Abstract
Cytokinins are classic plant hormones that orchestrate growth, development and the integrity of stem cell populations. Cytokinin receptors are eukaryotic sensor histidine kinases that are activated both by naturally occurring adenine-type cytokinins and by urea-based synthetic compounds. Crystal structures of the Arabidopsis histidine kinase 4 sensor domain in complex with different cytokinin ligands now rationalize the hormone-binding specificity of the receptor and may spur the design of novel cytokinin ligands.
Plants have evolved small molecule hormones that in chemical structure drastically differ from their animal counterparts1. The naturally occurring cytokinins for example are derivatives of adenine, and may carry either aliphatic or aromatic substitutions at the N6 position (Supplementary Results, Supplementary Fig. 1). Cytokinins play diverse roles in plant growth, development and in the interaction with the environment2. A set of eukaryotic sensor histidine kinases act as membrane receptors for cytokinins in plants3. A well-studied family member is the Arabidopsis histidine kinase 4 (AHK4)4-10. AHK4 contains a cytoplasmic histidine kinase module and a ∼270 residue sensor domain that harbours the cytokinin binding site11. Mutation of the conserved Thr278 in the sensor domain of AHK4 to Ile leads to a loss-of-function phenotype5. AHK4 can bind both natural cytokinins and the serendipitously discovered urea-type cytokinins12, such as the synthetic diphenylurea, with nanomolar affinity13 (Supplementary Fig. 1). At the same time AHK4 can efficiently discriminate between different cytokinin conjugates, naturally occurring enzymatic modifications that efficiently block cytokinin action in vivo14. How AHK4 can be activated by a set of chemically diverse cytokinins on one hand and discriminate between the conjugated forms on the other hand is not understood.
To address this question we expressed the sensor domain of AHK4 (residues 126-395) and determined its crystal structure to 1.7 Å resolution (Supplementary Methods and Supplementary Table 1). The N-terminus of the AHK4 sensor module folds into a long stalk helix followed by two PAS-like domains15 which are connected by a helical linker (Fig. 1a). The last β-strand of the membrane-proximal PAS domain is covalently linked to the N-terminus of the stalk helix by a disulphide bridge, bringing the flanking membrane helices into close proximity (Fig. 1a). The same structural arrangement has been seen previously with family 1 histidine kinase sensor domains16, despite a very low degree of sequence conservation between those bacterial receptors and AHK4 (Supplementary Fig. 2). AHK4 and bacterial sensor domains both form homodimers in crystals (Fig. 1a and Supplementary Fig. 2), and the full-length histidine kinases likely operate as homodimers in intact membranes17. AHK4 recognises cytokinins with its membrane-distal PAS domain as indicated by the presence of N6-isopentenyl adenine (iP) in our structure (Fig. 1a,b), and in agreement with earlier mutagenesis and modelling studies11,18. iP is a natural cytokinin also present in bacteria, where it is generated for example during tRNA degradation19,20. E. coli iP binds to AHK4 with nanomolar affinity13 and apparently co-purified with the isolated sensor domain during protein preparation. The AHK4 ligand binding pocket is occupied by both the adenine portion of iP and by its isopentenyl tail, which was found deeply inserted into the cavity (Fig. 1b). A similar mode of ligand binding has been observed with a cytokinin-binding protein from mung bean21. The lower half of the cytokinin binding site is formed by the central β-sheet of the PAS domain and is lined by small hydrophobic residues (Fig. 1b, Supplementary Fig. 3). Introducing bulkier amino-acids in this area inactivated AHK4, as judged by a functional assay in E. coli.22 (Supplementary Fig. 3, Supplementary Methods). Two β-strands form the upper half of the pocket and contribute additional hydrophobic contacts (Fig. 1b). Hydrogen bonds are established between Asp262 and the adenine ring and these interactions appeared critical for receptor function (Fig. 1b, Supplementary Fig. 3). Other polar interactions are mediated by water molecules, which in turn contact main-chain atoms (Fig. 1b). Mutation of Thr278 to Ile (the wooden leg allele5,8) likely restricts the overall size of the binding pocket, and thus our structure rationalises the associated loss-of-function phenotype (Fig. 1c and Supplementary Fig. 3).
Figure 1.
