and the control group is large.
In Scenario 1, we considered the parameter settings when the following two conditions are met: 1) there are difference in both the mixture probabilities πl 's and the mean of the normal distribution component μ between the case and control groups, 2) but the difference between the overall methylation levels of the case and control groups is small. This is the scenario when there is no much difference in the overall mean of the methylation percentage but there is difference in all three mixture components between the case and control groups. As we expected, the proposed method has almost 100% power for all settings considered while the t-test has very small power. The sample means provide in the table are the sample means over 1,000 simulations for case and control groups separately. Also expected, the power of the t-test increases as the difference between the overall methylation level in the two groups increases. More specifically, in parameter settings 1 and 2, when the sample difference between the case and control groups in methylation percentage is only about 0.014 and 0.027, there is no power at all with the t-test. But the proposed method has 100% power in both settings.
In Scenario 2, we considered the parameter settings when 1) there is difference in either the mixture probabilities πl 's or the mean of the normal distribution component μ between the case and control groups, 2) but the difference between the overall methylation levels of the case and control groups is small. This is the scenario when there is no much difference in the overall mean of the methylation percentage but there is difference in some of the mixture components. As we expected, the proposed method has much greater power than the t-test in all settings considered. More specifically, in parameter settings 1 and 3, case and control groups are different only in terms of mixture probabilities. In parameter settings 2 and 4, case and control groups are different only in terms of the mean of the normal distribution mixture component. The power of the proposed method is much higher than that of the t-test in all settings but the power difference is not as big as in Scenario 1.
In Scenario 3, we considered the parameter settings when 1) there is difference in both/either the mixture probabilities πl 's and/or the mean of the normal distribution component μ between the case and control groups, 2) and the difference between the overall methylation levels of the case and control groups is large. We can see that in this scenario, the t-test that tests for difference between overall means has good powers on all settings. Note that, under the parameter setting 1 when the difference between overall methylation levels between the case and control groups is large and comparable to the difference between either the mixture probabilities or the mean of the partially methylated group, the two-sample t-test has slightly higher power than the proposed method.
Simulations with Special Settings
We also investigated simulation settings when there is no data in one of the three components. Table 3 displays both Type I error rates and power under such scenarios. When we do not observe data in one of the three components, the Type I error rates are still well preserved by the proposed method and the t-test. In terms of power, the proposed method has much greater power than the t-test if the difference between the overall methylation levels of the case and control groups is small (scenarios 1 and 3). When the difference is big, the two methods have similar power (scenario 2).
Table 3.
Type I error and power to detect differentially methylated loci for the proposed method and the two-sample t-test under several special parameter settings.
| Parameter Settings | 1 | 2 | 3 | ||||
|---|---|---|---|---|---|---|---|
| π1 | 0.6 | 0.6 | 0 | ||||
| π2 | 0.4 | 0 | 0.3 | ||||
| π3 | 0 | 0.4 | 0.7 | ||||
| μ | 0.2 | 0 | 0.25 | ||||
| σ | 0.1 | 0 | 0.1 | ||||
| Type I error | t-test | 0.046 | 0.052 | 0.049 | |||
| Proposed | 0.011 | 0.055 | 0.039 | ||||
| Parameter Settings | 1 | 2 | 3 | ||||
| Case | Control | Case | Control | Case | Control | ||
| π1 | 0 | 0 | 0.6 | 0.4 | 0.6 | 0.7 | |
| π2 | 0.2 | 0.3 | 0 | 0 | 0.4 | 0.3 | |
| π3 | 0.8 | 0.7 | 0.4 | 0.6 | 0 | 0 | |
| μ | 0.3 | 0.2 | 0 | 0 | 0.2 | 0.3 | |
| σ | 0.1 | 0.1 | 0 | 0 | 0.1 | 0.1 | |
| Sample Mean* | 0.801 | 0.754 | 0.409 | 0.591 | 0.112 | 0.125 | |
| Power | t-test | 0.390 | 0.944 | 0.241 | |||
| Proposed | 1.0 | 0.954 | 0.978 | ||||
Total sample size was fixed at n1 = 250 cancer cases and n2 = 250 normal controls; two nuisance parameters τ1,τ2 were fixed at τ1 = 0.1 and τ2 = 0.9; simulation procedure was repeated 1,000 times.
Sample mean over 1,000 simulated data set with 250 cases, and 250 controls.