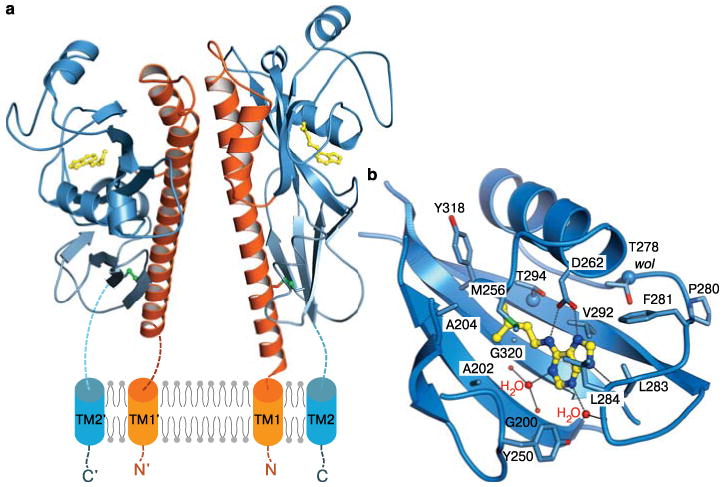
AHK4 binds cytokinins with its membrane-distal PAS domain. a, Ribbon diagram of the sensor domain homodimer (residues 126-391). The N-terminal stalk helix and the dimerisation interface is shown in orange, the membrane-distal cytokinin binding domain in dark-blue, and the membrane-proximal PAS domain is in light-blue, respectively. Disulphide bridges are depicted in green. One molecule of iP (in yellow) binds per AHK4 monomer. b, Close-up of the cytokinin binding pocket complexed with iP (in bonds representation).
In planta, cytokinins can be modified by N-glucosylation and N-alanine conjugation on the adenine ring and by O-glucosylation and O-acetylation on the isoprenoid tail for transport and storage (Supplementary Fig. 4)14. All these modifications render the respective cytokinin inactive13. This is in good agreement with our structure that reveals the N3 and N9, and especially the N7 position of the adenine ring buried in the binding pocket (Supplementary Fig. 4). In addition, the shape and predominantly apolar nature of the tail pocket restricts binding of larger tail groups as found in trans-zeatin (tZ) O-acetyl and tZ O-glucoside13 (Supplementary Fig. 4).
We next studied how AHK4 can perceive chemically diverse natural and synthetic cytokinins13 (Supplementary Fig. 1), by displacing iP in the binding pocket with an excess of cytokinin ligand during bacterial cell lysis and protein purification (see Supplementary Methods). A complex structure with N6-benzyladenine (BA) revealed that the hormone binding site can accommodate both isoprenoid and aromatic tail groups without undergoing major structural rearrangements (Fig. 2a,b and Supplementary Table 2). Next, we determined a 1.5 Å complex structure with tZ (Supplementary Table 2), the most potent endogenous cytokinin in Arabidopsis13. By comparison with the iP and BA bound structures, the hydroxylated isopentenyl side chain of tZ establishes an additional hydrogen bond with Thr294 (Fig. 2c). Because Thr294 is the only hydrogen bond acceptor in the tail binding pocket, our structure rationalises why AHK4 specifically recognises trans but not cis zeatin-type cytokinins with high affinity. Consistently, a complex structure with dihydrozeatin (DZ) revealed the hydroxyl group in the same orientation as found in the tZ complex (Fig. 2c and Supplementary Fig. 1 and Supplementary Table 2). A structure with the kinetin (KIN), the first cytokinin to be discovered23, (Supplementary Table 2) showed how the ligand binding pocket accommodates larger and charged tail groups. Again, Thr294 is involved in a polar interaction with the furfuryl group of KIN, in this case mediated by a water molecule (Fig. 2d).
Figure 2.
Structural plasticity in the AHK4 PAS domain allows for the binding of diverse cytokinins. a, Close-up of the cytokinin binding site occupied by iP (in bonds representation, Fo-Fc omit electron difference density map is contoured at 4.5σ). Interacting residues in the tail binding pocket are shown in blue. b, Structural superposition of the BA and iP complexes. c, The hydroxylated isopentenyl chain of tZ contacts Thr294 (in magenta). d, The tail binding pocket can accommodate the larger furfuryl group of KIN. A polar interaction (in magenta) with Thr294 now is mediated by a water molecule (in red). The chemical structures of the respective cytokinins are shown alongside.