We note that in all simulation settings considered, all parameter estimates are very close to the true values (data not shown).
Real Data Application
We applied the proposed method to the United Kingdom Ovarian Cancer Population Study (UKOPS) to select differentially methylated loci between ovarian cancer cases and age-matched healthy controls using Illumina Infinium Human Methylation27 Beadchip [Teschendorff et al., 2010]. The original data has 266 cases with 131 pre-treatment cases and 135 post-treatment cases, and 274 age-matched healthy controls. As whether or not patients have received treatment and age when blood samples were taken are known factors to affect DNA methylation levels, we chose to use a more homogenous population with 131 ovarian cancer cases who gave their blood at the time of their diagnosis prior to treatment and with age-matched controls to illustrate the feasibility and power of the proposed method. For quality control of the DNA methylation data, we removed samples with a low bisulfite (BS) conversion efficiency (BS control intensity values < 4,000); we also removed batches 10–12 due to over representation of controls on these batches and outliers using a quantile filtering [Teschendorff et al., 2010]; lastly, we required at least 95% CpG coverage per sample and at least 50 cases and 50 controls per locus. These quality control steps resulted in a β-valued data matrix of dimension of 22,951×(96 cases + 136 controls).
Because of the relatively small sample size in the real data application, we propose to assess the significance level associated with the proposed statistic and the t test using a permutation procedure rather than relying on the asymptotic distributions. We permuted the disease status among cases and controls, which generated a new data set in which the null hypothesis that no DNA methylation marker is associated with the disease status holds. We repeated the permutation procedure 1,000 times to generate the distribution of the test statistics under the null hypothesis. Thus, the p-value to testing if a marker is differentially methylated between case and control groups is the proportion of times the statistics from the permuted data are equal or greater than the statistics from the observed data [Westfall and Young, 1993]. We did not adjust for multiple comparisons as the purpose of the real data application is to demonstrate the feasibility and power of the proposed method rather than to identify DNA methylation loci for further study.
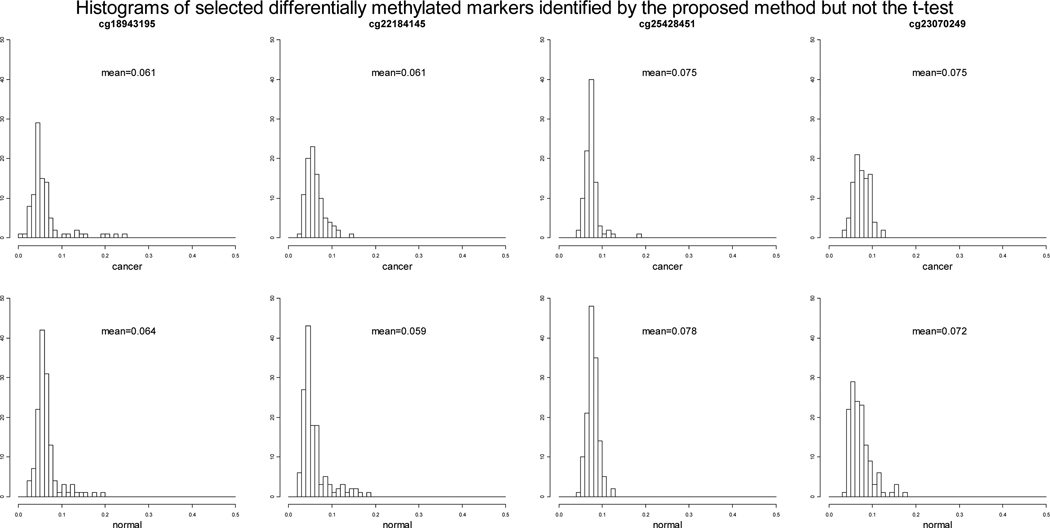
Of the 22,951 loci tested, 1,473 loci have permuted p-values 0.001 using the proposed method, while 1,655 loci have permuted p-values 0.001 with the t-test. Among those, there are overlapping 1,412 loci that have permuted p-values 0.001 using both methods. We further examined the loci that are identified by the proposed method but not by the t-test, and loci that are identified by the t-test but not by the proposed method. Out of the 61 loci that have permuted p-values 0.001 using the proposed methods but permuted p-values > 0.001 using the t-test, 16 loci have permuted p-values > 0.05 using the t-test. We selected 4 representative loci among the 16 and plotted the histograms of the β values of the case and control groups (Figure 2). Also included in the plots are overall mean methylation levels of case and control groups at each marker. It is clear that for the two loci from the left (cg18943195 and cg22184145) the percentage of unmethylated samples are different between case and control groups while the difference between the overall mean methylation levels of the case and control groups are as small as 0.002 or 0.003. Similarly, for the two loci from the right (cg25428451 and cg23070249) the average methylation levels of partially unmethylated samples are different between case and control groups while the difference between the overall mean methylation levels of the case and control groups are as small as 0.003. In these cases, the t-test is not able to detect the difference while the proposed method is. As we pointed out that the purpose of this ovarian cancer data application is to demonstrate the feasibility and power of the proposed method rather than to identify new DNA methylation loci for further study, we only want to comment on some potential interesting methylation loci among the 16 loci that have been identified by the proposed method but not the t-tests. For example, the identified locus cg18053505, located on chromosome 3, is on gene PCAF, which has been detected in primary esophageal squamous cell carcinoma (ESCC) tumors and ESCC cell lines as a candidate tumor suppressor gene [Qin et al., 2008]; the identified locus cg11375622, located on chromosome 8, is on gene BOP1, which has been suggested to play an oncogenic role in hepatocellular carcinoma by promoting epithelial-to-mesenchymal transition [Chung et al., 2011].
Figure 2.
Histograms of the DNA methylation β values of the cancer and normal groups at four selected differentially methylated loci identified by the proposed method but not the t-test.
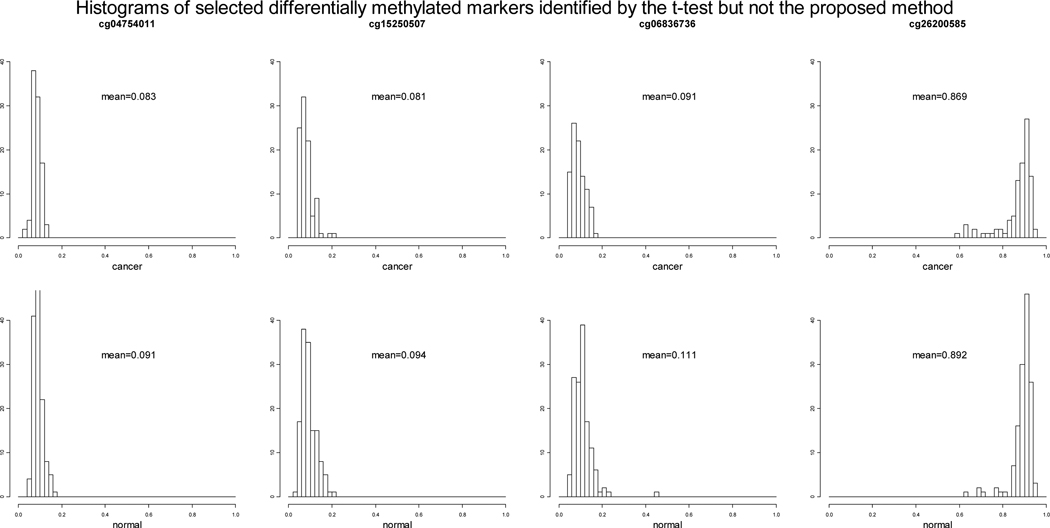
Out of the 158 loci that have permuted p-values 0.001 using the t-test but permuted p-values > 0.001 using the proposed method, 146 loci have permuted p-values < 0.005 using the proposed method, only 8 loci have permuted p-values > 0.05 using the proposed method. We selected 4 representative loci among the 8 and plotted the histograms of the β values of the case and control groups (Figure 3). We can see that the case and control groups are not much different in terms of the three components at the 4 loci (cg04754011, cg15250507, cg06836736 and cg26200585) but the difference between the overall mean methylation levels of the case and control groups are more than 0.01. The permuted p-values of the 16 markers that are identified by the proposed method but not the t-test and the permuted p-values of the 8 markers that are identified by the t-test but not the proposed method are displayed in Table 4.
Figure 3.
Histograms of the DNA methylation β values of the cancer and normal and groups at four selected differentially methylated loci identified by the t-test but not the proposed test.
Table 4.