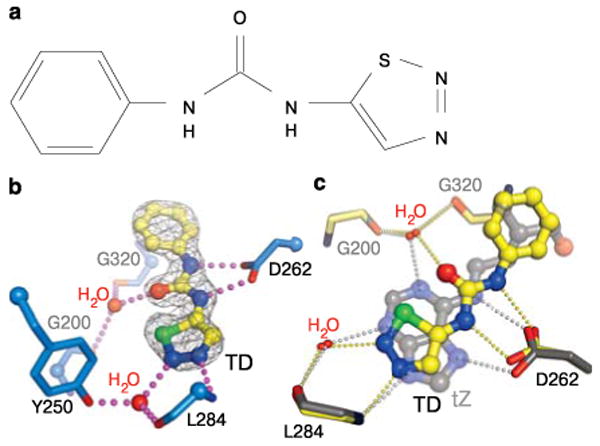
Finally, we solved a structure with the synthetic urea-type cytokinin thiadiazuron (TD; Fig. 3a), an important defoliant and herbicide. This structure revealed that both natural and synthetic cytokinins occupy the same binding site in AHK4 (Fig. 3b). The phenyl moiety of TD binds to the tail pocket and the thiadiazol group mimics the adenine ring. (Fig. 3c). Importantly, both the urea-moiety of TD and the thiadiazol group establish polar interactions (with Asp262 and Leu284, respectively) that are very similar to those observed in the adenine-type complexes (Fig. 3c). The TD structure thus rationalises why the serendipitously discovered urea derivatives12 are potent cytokinins.
Figure 3.
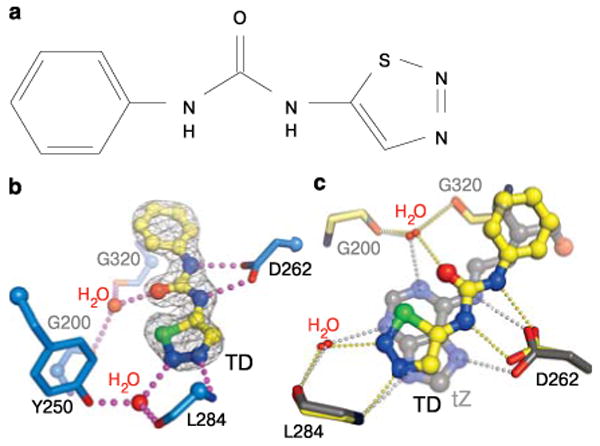
Urea-based synthetic cytokinins mimic adenine-type hormone-receptor interactions. a, Chemical structure of thiadiazuron (TD). a, Structure of the AHK4-TD complex with polar interactions to Asp262 and Leu284 included. The side-chain of Leu284 has been omitted for clarity. c, Structural superposition of the tZ (in gray) and TD (in yellow) complexes (r.m.s.d. is ∼0.l5 Å between 266 corresponding Cαatoms).
In summary, our work defines the molecular basis for the recognition of natural and synthetic cytokinins by eukaryotic sensor histidine kinases. We find that Arabidopsis HK4 shares significant structural homology with a family of bacterial histidine kinases and, like the bacterial sensor domains24, coordinates its ligand with the membrane-distal PAS domain. A simple size selectivity-filter (Supplementary Fig. 4) enables the receptor to discriminate between unmodified cytokinins and different cytokinin conjugates, enzymatic modifications that modulate cytokinin bioactivity in vivo14. The different hormone complexes described here reveal a highly adaptable binding pocket in which most receptor-hormone interactions are mediated by small hydrophobic residues. Discriminating polar contacts are contributed by Asp262 and by Thr294, which controls for the binding of the correct stereoisomer (Fig. 2c). Importantly, synthetic urea-based cytokinins establish, despite their distinct chemical structure, very similar polar contacts with the receptor (Fig. 3). Superposition of all AHK4 sensor domain structures unravels basic design principles for active cytokinins, i.e. the presence of a planar ring structure that occupies the adenine binding pocket, followed by a linker competent to establish hydrogen bonds with Asp262 and a planar aliphatic or aromatic tail group (Fig. 2,3). Thus our work may spur the rational design of novel synthetic cytokinins with potential applications in basic research and agriculture.