Permuted p-values of the markers identified by one of the methods (with permuted p-values 0.001) but not both methods using the ovarian cancer data.
| Marker | Proposed | t-test |
|---|---|---|
| Part I: Markers identified by the proposed method but not the t-test | ||
| cg02136132 | 0.001 | 0.380 |
| cg02508567 | 0.001 | 0.462 |
| cg05500015 | 0.001 | 0.164 |
| cg08876665 | 0.001 | 0.620 |
| cg11375622 | 0.001 | 0.879 |
| cg13682722 | 0.001 | 0.083 |
| cg13830624 | 0.001 | 0.537 |
| cg18053505 | 0.001 | 0.056 |
| cg18943195 | 0.001 | 0.562 |
| cg20812929 | 0.001 | 0.337 |
| cg22184145 | 0.001 | 0.523 |
| cg23070249 | 0.001 | 0.267 |
| cg23311628 | 0.001 | 0.223 |
| cg24530795 | 0.001 | 0.173 |
| cg25163476 | 0.001 | 0.209 |
| cg25428451 | 0.001 | 0.127 |
| Part II: Markers identified by the t-test but not the proposed method | ||
| cg03251079 | 0.058 | 0.001 |
| cg04754011 | 0.163 | 0.001 |
| cg05595345 | 0.803 | 0.001 |
| cg06836736 | 0.062 | 0.001 |
| cg14094960 | 0.074 | 0.001 |
| cg15250507 | 0.055 | 0.001 |
| cg18239753 | 0.058 | 0.001 |
| cg26200585 | 0.482 | 0.001 |
Discussion
With the importance of epigenetic changes on cancer development well understood, many research efforts have been devoted to searching for differentially methylated loci between cancer and normal patients to see the possible contribution of methlyation process on cancer development. However, the unique pattern we observed in methylation measurements generated by the Illumina methylation arrays that have been widely applied has been ignored by the existing methods to select differentially methylated loci. We observed that there is an enrichment in values close to “0” as unmethylated and an enrichment in values close to “1” as completely methylated. With the understanding that on the molecular level some loci are unmethylated, some are completely methylated, and some are hemi-methylated, i.e., the cytosine is only methylated in one strand but not in the other, we proposed a likelihood-based Uniform-Normal-mixture model to select differentially methylated loci between case and control groups. The proposed method models methylation loci as three types, unmethylated, completely methylated, and partially methylated. A three-component mixture model with two Uniform distributions and one truncated normal distribution is used to model the three types. To make inference about differential methylation, we compare the mixture probabilities and mean of the normal distribution between case and control groups. The rational underlying the proposed method is that it will be more sensitive to the difference in any methlyation patterns between two groups than the existing methods that search for overall difference. For example, in the situation when case and control groups differ in terms of the percentages of unmethylated or completely methylated but the mean methylation levels in case and control groups are similar, the proposed method is expected to be powerful to detect the difference in the distribution while existing methods such as the t-test that focus on detecting the difference in means do not have any power.
The simulation results illustrated the feasibility and power of the proposed method. The proposed method that takes into account the unique pattern observed in the methylation percentage measurements has higher power to detect differentially methylated loci than the two-sample t-test under almost all parameter settings considered. Especially under the settings when the difference between the overall methylation levels in case and control groups is small but there are differences in the proportions of unmethylated, partially methylated, or completely methylated, or there is difference in the mean methylation level of the partially methlyated component. Under this setting, the t-test has almost no power but the proposed method has almost perfect power. When the difference between the overall means of methylation proportions in case and control groups is large, both the t-test and the proposed method have good power. We need to note that when the difference between the overall methylation levels of case and control groups is large and comparable to the difference between either the mixture probabilities or the mean of the partially methylated group, the t-test has slightly higher power than the proposed method. Thus we believe the proposed method is a nice complement to the robust t-test. An application to a recently published study on ovarian cancer suggested that in the majority of the cases, the p-values generated using the proposed method and the t-test agree with each other. Among all the loci with permuted p-values 0.001 using both methods, 82.2% loci overlap. Among the non-overlapping loci, 15 loci have permuted p-values 0.001 with the proposed method but permuted p-values > 0.05 with the t-test. Further examination of those 15 loci suggested large differences in either the percentage of complete unmethylation or the level of partial methylation but small difference in the overall mean methylation level.
In this study, we have demonstrated the power and feasibility of the proposed three-component mixture model to detect differentially methylated loci with a case-control design. Although the results from the application on the United Kingdom Ovarian Cancer Population Study are promising, we note the limitations of the proposed three-component mixture model, and future studies are needed to improve the current method or to develop new methods for the detection of differentially methylated loci. With the mixture model, usually a large sample size is needed to obtain accurate estimates. Also the optimization of the two nuisance parameters τ1 and τ2 complicates the computation. We plan to improve estimates and computation and to take dependence among DNA methylation loci on a gene into account in our future research.
Acknowledgement
Support by the National Cancer Institute (grant R03 CA150140-01) is gratefully acknowledged. The author also wants to thank the Yale High Performance Computing center for providing computing facility.
Footnotes
No conflict of interest on this work is declared.
References
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - A mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Bediaga NG, Acha-Sagredo A, Guerra I, Viguri A, Albaina C, Ruiz Diaz I, Rezola R, Alberdi MJ, Dopazo J, Montaner D, de Renobales M, Fernández AF, Field JK, Fraga MF, Liloglou T, de Pancorbo MM. DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010;12:R77. doi: 10.1186/bcr2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, Goldmann T, Seifart C, Jiang W, Barker DL, Chee MS, Floros J, Fan JB. High-throughput DNA methylation profiling using universal bad arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Kushi LH, Kwan ML, Wiencke JK. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6(7):e1001043. doi: 10.1371/journal.pgen.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KY, Cheng IK, Ching AK, Chu JH, Lai PB, Wong N. Block of Proliferation 1 (BOP1) plays an oncogenic role in hepatocellular carcinoma by promoting epithelial-to-mesenchymal transition. Hepatology. 2011 doi: 10.1002/hep.24372. (In press) [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society Series B. 1977;39(1):1–38. [Google Scholar]
- Eberth S, Schneider B, Rosenwald A, Hartmann EM, Romani J, Zaborski M, Siebert R, Drexler HG, Quentmeier H. Epigenetic regulation of CD44 in Hodgkin and non-Hodgkin lymphoma. BMC Cancer. 2010;10(1):517. doi: 10.1186/1471-2407-10-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nature Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Houseman EA, Christensen BC, Yeh RF, Marsit CJ, Karagas MR, Wrensch M, Nelson HH, Wiemels J, Zheng S, Wiencke JK, Kelsey KT. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008;9:365. doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nature Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Kalari S, Pfeifer GP. Identification of driver and passenger DNA methylation in cancer by epigenomic analysis. Adv Genet. 2010;70:277–308. doi: 10.1016/B978-0-12-380866-0.60010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkel K, Schupf N, Hatta K, Pang D, Salas M, Kratz A, Minden M, Murty V, Zigman WB, Mayeux RP, Jenkins EC, Torkamani A, Schork NJ, Silverman W, Croy BA, Tycko B. Altered DNA Methylation in Leukocytes with Trisomy 21. PLoS Genet. 2010;6(11):e1001212. doi: 10.1371/journal.pgen.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan PF, Wang SJ, Zhou X, Chu HT. A statistical framework for Illumina DNA methylation arrays. Bioinformatics. 2010;26(22):2849–2855. doi: 10.1093/bioinformatics/btq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- Lugthart S, Figueroa ME, Bindels E, Skrabanek L, Valk PJ, Li Y, Meyer S, Erpelinck-Verschueren C, Greally J, Löwenberg B, Melnick A, Delwel R. Aberrant DNA hypermethylation signature in acute myeloid leukemia directed by EVI1. Blood. 2011;117:234–241. doi: 10.1182/blood-2010-04-281337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund AH, van Lohuizen M. Epigenetics and cancer. Genes & Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- Qin TR, Fu L, Sham PC, Kwong DL, Zhu CL, Chu KW, Li Y, Guan XY. Single-nucleotide polymorphism-mass array reveals commonly deleted regions at 3p22 and 3p14.2 associate with poor clinical outcome in esophageal squamous cell carcinoma. Int J Cancer. 2008;123:826–830. doi: 10.1002/ijc.23577. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Laird PW, Laird-Offringa IA. A comparison of cluster analysis methods using DNA methylation data. Bioinformatics. 2004;20:1896–1904. doi: 10.1093/bioinformatics/bth176. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T, Read A. Human Molecular Genetics – 3rd edition. New York: Garland Science; 2004. [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, Savage DA, Mueller-Holzner E, Marth C, Kocjan G, Gayther SA, Jones A, Beck S, Wagner W, Laird PW, Jacobs IJ, Widschwendter M. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20(4):440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based multiple testing: examples and methods for p-value adjustment. New York: Wiley; 1993. [Google Scholar]