Supplementary Material
Acknowledgments
We thank T. Mizuno for providing plasmid pCold IV – AHK4 and strain KMI001, M. Jinek and Y. Jaillais for comments on the manuscript and W. Kwiatkowski and staff at beamlines BL 8.2.1 and 8.2.2 of the Advanced Light Source, Berkeley for technical support. This work was supported by long-term fellowships from the European Molecular Biology Organisation, the International Human Frontier Science Program Organisation and by the Marc and Eva Stern foundation (M.H.) and by grants from the NIH (5R01GM52413), the NSF (IOS-0649389) and by the Howard Hughes Medical Institute (J.C.). Maintenance of the Salk X-ray equipment is supported by NIH grant P30 NS057096.
Footnotes
Accession codes. The atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 3T4J (AHK4-iP), 3T4K (-BA), 3T4L (-tZ), 3T4O (-DZ), 3T4Q (-tZr), 3T4S (-KIN) and 3T4T (-TD).
Author Contributions: M.H. designed the project. M.H. and T.D. expressed and purified proteins and carried out functional assays. M.H. crystallised, phased and refined the structures. M.H. analysed the data. J.C. supervised the project. M.H. wrote the paper.
Competing financial interests: The authors declare no competing financial interests.
Additional information: Reprints and permissions information are available online at http://www.nature.com/reprints/index.html.
References
- 1.Jaillais Y, Chory J. Nat Struct Mol Biol. 2010;17:642–645. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Werner T, Schmülling T. Curr Opin Plant Biol. 2009;12:527–538. doi: 10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Kakimoto T. Annu Rev Plant Biol. 2003;54:605–627. doi: 10.1146/annurev.arplant.54.031902.134802. [DOI] [PubMed] [Google Scholar]
- 4.Inoue T, et al. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- 5.Mähönen AP, et al. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki T, et al. Plant Cell Physiol. 2001;42:107–113. doi: 10.1093/pcp/pce037. [DOI] [PubMed] [Google Scholar]
- 7.Ueguchi C, Sato S, Kato T, Tabata S. Plant Cell Physiol. 2001;42:751–755. doi: 10.1093/pcp/pce094. [DOI] [PubMed] [Google Scholar]
- 8.Yamada H, et al. Plant Cell Physiol. 2001;42:1017–1023. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]
- 9.Riefler M, Novak O, Strnad M, Schmülling T. Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi M, et al. Proc Natl Acad Sci USA. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyl A, et al. BMC Evol Biol. 2007;7:62. doi: 10.1186/1471-2148-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amasino R. Plant Physiol. 2005;138:1177–1184. doi: 10.1104/pp.104.900160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romanov GA, Lomin SN, Schmülling T. J Exp Bot. 2006;57:4051–4058. doi: 10.1093/jxb/erl179. [DOI] [PubMed] [Google Scholar]
- 14.Bajguz A, Piotrowska A. Phytochemistry. 2009;70:957–969. doi: 10.1016/j.phytochem.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Ponting CP, Aravind L. Curr Biol. 1997;7:R674–677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Hendrickson WA. J Mol Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao R, Stock AM. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pas J, von Grotthuss M, Wyrwicz LS, Rychlewski L, Barciszewski J. FEBS Lett. 2004;576:287–290. doi: 10.1016/j.febslet.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 19.Caillet J, Droogmans L. J Bacteriol. 1988;170:4147–4152. doi: 10.1128/jb.170.9.4147-4152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray J, Gelvin SB, Meilan R, Morris RO. Plant Physiol. 1996;110:431–438. doi: 10.1104/pp.110.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasternak O, et al. Plant Cell. 2006;18:2622–2634. [Google Scholar]
- 22.Mizuno T, Yamashino T. Methods Enzymol. 2010;471:335–356. doi: 10.1016/S0076-6879(10)71018-1. [DOI] [PubMed] [Google Scholar]
- 23.Miller CO, Skoog F, Okumura FS, Von Saltza MH, Strong FM. JACS. 1956;78:1375–1380. [Google Scholar]
- 24.Zhou YF, et al. J Mol Biol. 2008;383:49–61. doi: 10.1016/j.jmb.2008.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


